Abstract
The GeoPCA package is the first tool developed for multivariate analysis of dihedral angles based on principal component geodesics. Principal component geodesic analysis provides a natural generalization of principal component analysis for data distributed in non-Euclidean space, as in the case of angular data. GeoPCA presents projection of angular data on a sphere composed of the first two principal component geodesics, allowing clustering based on dihedral angles as opposed to Cartesian coordinates. It also provides a measure of the similarity between input structures based on only dihedral angles, in analogy to the root-mean-square deviation of atoms based on Cartesian coordinates. The principal component geodesic approach is shown herein to reproduce clusters of nucleotides observed in an η–θ plot. GeoPCA can be accessed via http://pca.limlab.ibms.sinica.edu.tw.
INTRODUCTION
Multivariate statistics is widely applied to biological systems. It is used to unravel hidden trends in large data sets and to analyze the results of molecular dynamics simulations of biomolecules. Among the wide range of available multivariate techniques, principal component analysis (PCA) (1) is one of the most widely used methods. PCA transforms a data set consisting of several correlated variables into a new set of uncorrelated variables called principal components. By a linear orthogonal transformation, the first principal component represents the most variability in the data; the second principal component represents the second most variability in the data under the constraint that it is orthogonal to the first principal component, and so on. Thus, PCA rotates the axes of data variation, yielding a set of ordered orthogonal axes that represents decreasing proportions of the data variation. Using only the first few principal components, the dimensionality of the transformed data is reduced. For example, the first few principal components have been used to specify a set of representative coordinates of the free energy landscape for biological molecules containing many degrees of freedom (2). They have also been used to yield the dominant modes of structural variation in an ensemble of conformations for a given protein, derived from Nuclear Magnetic Resonance (NMR) and/or X-ray (3); i.e. structures of the free protein solved in different space groups or complexed with different ligands or from simulations (4,5).
In PCA of large biomolecules with many degrees of freedom, it is useful to replace the Cartesian coordinates of the atoms with a smaller set of internal coordinates to reduce the number of variables involved in PCA. A natural choice of internal coordinates would be dihedral angles that change much more than bond lengths and bond angles in structures of a given molecule. However, angular data pose difficulties in PCA and other multivariate statistical analyses due to their circular nature. For example, the arithmetic mean of 10° and 350° is (10° + 350°)/2 = 180° rather than the true mean of 0°. This difficulty remains even if the torsion angles are represented in the interval from −180° to 180°, as the arithmetic mean of −160° and 160° is 0° instead of 180°.
To circumvent the aforementioned difficulties with circular data, angles have been transformed into coordinates using cosine and sine values in PCA (referred to as dPCA in previous work) (2,6). For example, the two backbone dihedral angles ϕi and ψi of residue i have been replaced by four coordinates x4i−3 = cos(ϕi), x4i−2 = sin(ϕi), x4i−1 = cos(ψi) and x4i = sin(ψi). Thus, one disadvantage of the dPCA approach is an increased number of coordinates. Another disadvantage is the neglect of the cos2 +sin2 = 1 correlation (7,8); i.e. the coordinates are not independent since (x4i−3)2 + (x4i−2)2 = 1 and (x4i−1)2 + (x4i)2 = 1. Furthermore, there is no rigorous mathematical consideration of the applicability of dPCA (to the best of our knowledge). In justifying the transformation of angular data (6), the points are assumed to lie on a sphere in Euclidean space, subject to Euclidean geometry, whereas they should in fact be subjected to non-Euclidean geometry (9). Notably, some properties of non-Euclidean geometry are counterintuitive; e.g. the sum of the angles of a triangle is >180°. Furthermore, since all data point are on a sphere, the distance measured by the shortest path between points is an arc rather than a straight line.
Circular correlation and covariance matrices computed using known formulas for the circular correlation coefficient and the circular mean, respectively, can be used in PCA (10). PCA has been applied to torsion angles of a set of RNA trinucleotides using five different representations; viz., (i) angles between 0° and 360°, (ii) angles between −180° and 180°, (iii) angles represented by cosine and sine values (see dPCA above), (iv) circular correlation matrix and (iv) circular covariance matrix (11). The results were compared with those from PCA applied to Cartesian coordinates of the same data set of RNA trinucleotides. The outcome of the PCA results was found to depend on the choice of interval for representing the angles [(0°, 360°) or (−180°, 180°)]. Thus, for each torsion angle, its variance has to be analyzed a priori to determine if it should be represented by a (0°, 360°) or (−180°, 180°) interval. The interval that yields the larger total variance of the first principal component was assumed to be more accurate. Moreover, using a linear orthogonal transformation in PCA, the non-Euclidean nature of the circular data was not taken into account.
Various manifold (locally Euclidean space) learning and non-linear dimensionality reduction approaches may be considered as alternatives to linear PCA for angular data. These include self-organizing maps (12), principal curves (13), kernel PCA (14), isomap (15), diffusion maps (16) and principal geodesics (17). Most of them apply machine learning such as neural networks. For most of these methods, there is no simple interpretation of the results unlike linear principal components. Furthermore, these methods have not been used in lieu of linear PCA for dihedral angles (to the best of our knowledge).
Our aim is to develop a tool applying a generalization of PCA for angular data. Among the various manifold learning and non-linear dimensionality reduction approaches, geodesic PCA was chosen because (i) it is a straightforward generalization of PCA for manifolds that are generally only locally Euclidean and (ii) the mathematics underlying principal component geodesic has been described (17). Instead of determining a set of ordered orthogonal linear axes, which represents decreasing proportions of the data variation, we find a set of ordered orthogonal great circles (principal component geodesics) that minimizes the distances from the data points to their projections on the respective great circles. The distance between any two data points is an arc rather than a straight line, as in linear PCA.
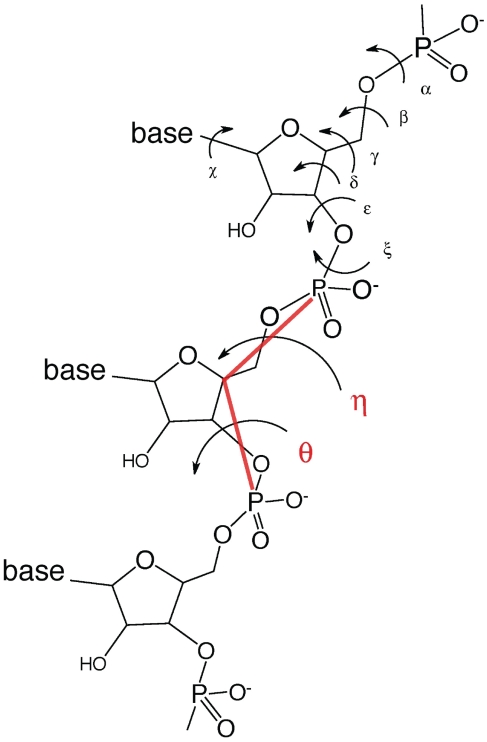
Below, we first present the essence of the principal component geodesic approach and the properties of principal geodesic components; we refer the reader to previous works for proofs of the necessary theorems (17). We then validate the principal component geodesic approach by using it to cluster a set of RNA conformations that had been classified as follows. Just as the protein backbone conformation can be described by two torsion angles (ϕ and φ), the RNA backbone conformation can be described by two pseudotorsion angles (η = C4′i−1−Pi−C4′i−Pi + 1 and θ = θ = Pi −C4′i−Pi + 1−C4′i + 1) and the sugar pucker, instead of the seven conventional torsion angles, α, β, γ, δ, ε, ζ and χ (Figure 1). A plot of the θ versus η angles of all nucleotides in a database containing 52 RNA structures revealed distinct clusters of nucleotides (18). Within a given cluster, the nucleotides share similar η and θ values as well as structural features such as A-platforms and GNRA tetraloops. These clusters of nucleotides have been statistically validated and refined using a larger data set containing 73 RNA structures (19). This work shows that the principal component geodesic approach provides a means of distinguishing clusters of nucleotides using seven conventional torsion angles per nucleotide. Its application is not limited to dihedral angles or nucleotides, but can be applied to analyze angular data of large, complex macromolecules.
Figure 1.
Structure of RNA nucleotides showing the seven conventional torsion angles, α, β, γ, δ, ε, ζ χ and two pseudotorsion angles, η, θ, which are defined as C4′i−1−Pi−C4′i−Pi + 1 and Pi−C4′i−Pi + 1−C4′i + 1, respectively. The bold red lines are pseudobonds connecting P and C4′ along the backbone.
METHODS
Embedding of an m-Sphere in (m + 1)-dimensional Euclidean space
Nash theorem (20) postulates that every Riemannian manifold M can be isometrically embedded in a Euclidean space of sufficiently higher dimension. Thus, an m-dimensional unit sphere can be embedded in a (m + 1)-dimensional Euclidean space. It is defined by
| (1) |
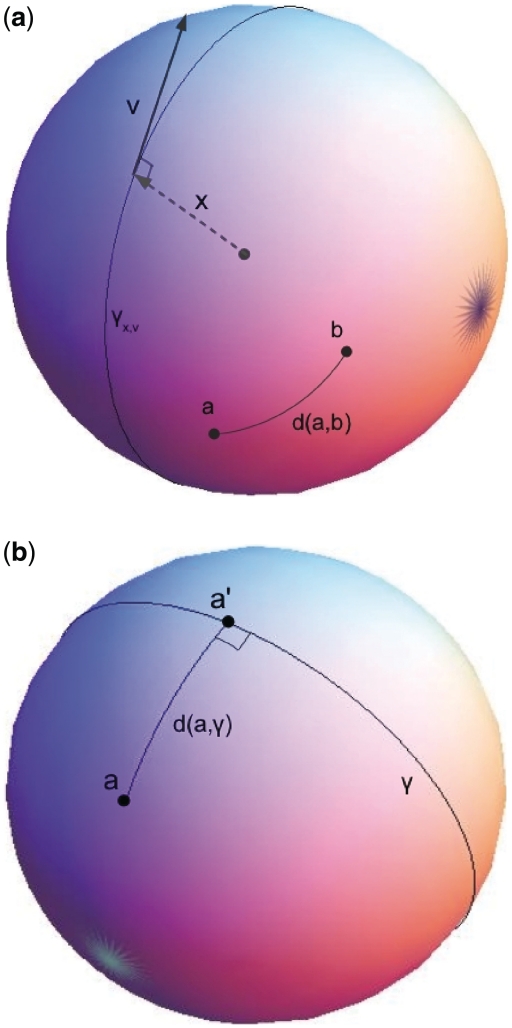
where x are points in the (m + 1)-dimensional Euclidean space and <•,•> denote the inner product. The inner product, <x, x>, in an m-dimensional sphere is equal to the scalar or dot product in the (m + 1)-dimensional Euclidean space. Thus, although the geometry on an m-sphere is non-Euclidean, it can be described in terms of (m + 1)-dimensional Euclidean space since the m-sphere has been embedded in (m + 1)-dimensional Euclidean space. The tangent space of the unit sphere S at x is defined as the set of all tangent unit vectors v (Figure 2a) that satisfy
| (2) |
Figure 2.
(a) The x,v vectors, the γx,v geodesic and the spherical distance d(a,b) between two points a and b are illustrated on a 2D unit sphere embedded in 3D Euclidean space. Although point x is on the sphere, its Euclidean radius-vector is in the direction from the sphere center to x (dashed line) and is therefore not on the sphere. The x vector is orthogonal to the Euclidean tangent vector v, which is in the direction of a path on the sphere and is on the sphere itself. Both x and v are vectors in (m + 1)-dimensional Euclidean space and are thus subjected to Euclidean geometry. (b) The spherical distance between point a and its projection onto the geodesic γx,v, point a′, are illustrated on a 2D unit sphere embedded in 3D Euclidean space.
Input data
Let P = (p1, p2, …, pn) denote a set of torsion angle measurements describing a molecule of interest. Each pi represents the ith conformation of the molecule. If P contains n conformations, there will be n observations for each torsion angle in the molecule. Let aik (k = 1, …, m) denote the value of kth torsion angle of pi. Each pi = (ai1, ai2, …, aim) can be treated as a point on the m-dimensional unit sphere, representing the ith conformation. For our test data set (see below), the nucleotides all have the same C3′-endo sugar pucker conformation. Hence, the input data consist of the seven conventional torsion angles in Figure 1 describing each nucleotide.
Geodesics
Instead of using straight-line axes as principal components, curves are used as principal components, so-called principal component geodesics. A geodesic is a curve on the m-dimensional sphere, which locally minimizes the distance between points on the surface. It is a straight line in the plane and a great circle (like the earth’s equator) on a sphere. Just as the distance between two points, a and b, in Euclidean space can be represented by a straight line of the form b + (t – 1)(b – a), 0 ≤ t ≤ 1, geodesics on spheres are great circles given by
| (3a) |
where 0 ≤ t ≤ 2π, a = x and b = v, and Equations (1) and (2) are satisfied; i.e.
| (3b) |
Spherical distance to a geodesic
The embedding of an m-dimensional sphere into a (m + 1)-dimensional Euclidean space induces a simple expression for a metric on the sphere. Since the inner product <a, b> is the standard scalar product of the (m + 1)-dimensional Euclidean space, the spherical distance d(a, b) between any two points a and b (Figure 2a) on the m-dimensional unit sphere is given by:
| (4) |
The projection of point a onto the geodesic γx,v is the point a′ (Figure 2b) given by
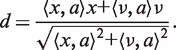 |
(5) |
Thus, the spherical distance between point a and its projection onto the geodesic γx,v, point a′, is given by
| (6) |
Principal component geodesics
Consider the following distance function that describes the mean distance between data points pi and their projections onto the geodesic γx, v.
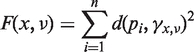 |
(7) |
where d(pi, γx,v) is given by Equation (6). Finding a first principal component geodesic that accounts for most of the data variability is equivalent to minimizing F(x, v) under the constraints given by Equations (1) and (2). Given the first principal component geodesic γx, v(1), the second principal component geodesic, γx, v(2), can be found as a geodesic that intersects γx, v(1) and is orthogonal to γx, v(1) by minimizing 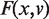 with the respective constraints.
with the respective constraints.
To obtain the other principal component geodesics, we define a principal component geodesic mean, 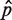 , as the point that minimizes the mean of
, as the point that minimizes the mean of 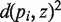 over all common points of γx, v(1)and γx,v(2). A principal component geodesic of higher order s (3 ≤ s ≤ n) minimizes the function
over all common points of γx, v(1)and γx,v(2). A principal component geodesic of higher order s (3 ≤ s ≤ n) minimizes the function 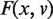 , passes through the principal component geodesic mean and is orthogonal to all geodesics of order ≤ s−1.
, passes through the principal component geodesic mean and is orthogonal to all geodesics of order ≤ s−1.
Geodesic variance
The variance explained by the sth principal component geodesic, obtained by projection of the data points pi on the sth principal component geodesic, is given by:
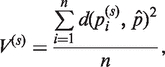 |
(8) |
where 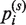 , the projection of pi on the sth principal component geodesic, is obtained using Equation (5), and n is the number of conformations of a given molecule (see above). As in conventional PCA, the first principal component geodesic represents the most variability in the data and has the smallest variance. However, if its variance was comparable with the variance of a randomly chosen geodesic on the sphere, then the principal component geodesic analysis would not help to reduce the dimensionality of the given input data.
, the projection of pi on the sth principal component geodesic, is obtained using Equation (5), and n is the number of conformations of a given molecule (see above). As in conventional PCA, the first principal component geodesic represents the most variability in the data and has the smallest variance. However, if its variance was comparable with the variance of a randomly chosen geodesic on the sphere, then the principal component geodesic analysis would not help to reduce the dimensionality of the given input data.
Output data
The above approach has been implemented in a program called GeoPCA. In the current version of GeoPCA, Lagrange multipliers are used to minimize F(x,v) under constraints, as described by Huckemann and Ziezold (17). This procedure yields fixed-point equations, y = f(y), which are solved by numerical iteration, yn + 1 = f(y). After solving the fixed-point equations, GeoPCA provides projection of the data onto the first two principal component geodesics and the corresponding Cartesian coordinates of the data points projected on the unit sphere in 3D space to enable plots to be made using standard plotting packages. Thus, GeoPCA allows visualization of the output data along the great circles, which accounts for most of the data variability.
Data set
To validate the principal component geodesic approach, it was used to cluster a set of RNA conformations derived from a published database of 73 RNA structures containing 7407 nt (19). We did not update this database so that clusters obtained from a plot of the first two principal component geodesics, which are characterized by two ‘principal’ angles, can be compared with the clusters found in an η–θ plot of all non-helical nucleotides with C3′-endo sugar pucker from the published database [see Figure 4 in Wadley et al. (19)]. The latter yielded six clusters of non-helical C3′-endo nucleotides, which were labeled as I, II, III, IV, V and VI by Wadley et al. (19). We chose to include in our data set non-helical C3′-endo nucleotides in clusters I and II, as they have the highest density in the η–θ plot, ensuring that they are statistically significant and should be detected by any effective clustering method. Furthermore, cluster I contains nucleotides that are often constituents of S1 and S2 motifs, while cluster II contains nucleotides that serve as the second bases in GNRA/GNRA-like tetraloops or in T-loop motifs. Unlike the nucleotides in cluster I or cluster II, nucleotides in cluster V do not belong to structural motifs. Hence, cluster V was also included in our data set to verify if the principal component geodesic approach, like the η/θ plot, also predicts this cluster.
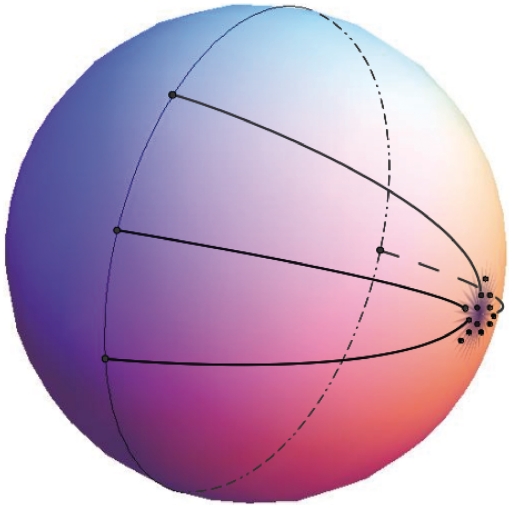
Figure 4.
An example of a possible arrangement of the cluster along the circle after projection of the 2D sphere onto 1D circle.
The non-helical C3′-endo nucleotides belonging to clusters I, II and V were extracted from the published data set of 7407 nt as follows: First, the η/θ values corresponding to the peak density of a given cluster was found from an initial guess of the ‘peak’ η/θ values from the η–θ plot of all non-helical C3′-endo nucleotides and refining them to yield the maximum number of nucleotides for a given cluster. Then, all C3′-endo nucleotides with η/θ values within ±15° of the ‘peak’ η/θ values were extracted; i.e. (147–187°)/(330–360°) for cluster I, (15–45°)/(225–255°) for cluster II and (299–329°)/(216–246°) for cluster V. This yielded 59, 88 and 43 nt for clusters I, II and V, respectively (Supplementary Table S1). This data set was used to test whether the principal component geodesic approach can yield the three clusters found in the η–θ plot.
RESULTS AND DISCUSSION
dPCA using dihedral angles
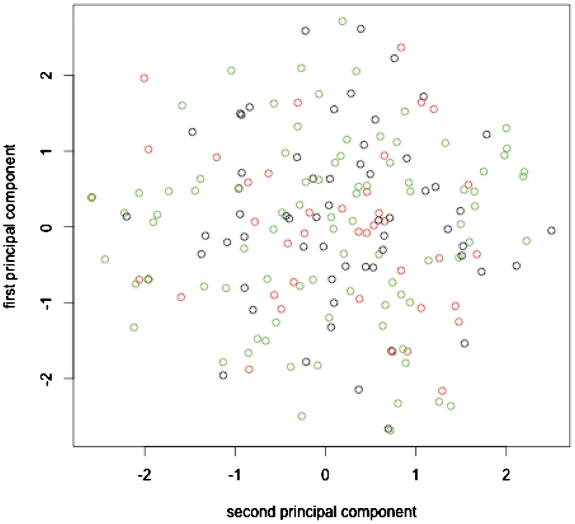
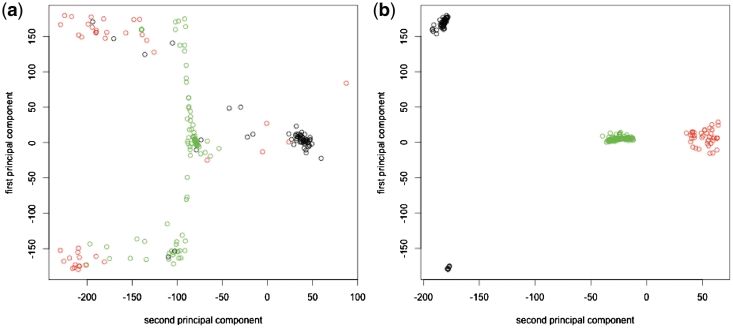
Using our database (see above), we first examined if dPCA (‘Introduction’ section) using the seven standard dihedral angles (α, β, γ, δ, ε, ζ and χ in Figure 1), represented by cosine and sine values but neglecting the cos2 + sin2 =1 correlation, could yield the three distinct clusters (I, II and V, see ‘Data set’ section) found in an η–θ plot in previous work (19). A 2D plot of the first two principal components in Figure 3 shows that dPCA cannot distinguish non-helical C3′-endo nucleotides belonging to cluster I (black circles), II (green circles) and V (red circles).
Figure 3.
Plot based on the angles describing the first and second principal components obtained using dPCA. The black, green and red circles denote non-helical C3′-endo nucleotides belonging to clusters I, II and V found in an η–θ plot of all non-helical C3′-endo nucleotides in previous work.
Clustering patterns in non-Euclidean space
Before presenting the clustering results using the principal component geodesic approach, we first highlight some differences between the clustering patterns in Euclidean and non-Euclidean space. Whereas data points belonging to a cluster appear close to one another in Euclidean space, they may be dispersed along a circle in non-Euclidean geometry. The latter becomes evident if data points clustered at the pole of a sphere are projected onto the big circle (equator) of that sphere. As shown in Figure 4, although points around the pole are close to each other, their projections on the equator may cover the entire big circle. Hence, points lying on a circle can be identified as forming a cluster. This property has the advantage that points lying on different circles can be unambiguously assigned to different clusters except for those at the intersection of circles.
Geodesic PCA using dihedral angles
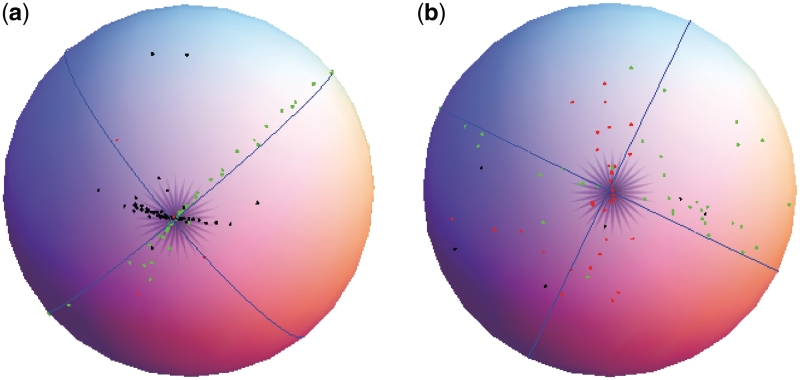
Geodesic PCA was performed using the seven standard dihedral angles for each nucleotide in our data set. Figure 5 shows the projection of the non-helical C3′-endo nucleotides on a sphere and the first two principal component geodesics (blue great circles). The results in Figure 5 show that geodesic PCA can separate the nucleotides into three distinct clusters, as observed in a η–θ plot. The C3′-endo nucleotides from cluster I (black points) and cluster II (green points) lie close to the first two principal geodesic components (Figure 5a) and are at different distances from the sphere center, so they are well separated from each other. However, the C3′-endo nucleotides from cluster V (red points) are not visible from this viewpoint, but become evident from another viewpoint (Figure 5b). Although the red circles do not form a compact cluster, they are nevertheless clearly separated from the nucleotides in the other two clusters.
Figure 5.
C3′-endo nucleotides on a sphere. The first two principal geodesic components are shown in blue. The (a) shows C3′-endo cluster I (black points) and cluster II (green points), while the (b) shows the C3′-endo cluster V (red points).
That the C3′-endo nucleotides form three clusters are also shown when the data points on the sphere are projected onto a plane. Figure 6a shows the non-helical C3′-endo nucleotides as a function of the first two principal component geodesics, which can be described by two ‘principal’ angles. The C3′-endo nucleotides from cluster V (red points) are located at the top and bottom of the 2D plot. They are well separated from the C3′-endo nucleotides from cluster II (green points) and cluster I (black points), which lie along two great circles.
Figure 6.
2D plot based on the angles describing the first and second principal component geodesics using (a) the seven conventional torsion angles and (b) the two pseudotorsion angles, η and θ (Figure 1). The black, green and red points denote non-helical C3′-endo nucleotides belonging to clusters I, II and V found in an η–θ plot of all non-helical C3′-endo nucleotides in previous work. Principal component geodesics are lines parallel to the axes, passing through zeros of the respective axes and described by dotted lines.
Geodesic PCA using pseudotorsion angles
Since the input data for the principal geodesics approach (seven torsion angles) differs from that for the η–θ plot (two pseudotorsion angles), the outcome from these two methods would not be expected to be identical. Indeed, the three distinct clusters found herein do not contain exactly the same nucleotides as clusters I, II and V from an η–θ plot in previous work (19). For example, C3′-endo nucleotides from clusters I (black circles) and V (red circles) are found along the big circle encompassing C3′-endo nucleotides from cluster II (green circles), as shown in Figure 6. To verify that this discrepancy is not due to limitations/errors in the GeoPCA program, geodesic PCA was performed with the two pseudotorsion angles, η and θ, used to derive clusters I, II and V in previous work (19). Note that the two ‘principal’ angles describing the first two principal component geodesics do not correspond to η and θ. Thus, although the 2D plot in Figure 6b is not the same as an η–θ plot, the same three clusters found in an η–θ plot are found.
SUMMARY
This work introduces a new tool, based on principal component geodesics, for conformational analysis using circular data such as bond, torsion and pseudotorsion angles. It shows how our approach could aid structural analysis like analyses of η–θ plots and counterintuitive consequences of non-Euclidean geometry (e.g. points lying on a circle belong to the same cluster). The web interface of GeoPCA, which implements the principal component geodesics approach described herein, requires as input, a file with angular data. It yields as output: (i) Cartesian coordinates of the data points projected on the first and second principal component geodesics of a sphere (orthogonal great circles on the sphere) and (ii) the values of two angles representing corresponding distances to first and second principal component geodesics for each data point. To the best of our knowledge, this is the first method to automatically reduce a multidimensional analysis of several angles to only two angles containing most of the information. GeoPCA thus provides a useful way of visualizing, analyzing and predicting conformations of complex macromolecules with many degrees of freedom.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1.
FUNDING
Funding for open access charge: National Science Council, Taiwan (grant NSC 95-2113-M-001-038 and NSC 95-2113-M-001-001 to C.L.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Kevin S. Keating and Anna Marie Pyle for providing information on the nucleotides used in our data set. The authors thank Karine Mazmanian for her help in preparation of the figures.
REFERENCES
- 1.Jolliffe IT. Principal Component Analysis. New York: Springer; 2002. [Google Scholar]
- 2.Mu Y, Nguyen PH, Stock G. Energy landscape of a small peptide revealed by dihedral angle principal component analysis. PROTEINS: Structure, Function, and Bioinformatics. 2005;58:45–52. doi: 10.1002/prot.20310. [DOI] [PubMed] [Google Scholar]
- 3.Yang LW, Eyal E, Bahar I, Kitao A. Principal component analysis of native ensembles of biomolecular structures (PCA NEST): insights into functional dynamics. Bioinformatics. 2009;25:606–614. doi: 10.1093/bioinformatics/btp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ichiye T, Karplus M. Collective motions in proteins: a covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins. 1991;11:205–271. doi: 10.1002/prot.340110305. [DOI] [PubMed] [Google Scholar]
- 5.Bahar I, Rader A. Coarse-grained normal mode analysis in structural biology. Curr. Opin. Struct. Biol. 2005;15:586–592. doi: 10.1016/j.sbi.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altis A, Nguyen PH, Hegger R, Stock G. Dihedral angle principal component analysis of molecular dynamics simulations. J. Chem. Phys. 2007;126:244111. doi: 10.1063/1.2746330. [DOI] [PubMed] [Google Scholar]
- 7.Hinsen K. Comment on: energy landscape of a small peptide revealed by dihedral angle principal component analysis. Proteins. 2006;64:795–797. doi: 10.1002/prot.20900. [DOI] [PubMed] [Google Scholar]
- 8.Mu Y, Nguyen P, Stock G. Reply to the comment on Energy landscape of a small peptide revealed by dihedral angle principal component analysis. Proteins. 2006;64:798–799. doi: 10.1002/prot.20310. [DOI] [PubMed] [Google Scholar]
- 9.Petersen P. Riemannian Geometry. Berlin: Springer; 2006. [Google Scholar]
- 10.Mardia KV, Jupp P. Directional Statistics. 2nd edn. Chichester: John Wiley & Sons Ltd; 2000. [Google Scholar]
- 11.Reijmers TH, Wehrens R, Buydens LMC. Circular effects in representations of an RNA nucleotides data set in relation with principal components analysis. Chemometrics and Intelligent Laboratory Systems. 2001;56:61–71. [Google Scholar]
- 12.Kohonen T. Self-organizing Maps. Berlin: Springer; 2001. [Google Scholar]
- 13.Kégl B. Principal Curves: Learning, Design, and Applications. Canada: Concordia University; 1999. [Google Scholar]
- 14.Schölkopf B, Smola A, Müller K-R. Kernel principal component analysis. Artificial Neural Networks—ICANN'97, Lecture Notes in Computer Science. 1997;1327:583–588. [Google Scholar]
- 15.Tenenbaum JB, de Silva V, Langford JC. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290:2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- 16.Coifman RR, Lafon S, Lee AB, Maggioni M, Nadler B, Warner F, Zucker SW. Geometric diffusions as a tool for harmonic analysis and structure definition of data: diffusion maps. Proc. Natl Acad. Sci. USA. 2005;102:7426–7431. doi: 10.1073/pnas.0500334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huckemann S, Ziezold H. Principal component analysis for Riemannian manifolds, with an application to triangular shape spaces. Adv. Appl. Prob. 2006;38:299–319. [Google Scholar]
- 18.Duarte CM, Pyle AM. Stepping through an RNA structure: a novel approach to conformational analysis. J. Mol. Biol. 1998;284:1465–1478. doi: 10.1006/jmbi.1998.2233. [DOI] [PubMed] [Google Scholar]
- 19.Wadley LM, Keating KS, Duarte CM, Pyle AM. Evaluating and learning from RNA pseudotorsional space: quantitative validation of a reduced representation for RNA Structure. J. Mol. Biol. 2007;372:942–957. doi: 10.1016/j.jmb.2007.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nash J. The imbedding problem for Riemannian manifolds. Ann. Math. 1956;63:20–63. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.