Abstract
Transcription factors play a central role in cell development, differentiation and growth in biological systems due to their ability to regulate gene expression by binding to specific DNA sequences within the nucleus. The dysregulation of transcription factor signaling has been implicated in the pathogenesis of a number of cancers, developmental disorders, inflammation and autoimmunity. There is thus a high demand for convenient high-throughput methodologies able to detect sequence-specific DNA-binding proteins and monitor their DNA-binding activities. Traditional approaches for protein detection include gel mobility shift assays, DNA footprinting and enzyme-linked immunosorbent assays (ELISAs) which tend to be tedious, time-consuming, and may necessitate the use of radiographic labeling. By contrast, luminescence technologies offer the potential for rapid, sensitive and low-cost detection that are amenable to high-throughput and real-time analysis. The discoveries of molecular beacons and aptamers have spearheaded the development of new luminescent methodologies for the detection of proteins over the last decade. We survey here recent advances in the development of luminescent detection methods for DNA-binding proteins, including those based on molecular beacons, aptamer beacons, label-free techniques and exonuclease protection.
INTRODUCTION
Many proteins are involved in regulating cellular processes through their ability to bind and interact with DNA. Transcription activators or repressors regulate gene expression by binding to specific DNA sequences within the regulatory regions of genes (1). Due to their important role in the regulation of the gene expression, transcription factors play a central role in cell development, differentiation and growth in biological systems (2–4). Transcription factors typically exist in the cell in an inactive state, and become activated by the presence of a specific ligand, leading to the expression of target gene(s). The inhibition or undesired activation of transcription factors can lead to a number of diseases including cancer, developmental disorders, abnormal hormone responses, autoimmunity and inflammation (5–10). The cellular levels of such proteins can thus be used as diagnostic markers for many diseases, and the targeting of transcription factor activity represents a potential avenue for therapeutic intervention (11–13). Therefore, rapid and convenient high-throughput methodologies to detect sequence-specific DNA-binding proteins and monitor their DNA-binding activities are in high demand. Such technologies may be used for the development of inhibitors for the diagnosis or treatment of diseases related to dysfunctional transcription factor activity.
Traditional approaches for protein detection include gel mobility shift assays (5), DNA footprinting (6) and enzyme-linked immunosorbent assays (ELISAs) (7). In the gel mobility shift assays, the protein-bound and free DNA species are separated in non-denaturing polyacrylamide or agarose gel, and usually employ radioisotopes for visualization. These assays thus tend to be time-consuming, tedious and necessitate stringent safety measures to control radiographic exposure. Several DNA footprinting methods have been introduced including DNase I, exonuclease III, methidiumpropyl-EDTA-Fe(II), 1,10-phenanthroline-Cu(I) and EDTA-Fe(II) footprinting (6,8–10). Complementary approaches based on disrupting binding (interference footprinting) by specific base or phosphate modifications have also been developed (11). Gel mobility shift assays and DNA footprinting methods provide information on the highest affinity sites within a segment of DNA, but they do not provide quantitative binding information. ELISAs (7) are more sensitive and offer higher throughput detection, but they require multiple preparation and signal-amplification steps for the detection of low abundance transcription factors. Additionally, none of the traditional methods are amenable for real-time analysis.
Luminescence-based methodologies have emerged as an attractive alternative to the traditional methods of transcription factor activity detection due to their simplicity, low cost, high sensitivity and amenability to high-throughput screening (12). The use of luminescence also negates concerns related to radiographic exposure and/or the disposal of radioactive waste. The selectivity of luminescence detection can be easily achieved through modulation of the excitation and emission wavelengths. In addition, luminescence provides a rapid and continuous signal, allowing real-time monitoring of processes and cellular imaging by microscopy.
Over the last decade, there have been extraordinary advances in the field of luminescent methodologies for the detection of DNA-binding proteins. The discovery of molecular beacons (MBs) and aptamers have spurred the development of novel sensing platforms based on the myriad of DNA topologies and luminescent dyes available for the construction of the nucleic acid probe. We survey here recent advances in the development of luminescent methods for sensing DNA-binding proteins. For ease of access, we have divided the survey into several categories: (i) early methods (pre-2000) based on fluorescence anistropy or rudimentary FRET methods; (ii) the MB strategy; (iii) the use of aptamers for protein detection, including aptamer beacons; (iv) label-free detection of proteins using MBs or aptamers; and (v) methods based on exonuclease protection. We give a brief description for each assay and summarize their main findings, and we evaluate the advantages and disadvantages of each technique particularly with regards to sensitivity, selectivity, robustness and generalizability.
EARLY METHODS FOR LUMINESCENT PROTEIN DETECTION
The first methods for the luminescent detection of DNA-binding proteins were based on the recognition of the cognate sequence by the transcription factor, resulting in a change in the fluorescent properties of the labeled oligonucleotide or protein. As these methods are not the focus of our survey, we highlight here only a few examples of the early DNA-binding protein assays before they were superseded by the more modern FRET techniques discussed later. In 1990, Heyduk and Lee developed a novel fluorescence method to study the binding of Escherichia coli cAMP receptor protein (CRP) to the lac promoter (13). CRP, also known as catabolite gene activator protein (CAP), is a cAMP-binding regulatory protein in bacteria (14–16). A 32-bp duplex fragment 5′-CGCAATTAATGTGAGTTAGCTCACTCATTAGG-3′ of the lac promoter containing the primary binding site for CRP was labeled with 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (CPM) at the 5′-end via a short thiol linker. The cAMP-mediated binding of CRP to this duplex fragment can be followed by fluorescence anisotropy or fluorescence resonance energy transfer (FRET) from the protein tryptophan residues to CPM. The fluorescence titration results showed that a linear increase in the anisotropy or fluorescence intensity was observed with increasing CRP/DNA ratio in the presence of cAMP until ∼1.0, after which both signals plateaued. However, the maximal fold-change in the fluorescence emission response was only ∼20%, presumably due to unoptimized nature of the FRET interaction between the tryptophan residues and CPM. No detection limit was reported in the paper, but the data presented showed an observable increase in the fluorescence emission at CRP/DNA = 0.2, corresponding to ∼20 nM of CRP (13).
A FRET approach was employed by Parkhurst and co-workers in 1996 to study the interaction of Saccharomyces cerevisiae TATA-binding protein (TBP) with the TATA-promoter element (17). A 14-bp 5′-GGGCTATAAAAGGG-3′ based on the adenovirus major late promoter was doubly labeled with tetramethylrhodamine (TMR) and fluorescein at the 5′- and 3′-termini, respectively. In the absence of TBP, the relatively rigid nature of the DNA duplex holds the fluorophores apart, resulting in a low FRET response. However, addition of TBP bends the DNA and brings the 3′- and 5′-ends of the duplex closer together, resulting in a 37% decrease in the emission at 520 nm of fluorescein (donor) at a saturating 14:1 ratio of protein: DNA. FRET to the rhodamine (acceptor) moiety resulted in a concomitant increase in the emission at 580 nm. While not described as such, this early approach could be considered a prototype of the MB strategy.
In 1997, Gorski and co-workers (18) utilized fluorescence anisotropy to investigate the binding of estrogen response element (ERE) sequences to the estrogen receptor (ER) (19). A 35-bp duplex sequence 5′-AGCTTCGAGGAGGTCACAGTGACCTGGAGCGGATC-3′ containing the palindromic chicken vitellogenin ERE (20) was labeled with fluorescein at the 5′-end. Interestingly, both estradiol-bound and unbound ER was found to interact with the consensus ERE sequence with similar affinity, indicating that estrogen affects ER activity at a step other than DNA binding. However, at saturation, the fluorescence anisotropy of the DNA label increased by only ∼15%. While no detection limit was reported by the authors, the data suggest that ∼1 nM of ER could be readily detected by the fluorescence anisotropy method (18).
Even up until 2000, fluorescence anistropy was still being utilized for analysis of protein–DNA interactions, such as the interactions of herpes simplex virus type I protein (21) or human papillomavirus E2 protein (22) with DNA. The chief advantage of fluorescence anistropy methods is that only one fluorescent label on the DNA is required. However, fluorescence polarization is generally limited by a small dynamic range and is strongly dependent on the sizes of the DNA and protein molecules. Additionally, fluorescence anistropy is unable to give a visual signal, and this limits the applicability of such methods for the optical detection of DNA-binding proteins. Consequently, the use of fluorescence anistropy for protein detection has fallen somewhat out of favor, and its current primary use is for the direct measurement of binding isotherms for protein–DNA interactions (23). In the work by Heyduk and Lee (13), a change in the emission of the fluorophore results from resonance energy transfer between endogenous tryptophan residues in the protein to the fluorescent label on the oligonucleotide. The intensity of the fluorescence signal thus depends on several factors, such as the number of tryptophan residues in the protein as well as their proximity to the DNA-binding site in the protein. This method therefore is not generally applicable for the detection of DNA-binding proteins. Interest in these early methods for the detection of DNA-binding proteins waned soon after the emergence of MBs, as described in the next section.
METHODS FOR PROTEIN DETECTION USING MOLECULAR BEACONS
MBs have been widely used for the detection of nucleic acids due to their strong affinity and selectivity for their complementary sequences (24–26). The classical MB approach employs a DNA hairpin (stem-loop) oligonucleotide doubly-labeled with a fluorophore and a quencher at its two termini. In solution, the oligonucleotide exists in a hairpin conformation and is weakly fluorescent in solution due to effective FRET. Addition of the target DNA sequence ‘opens up’ the MB through hybridization, forming a duplex structure releases the fluorophore resulting in the restoration of fluorescence emission. Alternatively, if a fluorescent acceptor is used in place of a quencher, the emission of the acceptor label can also be measured to monitor the target-binding event. Various combinations of fluorescent labels and nucleic acid conformations have been employed to develop a range of switch-on or switch-off luminescent MBs for DNA-binding proteins.
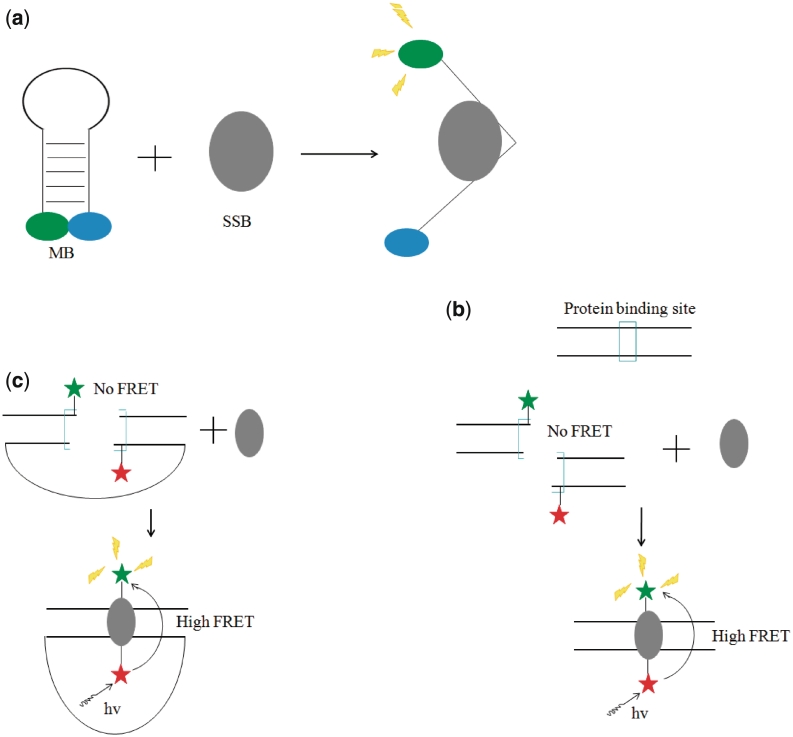
One of the first approaches employing the MB for the detection of DNA-binding proteins was reported by Tan and co-workers in 2000 (27). A 29-bp DNA sequence 5′-GCTCGTCCATACCCAAGAAGGAAGCGAGC-3′ was labeled with tamra (TMR) and dabcyl [4-(4′-dimethylaminophenylazo)benzoic acid] at the 5′- and 3′-termini, respectively. This oligonucleotide was utilized to detect the presence of E. coli single-stranded DNA-binding protein (SSB) (28). The binding of SSB to the MB was expected to disrupt the hairpin conformation of the oligonucleotide, resulting in the release of the fluorophore and the restoration of fluorescence (Figure 1a). Fluorescence titration experiments with SSB indicated a 1:1 binding ratio of the protein to the DNA, and a maximum fold-change of ∼30-fold was reached at ∼2:1 ratio. The detection limit for SSB for this system was determined to be 0.2 nM (27). This pivotal work by Tan and co-workers demonstrated that the MB approach could be successfully utilized for the switch-on detection of SSB with high sensitivity and selectivity (27). However, this system would be unable to differentiate between different SSB due to the lack of sequence recognition involved in the target-binding event. Later, the group utilized a similar MB strategy to analyze the non-specific binding of lactate dehydrogenase to single-stranded (ss) DNA (29).
Figure 1.
(a) Schematic representation of the MB strategy for the switch-on detection of prokaryotic ssDNA binding protein via the target-induced conversion of the MB hairpin into an ss conformation (27). (b) Schematic representation of MB strategy for the switch-on detection of prokaryotic CAP via the target-promoted association of the half-site duplexes (30). (c) Schematic representation of the unimolecular beacon strategy for the switch-off detection of thrombin via the target-induced formation of the closed state (31).
In 2002, Heyduk and Heyduk developed a general strategy for the detection of sequence-specific DNA-binding proteins (30). In this variation of a MB approach, the binding sequence for the transcription factor was split into two half-sites distributed between two labeled duplex fragments containing partially complementary overhangs. The sequence-specific binding of the transcription factor drives the formation of the DNA duplex, bringing the fluorochromes into close proximity and resulting in a high FRET signal (Figure 1b). For the detection of CAP, a 38-bp sequence corresponding to the consensus CAP site was used to generate the two half-site duplexes, each containing a 6-nt overhang of which only two were complementary to the other strand. The presence of CAP promoted the association of the two half-site duplexes, causing the emission of fluorescein (fluorophore) to be quenched by dabcyl (quencher). A maximum quenching efficiency of ∼70% was reached at 3:1 ratio of the CAP to the half-site duplexes (used at 50 nM). While no detection limit for this switch-off assay was reported by the authors, the data showed that nanomolar CAP could be readily detected (30). To achieve a switch-on fluorescence detection mode, Heyduk and Heyduk employed coumarin as a fluorescence donor and fluorescein as the acceptor. An ∼3-fold increase in the acceptor: donor emission ratio was observed at a saturating level of the protein. While the fold-change increase of this system was modest, the ratiometric detection of the signal could potentially mitigate the effects of a high background and/or non-selective quenching mechanisms due to interfering substances in the solution. The authors also adapted the MB for other prokaryotic transcription factors including trp repressor and lac repressor, and the human cancer-related DNA-binding protein p53, and for the detection of p53 in HeLa nuclear cell extracts. Furthermore, this MB approach was demonstrated for the multiplex detection of DNA-binding proteins. This important work by Heyduk and Heyduk demonstrated the feasibility of this alternative MB strategy as a general method for the selective detection of DNA-binding proteins (30). However, one drawback to this approach was that the rate of association of the half-site duplexes was relatively slow. Kinetic analysis performed by the group in a later work suggested that the rate of association of the duplex half-sites was the rate-limiting step of the overall reaction (31).
Heyduk and co-workers later developed a variant of the MB approach for CAP detection by tethering the two half-site duplexes together with a polyethylene glycol linker (32). The rationale for developing the unimolecular beacon was that the response time of the assay could be improved by enhancing the association of the two fragments, the rate-limiting step of the overall reaction (31). In this study, the addition of the target would be expected to convert the beacon from an ‘open’ formation to the ‘closed’ state, resulting in quenching of fluorescein by dabcyl (Figure 1c). When the protein was rapidly mixed with the MB, stopped-flow fluorescence measurements revealed that the reaction was complete in ∼0.5 s, which was ∼1200-fold faster than that with the bimolecular beacon (∼10 min) (30), verifying the authors’ hypothesis. ∼60% quenching was observed at a saturating concentration of CAP, and the data indicated a nanomolar detection limit. The authors also demonstrated the application of the unimolecular MB for immobilization onto a solid support, which opens up possibilities for the high-throughput detection of multiple protein targets. The immobilized MB was also able to sense nanomolar thrombin with a similar performance compared to the free beacon (32).
The chief advantages of MBs over the fluorescence polarization and early FRET methods described in the previous section are increased dynamic range and independence of the fluorescent signal to the size of the molecules or number and position of endogenous fluorescent amino acid residues, respectively. As a result, the MB can be readily adapted for sensing different DNA-binding proteins simply by changing the binding sequence of the oligonucleotide. However, these methods are still limited to the detection of proteins with intrinsic DNA-binding activity. The development of nucleic acid aptamers has extended the range of protein targets that can be detected, as discussed below.
METHODS FOR PROTEIN DETECTION USING APTAMERS
Aptamers can be regarded the nucleic acid equivalents of protein antibodies, and both DNA and RNA aptamers have been developed for a range of different targets including organic dyes, nucleotides, biological cofactors, amino acids, peptides and enzymes (33,34). These aptamers have been generally developed by the systematic evolution of ligands by exponential enrichment (SELEX) (35–38), a selection process that allows the discovery of high affinity ligands for any chemical or biological target. Compared to protein antibodies, DNA aptamers are generally smaller, less expensive to manufacture and modify, stable over a greater range of temperatures and pH values, and more readily reusable. Also, while the fluorescence signal of fluorescently-labeled antibodies generally remains the same whether or not the antibody is bound to the protein, the ligand-mediated transition of the aptamer secondary structure can be conveniently monitored using covalently-attached luminescent labels or non-covalent luminescent probes. These promising features of DNA aptamers have made them highly useful for sensing and biotechnology applications (39–44).
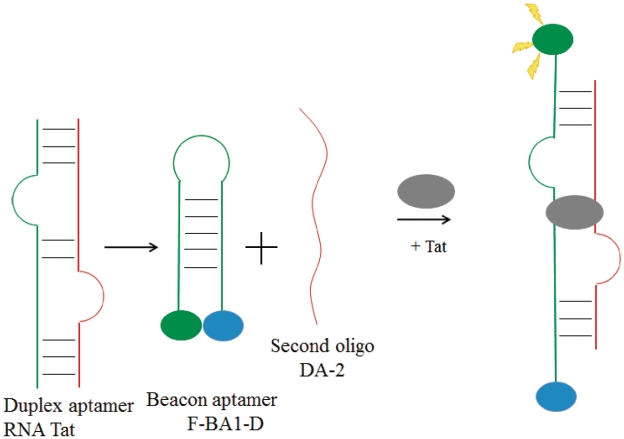
One of the first methods employing aptamers for the detection of nucleic acid-binding proteins was reported by Kumar and co-workers in 2000 (45). The HIV Tat protein RNA aptamer RNATat (46) was split into two separate strands, and one of the oligomers was elaborated into a MB by extending the sequences flanking the binding site with complementary nucleotides that was labeled with fluorescein and dabsyl at the 5′- and 3′-termini, respectively. Thus, the MB 5′-CGCGAAGCUUGAUCCCGAGAGCUUA-3′ (termed F-BA1-D) forms a stem–loop structure in solution with the binding residues located in the loop of the hairpin. Due to only partial complementarity of the beacon with the other half of the RNA aptamer 5′-CUCGGUCGAUCGCUUC-3′ (termed DA-2), the RNA duplex is not formed in the absence of Tat and the fluorescence of system remains low. The addition of the protein target causes the dissociation of the hairpin and promotes the formation of the Tat: F-BA1-D: DA-2 ternary complex, releasing the fluorophore and resulting in a switch-on fluorescence enhancement (Figure 2). In the presence of the minimal recognition region of Tat-1, termed CQ peptide, the emission of fluorescein was increased by 4.9-fold at a 10:1 ratio of the peptide to the beacon. Despite the relatively complex duplex nature of the RNA aptamer, necessitating the formation of a ternary complex with the protein target, this early work by Kumar demonstrated that the aptamer concept could be successfully utilized in conjunction with MBs to engineer novel oligonucleotide-based sensing platforms for nucleic-acid binding proteins.
Figure 2.
Schematic representation of molecular aptamer beacon strategy for the switch-on detection of viral Tat protein via the target-induced conversion of the MB hairpin into the aptamer duplex (45).
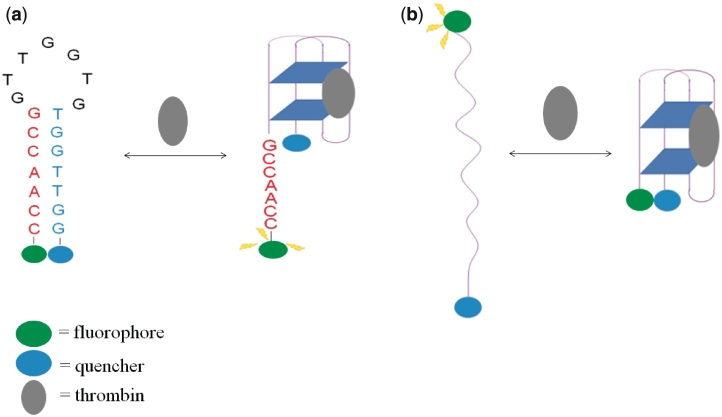
Hamaguchi and co-workers (47) designed a MB for thrombin (48) detection based on the thrombin aptamer in 2001. Intriguingly, the guanine-rich thrombin aptamer 5′-GGTTGGTGTGGTTGG-3′ (termed G15D) binds thrombin in an antiparallel G-quadruplex conformation (49,50). The aptamer was converted into a MB by extending the 5′-end of the aptamer by 5 nt, such that a hairpin conformation would be favoured in the absence of the target, and the aptamer beacon 5′-CCAACGGTTGGTGTGGTTGG-3′ (G15D5d) was labeled with fluorescein and dabcyl at the 5′- and 3′-ends, respectively (Figure 3a). Since thrombin only binds to the G-quadruplex conformation, addition of the target shifts the equilibrium towards the quadruplex form, resulting in the restoration of fluorescence. A maximum 2.5-fold increase in the fluorescence emission intensity was recorded at a saturating concentration of thrombin (45). No detection limit was reported by the authors, but 10 nM of thrombin could be readily detected using this assay. This early work by Hamaguchi demonstrated the application of molecular aptamer beacons for the detection of thrombin detection, and showed that the design of the molecular aptamer beacon was very important for the assay principle due to the strong dependence of the duplex–quadruplex equilibrium on the nucleotide sequence (45).
Figure 3.
(a) Schematic representation of the molecular aptamer beacon strategy for the switch-on detection of thrombin via the target-induced conversion of the MB hairpin into the G-quadruplex aptamer (45). (b) Schematic representation of the molecular aptamer beacon strategy for the switch-on (FRET) or switch-off (quenching) detection of thrombin via the target-induced conversion of the random coil into the G-quadruplex aptamer (51).
Tan and co-workers developed an alternative molecular aptamer beacon strategy for thrombin detection in 2002 (51). The 17-nt thrombin aptamer sequence 5′-TGGTTGGTGTGGTTGGT-3′ was labeled according to quenching or FRET detection modes. In this approach, the molecular aptamer initially exists as a random coil conformation, in which the fluorochromes are far apart (Figure 3b). The addition of the target protein shifts this equilibrium towards the G-quadruplex, resulting in quenching or an increase in acceptor emission for the quenching and FRET detection modes, respectively. For the quenching mode, a 60% decrease in the emission of fluorescein was observed at a saturating concentration of thrombin, and a detection limit of 0.37 nM of thrombin was reported (51). For the FRET mode of detection, the addition of thrombin resulted in ∼1-fold increase in the fluorescence intensity of fluorescein upon excitation of coumarin, with a detection limit of 0.43 nM. The performance of the sensor could be improved by ratiometric analysis of the acceptor and donor emission intensities. With ratiometric measurement, the detection limit was lowered to 0.11 nM (51).
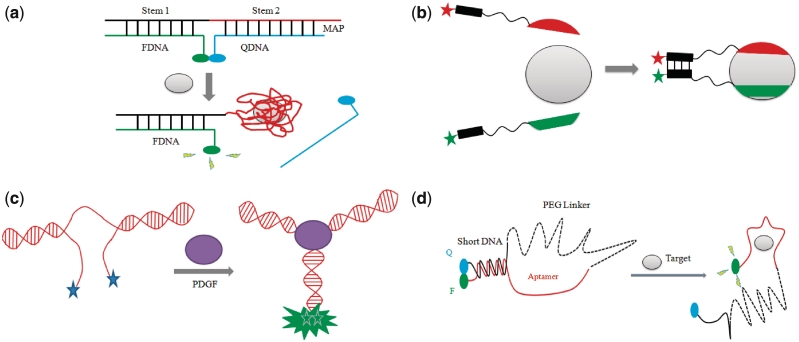
Li and co-workers designed a general aptamer beacon method for the detection of small molecules and proteins based on the target-induced disruption of a MB duplex (52). Unlike previous approaches, this strategy is less dependent on the size and structure of the aptamer. In the present study, the aptamer beacon consists of a tripartite duplex structure consisting of a 5′-fluorescein-labeled oligomer (FDNA), a 3′-dabcyl-labeled oligomer (QDNA) and a longer oligonucleotide sequence comprising Stem-1 and Stem-2 that are complementary to FDNA and QDNA, respectively. Stem-2 also contains the thrombin binding sequence 5′-GGTTGGTGTGGTTGG-3′ in a partial overhang. The addition of thrombin promotes the formation of the aptamer-target complex, leading to the dissociation of QDNA and the release of the fluorescein fluorophore, resulting in a fluorescence enhancement (Figure 4a). Titration experiments with thrombin revealed a linear response to thrombin concentrations over the range of 10–1000 nM with a maximum 12-fold enhancement in the fluorescence intensity (52). Although no detection limit was reported, 10 nM of thrombin could be readily sensed using this assay. Interestingly, reducing the number of blocked nucleotides in the aptamer sequence by QDNA also improved the performance of the ATP aptamer beacon reported in the same work, as this promotes the formation of the aptamer–target complex (52).
Figure 4.
(a) Schematic representation of the structure-switching aptamer strategy for the switch-on detection of thrombin via the target-induced dissociation of the duplex structure (52). (b) Schematic representation of molecular aptamer beacon design for the switch-on (FRET) or switch-off (quenching) detection of thrombin based on the target-induced co-association of two distinct aptamer recognizing distinct epitopes (53). (c) Schematic representation of aptamer beacon design for the excimer-based detection of PDGF via the target-induced hybridization of the terminal arms (54). (d) Schematic representation of the polyethylene glycol-linked aptamer beacon strategy for the switch-on detection of thrombin via the target-induced dissociation of the duplex conformation (58).
In 2005, Heyduk and co-workers developed a molecular aptamer beacon strategy for the detection of thrombin based on the protein-induced co-association of two aptamers recognizing distinct epitopes of the protein (53). The 60–18 5′-AGTCCGTGGTAGGGCAGGTTGGGGTGACT-3′ aptamer recognizing the fibrinogen-binding exosite was labeled with fluorescein at the 5′-end while the G15D aptamer recognizing the heparin-binding site was labeled with dabcyl at the 3′-terminus. The fluorophores were connected to their respective aptamers via a polyethylene glycol linker and a 7-nt complementary sequence, allowing the hybridization of the aptamers when both are bound to the target and resulting in efficient FRET (Figure 4b). A maximum quenching of ∼40% was observed at a saturating concentration of thrombin, with a detection limit of 1 nM of thrombin (53). With FRET fluorophores, up to 15-fold enhancement in the fluorescent emission of the acceptor could be observed, greatly improving the dynamic range of the sensor and allowing a favorable switch-on mode of detection. A further modification could be made by employing a europium chelate-Cy5 pair, which allows for a time-resolved FRET (TR-FRET) detection mode. In TR-FRET, the effects of background due to light scattering are avoided, allowing a further improvement of the signal-to-noise ratio of the beacon. With the Eu3+-Cy5 pair, a maximum 22-fold change in the fluorescence signal was registered. Additionally, an improved detection limit of 50 pM for thrombin was achieved (53). One advantage of this method over previously reported assays utilizing a single aptamer beacon is that the co-association of three molecular ‘contacts’ is required to generate a fluorescence signal. Due to the exponential relationship between the free energy and equilibrium dissociation constant of the complex, the overall stability of the complex will be greatly decreased if any of the three components were absent. Thus, the dual aptamer MB exhibited a greater specificity for thrombin detection compared to the single aptamer beacons. The authors also demonstrated that the dual aptamer beacon could detection nanomolar thrombin in spiked HeLa cellular extract and in factor Xa-treated plasma.
In 2005, the group of Turro and Tan (54) reported a molecular aptamer beacon for the detection of platelet-derived growth factor (PDGF)-BB (55,56) based on pyrene excimer emission. In the absence of protein, the PDGF-BB aptamer assumes a partial duplex structure where the 5′- and 3′-ends of the aptamer are free in solution. The binding of PDGF-BB promotes the hybridization of the terminal strands, forming a three-helix junction (Figure 4c). Where the ends of the aptamer are labeled with pyrene units, this causes a change of the fluorescence profile of pyrene from monomer emission (two signals at 375 and 398 nm) to excimer emission (485 nm) due to the close proximity of an excited state molecule with another ground state molecule. The 3-mer stem aptamer 5′-AGGCTACGGCACGTAGAGCATCACCATGATCCT-3′ gave the maximal signal response of the beacon to PDGF, ∼40-fold at saturation. The results revealed a linear response to PDGF over the range 0–40 nM, with a detection limit in the picomolar range (54). The authors also investigated the application of the excimer beacon for the detection of PDGF-BB in cell media. Taking advantage of the very long fluorescence lifetime (∼100 ns) of the pyrene excimer compared to most of the endogenous biological background fluorescence species (<5 ns), time-resolved fluorescence measurements were used to improve the sensitivity of the probe for PGDF in cell media, and a linear response to PDGF-BB concentration was observed over the range of 0–100 nM in cell media (54). As a proof-of-concept, the TNF-α-induced stimulation of PDGF production in human prostate cancer cells was readily detected using the excimer beacon. Although this MB relies on the specific conformation change of the PDGF aptamer and is not readily generalizable to other proteins, the authors demonstrated that the pyrene excimer emission can be a powerful tool for the sensitivity detection of proteins in the presence of interfering biological fluorophores in cell media.
Katilius and co-workers developed a variation of the molecular aptamer beacon strategy for the detection of proteins by incorporation a fluorescent nucleotide anologs within the aptamer sequence (57). The fluorescent quantum yields of these nucleotide anologs, such as 2-aminopurine (2AP), 4-amino-6-methylpteridone (6MAP) or 3-methylisoxanthopterin (3MI), are strongly dependent on the base-stacking interactions between the anolog and its neighbors in the SS or duplex chain. Consequently, if the fluorescent nucleotide is incorporated at a position that undergoes conformational change upon aptamer binding, the binding event can be monitored by measuring the emission signal of the nucleotide anolog. According to the published structure of the thrombin–aptamer complex, the thymine residue at position 7 becomes unstacked relative to its neighboring bases upon formation of the G-quadruplex, where it occupies part of a loop that does not interact with the protein. Thus, the authors incorporated a fluorescence nucleotide at T7, and a maximal 30-fold increase in the fluorescence intensity of 6MAP was registered upon addition of a saturating concentration of thrombin (57). While no detection limit was reported by the authors, the data revealed that ∼20 nM of thrombin could be readily detected using the 6MAP-modified aptamer. With PDGF and immunoglobulin E (IgE), no information on the tertiary structure of the aptamer–protein complex was available to the authors. However, the authors were able to calculate the secondary structure of the aptamer using the program mfold and predict which residues of the oligonucleotide would undergo major conformational changes upon target binding. The results showed that 6MAP modification at position 22 of the PDGF aptamer 5′-CACAGGCTACGGCACGTAGAGCATCACCATGATCCTGTG-3′ produced a 6.7-fold fluorescence increase upon saturating concentrations of the protein, with a detection limit of 10 nM. For the aptamers modified at the other positions, the addition of protein produced either no change or a slight decrease. This demonstrates that the actual position of the fluorescent nucleotide is of critical importance in the performance of the modified aptamer for protein detection. Using similar methods, replacement of position 18 of the IgE aptamer 5′-GGGGCACGTTTATCCGTCCCTCCTAGTGGCGTGCCCC-3′ with 2AP was found to give the highest response to target binding, and a 5.6-fold increase in the emission of the fluorescent nucleotide was observed at a saturating concentration of IgE. Interestingly, 6MAP exhibited the highest dynamic range for the thrombin and PDGF aptamers, whereas 2AP gave the highest fold change for the IgE aptamer (57). This suggests that the fluorescent nucleotides exhibit differential fluorescent behavior upon target binding when incorporated into the different aptamers, and that optimization of the fluorescent label can yield superior probe performance.
In 2008, Tan and co-workers developed a variation of the molecular aptamer beacon approach described previously (58). A polyethylene glycol linker was inserted into the G15D5d thrombin aptamer to give an aptamer beacon containing a longer loop region. The aptamer beacon 5′-CCAAC-(CH2CH2O)30-GGTTGGTGTGGTTGG-3′ was labeled with Chlorin e6 (Ce6) and Black Hole Quencher 2 (BHQ2) at the 5′- and 3′-ends of the oligomer, respectively. In the absence of the protein, the aptamer beacon forms a hairpin conformation containing a 5-mer stem and the emission intensity of the fluorophore is quenched. The addition of thrombin promotes the dissociation of hairpin, releasing the fluorophore and resulting in a fluorescence enhancement (Figure 4d). A 17.6-fold enhancement in fluorescence was reported after the addition of 300 nM of thrombin (58). Furthermore, the beacon was selective for thrombin over IgG, IgM and BSA added at the same concentration as thrombin. Mechanistically, the molecular aptamer beacon for thrombin detection operates in a similar fashion to that reported by Hamaguchi and co-workers (47). The higher maximal signal response reported in the present study may be attributed to the long polyethylene glycol linker which holds the quencher much further away from the aptamer strand upon target binding, enhancing the fluorescence intensity of the ‘on’ state. The use of the highly efficient BHQ2 as the quencher may also have contributed to the superior fold-change response by lowering the background fluorescence signal. The authors also demonstrated the application of this polyethylene glycol-linked aptamer beacon for ATP detection.
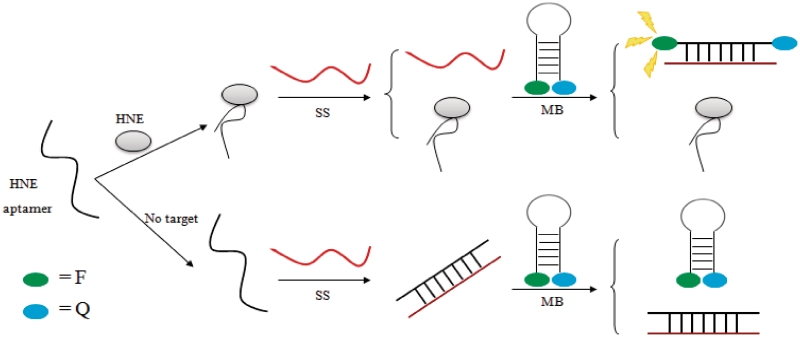
In 2010, Yu and co-workers developed an interesting aptamer probe for human neutrophil elastase (HNE) using a three-component system (59). The HNE aptamer 5′-TAGCGATACTGCGTGGGTTGGGGCGGGTAGGGCCAGCAGTCTCGT-3′ forms a G-quadruplex structure with duplex ends (60). The probe consists of the HNE aptamer, a MB labeled with fluorescein-dabcyl and an ss that is complementary to both the aptamer and the beacon. In the absence of the target, the free aptamer forms a duplex with ss, and the MB remains in the closed state with a low fluorescence. HNE competes with ss for the DNA aptamer, and free ss is instead able to interact with and open up the MB, resulting in the release of the fluorophore moiety and an enhancement of fluorescence emission (Figure 5). A linear response to HNE in the concentration range of 0.34–68 nM was observed, and a detection limit of 47 pM was reported (59). The advantage of this three-component system is that the aptamer does not have to be fluorescently labeled, so that the maximum affinity of the aptamer can be retained. Furthermore, this system is readily generalizable to any protein–aptamer pair as the precise structure of the aptamer is irrelevant to the working principle of the assay.
Figure 5.
Schematic representation of the three-component aptamer strategy for the switch-on detection of HNE (59).
LABEL-FREE METHODS FOR PROTEIN DETECTION
MBs and molecular aptamer beacons tend to be relatively easy to operate due the fixed arrangement of the fluorophore moieties on the oligonucleotide. However, oligonucleotide labeling is typically quite expensive, and can comprise up to 90% of the total cost of the oligonucleotide. Consequently, this has spurred the development of ‘label-free’ detection methods, where the luminescent probe is not covalently bound to the oligonucleotide. The key feature of label-free methods is that the emissive properties of the luminescent probes are highly sensitive to the local environment, such that the target-induced rearrangement of the MB or molecular aptamer beacon can be readily transduced into a luminescent signal output. The much lower cost of unlabeled oligonucleotides compared to their fluorescent counterparts makes them more attractive for low-cost applications and/or high-throughput detection. Furthermore, with fluorescently-labeled oligonucleotides, it is always necessary to consider the effect of the fluorophore on the secondary structure of the aptamer and/or the stability of the aptamer–protein complex. This factor is not as important with the label-free methods since most of the luminescent probes are able to bind to multiple locations on the aptamer. However, drawbacks to label-free method include a sometimes lower sensitivity and a lack of multiplex detection capability.
In 2004, the group of Fang and Bai developed a label-free detection method (61) for protein detection using aptamers in conjunction with the ‘molecular light switch’ complex [Ru(phen)2(dppz)]2+ (phen = 1,10-phenanthroline, dppz), dipyrido[3,2-a:2′,3′ c]phenazine) (62). The ruthenium complex is only weakly emissive in aqueous solution due to the coordination of the phenazine nitrogen with the water molecules, resulting luminescent quenching of the triplet MLCT excited state through non-radiative decay modes. Intercalation of the ruthenium complex into the hydrophobic environment within the DNA duplex protects the phenazine ligand from solvent quenching, leading to an emission enhancement. The phosphorescence emission of the ruthenium complex at 610 nm increased by 20-fold in the presence of the IgE aptamer, which is predicted to form a hairpin conformation in solution. However, upon addition of IgE, the secondary structure of the aptamer was disrupted, resulting in a maximal luminescent decrease of ∼35% at a 14:8:1 ratio of protein:complex:aptamer. The response was linear in the range of 0–0.8 nM, and a detection limit of 0.1 nM was reported (61). The probe was also able to function with the same performance in 1% serum compared to physiological buffer. The presence of non-specific duplex DNA could also be overcome by increasing the concentration of the ruthenium complex to ensure saturation of the intercalating dye into the aptamer. However, this would likely decrease the dynamic range of the system due to strong emission of the ruthenium complex when bound to the non-specific DNA. In the same work, similar label-free probes were developed for PDGF-BB and thrombin. For PDGF-BB, the authors employed the 35-nt three-way helix aptamer and achieved a detection limit of 1.0 nM for PDGF in both physiological buffer and 1% serum. A maximum decrease of 30% was observed at the highest concentration of PDGF tested. For thrombin detection, the authors interestingly chose a hairpin RNA aptamer instead of the DNA aptamers that have been described previously. Presumably, the ruthenium complex may not have exhibited strong binding to the well-known G15D thrombin G-quadruplex aptamer. With the RNA aptamer, the luminescence intensity of the probe decreased by ∼35% at saturation, and a detection limit of 10 pM was reported. This detection limit was superior to those achieved with DNA aptamers, and represents the lowest detection limit for thrombin using these methods reported to date. This work by the group of Fang and Bai successfully demonstrated the feasibility of using label-free detection for DNA-binding proteins (61). However, the relatively small dynamic range of the assay (∼35% maximal signal decrease) and the switch-off mode of detection are the drawbacks to this assay.
In 2006, the group of Li and Fang reported a modification of their earlier strategy by employing TOTO (63) instead of a ruthenium light switch complex (64). TOTO is an unsymmetrical cyanine dye that is non-fluorescent in solution but forms a highly fluorescent complex with DNA, with a 3000-fold increase in fluorescence quantum yield. Here, TOTO binds strongly to the PDGF-BB aptamer 5′-CAGGCTACGG CACGTAGAGCATCACCATGATCCTG-3′ in the absence of the protein, and is strongly fluorescent. However, the PDGF was found to decrease the fluorescence emission intensity of TOTO, presumably due to the protein-induced conformational change of the aptamer and the blocking of TOTO intercalation by the protein. The fluorescence of TOTO decreased by a maximum of ∼50% at a 20:6:1 ratio of protein: TOTO: aptamer, with a detection limit of 0.1 nM was reported for this method. By comparison, the use of the ruthenium complex [Ru(phen)2(dppz)]2+ as the intercalating dye for this particular system gave inferior results. This suggests that the response of the luminescent probes towards target binding may be different for different aptamer–protein complexes, and that a variety of luminescent probes should be tested for each system to achieve the best performance.
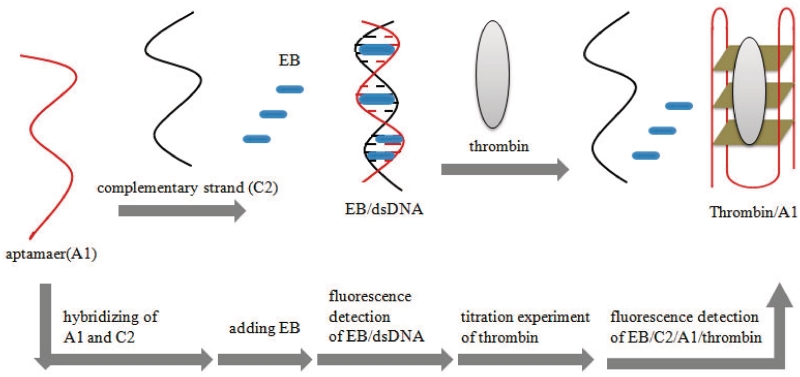
Dong and co-workers reported a more general method for the label-free luminescent detection of thrombin that does not rely on the nature of the target-induced conformational rearrangement of the aptamer (65). Here, the 15-nt thrombin aptamer 5′-GGTTGGTGTGGTTGG-3′ was hybridized with its complementary sequence 5′-CCAACCACACCAACC-3′, allowing intercalation of the well-known DNA-binding dye ethidium bromide (EB) (66) and resulting in a strong fluorescence signal. Thrombin competes with the complementary strand for the aptamer strand, leading to partial unwinding of the duplex and shifting the equilibrium towards the aptamer-thrombin complex (Figure 6). Since EB does not interact as well with the released ss complementary sequence or the aptamer G-quadruplex, a concentration-dependent reduction in the fluorescence intensity of EB was observed as the amount of thrombin added was increased. The titration results revealed a nearly linear response of the probe to thrombin in the range 0–22.8 nM, and a detection limit of 2.8 nM was reported (65). However, the maximum decrease in the emission intensity at a saturating concentration of thrombin was only 9%. This could be due to the non-specific binding of EB to ssDNA and/or the G-quadruplex, resulting in a lower sensitivity for the assay. Using a more selective DNA-binding dye may thus improve the performance of this probe for thrombin. Mechanistically, this duplex-to-quadruplex conversion resembles the thrombin molecular aptamer beacon previously described (47), except that in the present case, the aptamer binding site is buried within a duplex arrangement instead of mainly residing within the unhybridized loop region of the aptamer beacon. As a consequence, the aptamer binding sequence is less exposed to the target, resulting in a longer incubation time (30 min) required for binding.
Figure 6.
Schematic representation of the label-free aptamer strategy for the switch-off detection of thrombin via the target-induced conversion of the duplex structure into a G-quadruplex (65).
METHODS FOR PROTEIN DETECTION BASED ON EXONUCLEASE PROTECTION
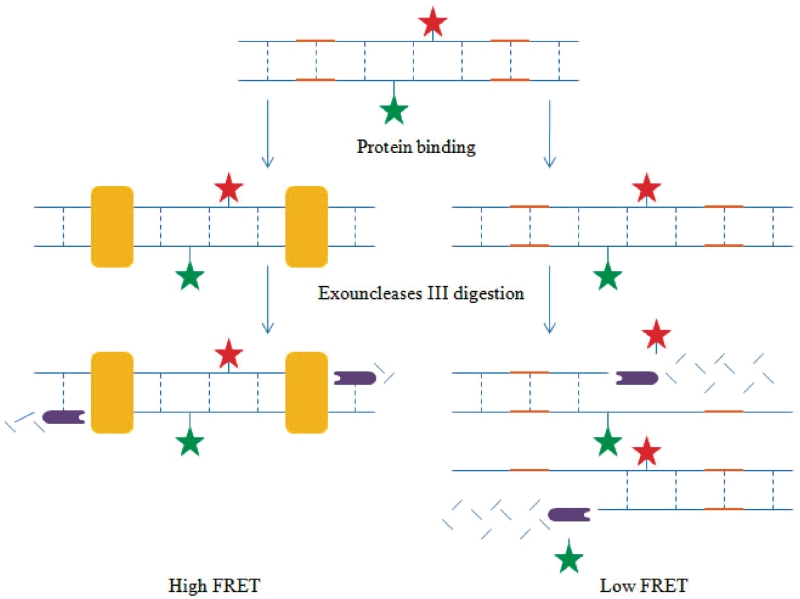
DNase footprinting assays (6) tend to be unwieldy and time-consuming, and necessitate stringent safety measures to control radiographic exposure. Wang and co-workers pioneered a new method for the detection of specific DNA-binding proteins by combining the principles of DNA footprinting with FRET detection (67). In this assay, the FRET probe consists of two complementary DNA oligonucleotides, one of which was labeled with a donor fluorochrome and the other with an acceptor, in the central region of the oligomers (Figure 7). Flanking the donor–acceptor pair are two identical complete protein-binding sites. If the cognate protein is present, the formation of the protein–DNA complex prevents the exonuclease III (ExoIII)-mediated nucleolytic degradation of the probe, and the FRET pair remains intact. In the absence of protein, the digestion of the probe by ExoIII will release the fluorochromes as mononucleotides, resulting in a low FRET signal. As a proof-of-concept, the complementary oligonucleotides 5′-AGTTGAGGGGACTTTCCCAACTAGGAAFTCTACCTGGGGACTTTCCCAGGC-3′ (F = dT-fluorescein) and 3′-TCAACTC CCCTGAAAGGGTTGATCCTDAAGATGGACCCCTGAAAGGGTCCG-5′ (D = dT-dabcyl) containing the NF-κB (68–70) consensus sequence GGGACTTTCC were prepared for p50 transcription factor detection. The results revealed that as the concentration of p50 protein was progressively increased, the emission intensity of the fluorescein at the end of the digestion reaction was gradually reduced, due to the protection of the FRET probe by the DNA-binding protein which preserved the close proximity of the fluorophore to the quencher. While no detection limit was reported by the authors, the 5 nM of p50 could be readily detected using this probe (67). Similar FRET probes were prepared for SP1 and p53. The authors also applied the probe for the detection of NF-κB activity in TNF-α-stimulated HeLa nuclear cell extracts. Furthermore, multiplex protein detection was demonstrated using this assay by utilizing fluorophores with distinct excitation and emission wavelengths (67). The chief advantage of the exonuclease protection assay method for the detection of DNA-binding proteins compared to the MB or aptamer beacon methods described previously is that it is no longer necessary to consider the equilibria between the multiple conformations of the oligonucleotide and the bound and free states of the beacon or aptamer. Thus, the present method is generally applicable for the detection of proteins with intrinsic DNA-binding activity. Note that there should be sufficient distance between the fluorophore pair and each of the protein binding sites, to avoid steric hindrance to protein binding. Furthermore, the exonuclease protection assay method may also be susceptible to false positives arising from non-specific DNA binding.
Figure 7.
Schematic representation of the exonuclease protection assay for the switch-off detection of transcription factors (67).
A similar principle was employed by the group of Levine and Mirkin to develop a variant of the exonuclease protection assay termed ‘fluorescence recovery’ (71). Here, the fluorophore (fluorescein) and quencher (dabcyl) were incorporated at the end of the duplex probe rather than in the middle. Since ExoIII preferentially recognizes the free 3′-hydroxyl on the double-stranded DNA and cleaves in the 3′ → 5′ direction, the digestion of the duplex can only occur from one terminus since the other is blocked by the fluorescent dyes. Consequently, only one protein binding site is required. In the absence of the protein, digestion of the strand containing the fluorophore proceeds freely, releasing the fluorophore from the quencher and resulting in the recovery of fluorescence. In the presence of the target, ExoIII digestion cannot proceed past the protein-binding site, and the proximity of the fluorophore pair is preserved. To demonstrate this, a 26-bp Xenopus laevis vitellogenin A2 ERE sequence 5′-CAAAGTCAGGTCACAGTGACCTGATC-3′ was labeled with fluorescein at the 5′-end, while its complementary sequence 5′-GATCAGGTCACTGTGACCTGACTTTG-3′ was labeled with dabcyl at the 3′-terminus. As the concentration of ER was increased, the percentage of fluorescence recovered was reduced due to the protection of the DNA duplex by the transcription factor. At a 2.6:1 ratio of protein to DNA, only ∼42% of the emission of fluorescein was recovered after incubation with ExoIII for 20 min (71). The authors demonstrated the multiplex detection capability of the system by constructing distinct probes that contained different fluorophore–quencher pairs (such as fluorescein–dabcyl or Cy5–dabcyl). When the probes were incubated in the same solution in the presence of ERα and ExoIII, the fluorescence recovery for the Vit ERE sequence (65%) was lower than that for the VEGF sequence (90%), suggesting that the transcription factor binds more strongly to the ERE sequence than VEGF (71). This study demonstrates that this system can be employed to determine the relative binding affinity of a transcription factor for a variety of sequences in a multiplexed format.
In 2011, our research group reported a label-free variation of the fluorescence recovery assay for the detection of NF-κB p50 DNA-binding activity (72). Here, the ‘molecular light switch’ complex [Ru(phen)2(dppz)]2+ was used to discriminate between bound and unbound DNA products resulting from exonuclease digestion. In the absence of p50, the duplex probe becomes digested by ExoIII. The ruthenium complex only interacts weakly with the resulting ss fragments, and the final phosphorescence intensity of the solution is low. The addition of p50 protects the probe from exonucleolytic cleavage, allowing the intercalation of the metal complex and resulting in a high luminescence signal (Figure 8). The results revealed that the duplex substrate 5′-TTGAGGGACTTTCCGAACATGCAGGCAAGCTGGGGACTTTCCAGG-3′ containing two binding sites exhibited the best performance, with an 8-fold maximal luminescence increase at a 50:20:1 ratio of the ruthenium complex:protein:DNA after incubation with ExoIII (72). Significantly, this assay produces a ‘switch-on’ response to the transcription factor target, in contrast to the switch-off mode of detection reported for the previously described exonuclease protection assays. A switch-on detection mode is generally more desirable for analytical purposes as they are less susceptible to false positives arising from non-specific quenching mechanisms from interfering species in the sample matrix. In the presence of NF-κB inhibitor oridonin (73,74), a marked reduction in the luminescence response of the probe was observed, indicating that this assay may be used to screen for modulators of transcription factor activity.
Figure 8.
Schematic representation of the label-free exonuclease protection assay for NF-κB p50 using molecular light switch complex [Ru(phen)2(dppz)]2+.
CONCLUSIONS AND OUTLOOK
Over the last 10 years, there have been tremendous strides in the development of luminescent assays for the detection of DNA-binding proteins. Early methods were based on fluorescence anistropy or rudimentary FRET methods relying on the interaction between endogenous tryptophan residues or fluorescent labels in the protein with fluorescent oligonucleotides. However, disadvantages of these approaches include a strong dependence on molecular size (for anistropy) and lack of generalizability of the assay. The discovery of the MB strategy mitigated many of these drawbacks as the conformational change of the oligonucleotide upon target binding could be rationally predicted. Thus, labeled probes could be engineered that were able to respond to this structure-switching event with a measurable luminescence response, based largely on FRET principles. We have described here several approaches that employ MBs for the detection of DNA binding probes, including those based on the association of duplex half-sites (30–32) or the disruption of the MB hairpin structure by non-specific SSB (27,29). The most sensitive MB in this class was that for prokaryotic SSB reported by Tan and co-workers, which achieved a detection limit of 0.2 nM (27). Heyduk and Heyduk also demonstrated that the use of a donor–acceptor combination for ratiometric FRET detection could give a higher fold-change luminescence response for the MB compared to the quenching mode using a fluorophore–donor pair (30). However, these beacons were limited only to the detection of non-specific DNA-binding proteins or those with endogenous sequence-recognition capabilities. The discovery of aptamers, which are the nucleic acid equivalents of proteins that are obtained through the SELEX, has greatly expanded the repertoire of possible proteins that can be detected through this approach. Some of these molecular aptamer beacons were based on the protein-induced conformational change of the aptamer resulting in the joining or separation of fluorophores in space, producing a change in the fluorescence signal. Others were based on the displacement of a competitor strand from a DNA duplex by the target. The advantage of the latter approach is that the precise conformational change of aptamer is irrelevant, allowing the probe design to be generalized for the detection of other DNA-binding proteins. However, the drawback is that the partial blocking of the protein binding site on the aptamer within a duplex environment necessitates an increased incubation time to allow the equilibrium to gradually shift towards the dissociated aptamer strand, which could then be progressively captured by the protein target (52,59). Both quenching and ratiometric modes of detection have been utilized for the molecular aptamer beacon approach. Furthermore, the group of Turro and Tan showed that excimer emission could be an excellent alternative to FRET probes due to their longer fluorescence lifetime compared to that of background biological species, allowing the sensitive detection of PDGF in biological fluids using a pyrene-labeled aptamer and employing time-resolved fluorescence (54). We also highlighted one example where the incorporation of a fluorescent nucleotide anolog into the aptamer sequence could be used to produce a probe for DNA-binding proteins exhibiting up to 30-fold increases in emission intensity (58). The lowest detection limit in the class of molecular aptamer beacons reported was the three-component aptamer probe for human neutrophil elastase reported by Yu and co-workers, which could sense 47 pM of protein at a signal-to-noise cut-off ratio of 3 (59). However, the expensive cost of fluorescently-labeled oligonucleotides has engendered the development of label-free luminescence detection methods that are more attractive from an economical standpoint for high-throughput or low-cost applications. Based on the selectivity of the external dyes for different nucleic acid conformations, the target-induced structure switching of the MB or aptamer beacon can be readily transduced into a luminescence response. The dyes that have been used for the luminescence detection of DNA-binding proteins include both organic and inorganic molecules, such as TOTO (64), EB (65) and the molecular light switch complex [Ru(phen)2(dppz)]2+ (61). However, a significant drawback to the label-free approach is the lack of multiplex capability. The lowest detection limit for the label-free method was 10 pM for thrombin using [Ru(phen)2(dppz)]2+ and the thrombin aptamer (61). Finally, we described luminescence methods for the detection of DNA-binding proteins based on exonuclease protection. Here, the binding of the protein target to its cognate sequence protects the DNA from degradation by exonuclease III. Both labeled oligonucleotides (67,71) and label-free approaches using [Ru(phen)2(dppz)]2+ (72) have been applied towards the luminescence exonuclease protection assay for the detection of proteins. The advantages and disadvantages of the different luminescent detection methods reviewed in this work are summarized in Table 1.
Table 1.
Advantages and disadvantages of different luminescent detection methods for DNA-binding proteins
| Methods | Advantages | Disadvantages |
|---|---|---|
| Fluorescence anistropy or early FRET methods | Only one fluorescent label on the DNA is required. |
|
| MB strategy |
|
|
| Aptamer beacon strategy |
|
|
| Label-free strategy | Economical, suitable for high-throughput or low-cost applications. |
|
| Exonuclease protection strategy | No need to consider the equilibria between multiple oligonucleotide conformations and the bound and free states of the beacon or aptamer. | Can be more susceptible to false positives. |
In our opinion, the outlook for the further development of luminescent detection methods for DNA-binding proteins is highly promising. The MB strategy should be generally applicable to all transcription factors since these proteins recognize specific DNA sequences in order to regulate gene expression. Aptamers obtained through SELEX have already been discovered for a whole host of molecular entities such as amino acids, proteins, cofactors and small molecules. Similar to antibodies, it should be theoretically possible in principle to raise high-affinity aptamers to any protein target, which could allow these aptamer-based methods to be applied to the detection of proteins that have high diagnostic or therapeutic importance, regardless of their intrinsic DNA-binding activity. Methods based on exonuclease protection also offer new possibilities for the design of novel luminescent detection assays, as these do not rely on specific conformational changes for the beacon or aptamer and thus can be readily generalizable for the detection of any transcription factor. Furthermore, the principles of the exonuclease protection assay can be adapted for the detection of any enzyme with DNA-modulating activities (75,76).
In terms of the luminescent labels or dyes, there are several potential areas of improvement. First, new fluorescence labels are being developed all the time (77,78), including new technologies such as quantum dots (79), which may pave the way for improved detection technologies that exhibit lower background signals and higher dynamic ranges. Another exciting recent development is the use of surface plasmon resonance (SPR) technologies to analyze protein–DNA interactions, which features an optical readout, high sensitivity, semi-automated detection protocols and ability to determine binding rate constants and equilibrium constants. For example, SPR has been used to analyze the interaction between DNA and E. coli SSB (80), tobacco transcription factor NtERF2 (81), methyl-CpG binding domain protein (82), human ER (83), HIV-1 reverse transcriptase (84), streptavidin (85), human IgE (86) and yeast transcription factor Gal4 (87). The ultimate objective of luminescence assays is to be able to image specific DNA–protein interactions directly in living tissues. Imaging in the near infra-red (NIR) range is a step towards that goal due to lower scattering and absorption of NIR photons in the tissue. Bogdanov and co-workers have recently developed a FRET-based NIR probe for NF-κB detection. Using a Cy5.5–Cy7 donor–acceptor pair, the authors were able to detect nanomolar p50 by measuring the ratio of the fluorescence emission intensities at 700 (donor) and 800 (acceptor) nm (88). However, it is likely that additional modifications will also have to be made to the nucleic acid probes to protect them from degradation by endogenous nucleases in living tissue. For the label-free techniques for the detection of DNA-binding proteins, all of the methods described here have utilized an intercalating organic or inorganic dye for signal generation. Potentially, the use of a more selective DNA-binder with the ability to interact specifically with a particular aptamer conformation (for example, the thrombin aptamer G-quadruplex) could give rise to new detection methodologies with improved selectivity in the presence of excess genomic DNA. The methods described in this review have paved a firm foundation for the future development of novel luminescent DNA-based sensors for the detection of proteins. Issues of biostability and cellular delivery will likely be of paramount importance for the sensing of proteins in living cells or tissues. Given the tremendous advances achieved in this field over the last decade, it is exciting to consider the realm of possibilities as researchers take these developments to the next level.
FUNDING
Funding for open access charge: Hong Kong Baptist University (FRG2/10-11/008); Environment and Conservation Fund (ECF Project 3/2010).
Conflict of interest statement. None declared.
REFERENCES
- 1.Papavassiliou AG. Molecular medicine. Transcription factors. N. Engl. J. Med. 1995;332:45–47. doi: 10.1056/NEJM199501053320108. [DOI] [PubMed] [Google Scholar]
- 2.Tenen DG, Hromas R, Licht JD, Zhang D-E. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 3.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 4.Costa RH, Kalinichenko VV, Holterman A-XL, Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38:1331–1347. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 5.Garner MM, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galas DJ, Schmitz A. DNAase footprinting a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engvall E, Perlmann P. Enzyme-Linked Immunosorbent Assay, Elisa. J. Immunol. 1972;109:129–135. [PubMed] [Google Scholar]
- 8.Kuwabara M, Sigman DS. Footprinting DNA-protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry. 1987;26:7234–7238. doi: 10.1021/bi00397a006. [DOI] [PubMed] [Google Scholar]
- 9.Van Dyke MW, Hertzberg RP, Dervan PB. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II) Proc. Natl Acad. Sci. USA. 1982;79:5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tullius TD, Dombroski BA, Churchill MEA, Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- 11.Hayashibara KC, Verdine GL. Template-directed interference footprinting of cytosine contacts in a protein-DNA complex: potent interference by 5-aza-2′-deoxycytidine. Biochemistry. 1992;31:11265–11273. doi: 10.1021/bi00161a002. [DOI] [PubMed] [Google Scholar]
- 12.Hill JJ, Royer CA. In: Methods in Enzymol. Ludwig Brand MLJ, editor. Vol. 278. Amsterdam: Academic Press; 1997. pp. 390–416. [DOI] [PubMed] [Google Scholar]
- 13.Heyduk T, Lee JC. Application of fluorescence energy transfer and polarization to monitor Escherichia coli cAMP receptor protein and lac promoter interaction. Proc. Natl Acad. Sci. USA. 1990;87:1744–1748. doi: 10.1073/pnas.87.5.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harman JG. Allosteric regulation of the cAMP receptor protein. Biochim. Biophys. Acta. 2001;1547:1–17. doi: 10.1016/s0167-4838(01)00187-x. [DOI] [PubMed] [Google Scholar]
- 15.Bott M. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch. Microbiol. 1997;167:78–88. [PubMed] [Google Scholar]
- 16.Weickert MJ, Adhya S. The galactose regulon of Escherichia coli. Mol. Microbiol. 1993;10:245–251. doi: 10.1111/j.1365-2958.1993.tb01950.x. [DOI] [PubMed] [Google Scholar]
- 17.Parkhurst KM, Brenowitz M, Parkhurst LJ. Simultaneous binding and bending of promoter DNA by the TATA binding protein: real time kinetic measurements. Biochemistry. 1996;35:7459–7465. doi: 10.1021/bi9530301. [DOI] [PubMed] [Google Scholar]
- 18.Ozers MS, Hill JJ, Ervin K, Wood JR, Nardulli AM, Royer CA, Gorski J. Equilibrium binding of estrogen receptor with DNA using fluorescence anisotropy. J. Biol. Chem. 1997;272:30405–30411. doi: 10.1074/jbc.272.48.30405. [DOI] [PubMed] [Google Scholar]
- 19.McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296:1642–1644. doi: 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- 20.Burch JB, Evans MI, Friedman TM, O'Malley PJ. Two functional estrogen response elements are located upstream of the major chicken vitellogenin gene. Mol. Cell. Biol. 1988;8:1123–1131. doi: 10.1128/mcb.8.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gourves AS, Tanguy Le Gac N, Villani G, Boehmer PE, Johnson NP. Equilibrium binding of single-stranded DNA with herpes simplex virus type I-coded single-stranded DNA-binding protein. ICP8. J. Biol. Chem. 2000;275:10864–10869. doi: 10.1074/jbc.275.15.10864. [DOI] [PubMed] [Google Scholar]
- 22.Lima LMTR, Foguel D, Silva JL. DNA tightens the dimeric DNA-binding domain of human papillomavirus E2 protein without changes in volume. Proc. Natl Acad. Sci. USA. 2000;97:14289–14294. doi: 10.1073/pnas.250352197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LiCata VJ, Wowor AJ. In: Methods Cell Biol. John JC, Detrich HW III, editors. Vol. 84. Amsterdam: Academic Press; 2008. pp. 243–262. [Google Scholar]
- 24.Leone G, vanGemen B, Schoen CD, van Schijndel H, Kramer FR. Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res. 1998;26:2150–2155. doi: 10.1093/nar/26.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan W, Wang K, Drake TJ. Molecular beacons. Curr. Opin. Chem. Biol. 2004;8:547–553. doi: 10.1016/j.cbpa.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 26.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 27.Li JJ, Fang X, Schuster SM, Tan W. Molecular beacons: a novel approach to detect protein – DNA interactions. Angew. Chem. Int. Ed. 2000;39:1049–1052. doi: 10.1002/(sici)1521-3773(20000317)39:6<1049::aid-anie1049>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 28.Lohman TM, Ferrari ME. Escherichia Coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Ann. Rev. Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 29.Fang X, Li JJ, Tan W. Using molecular beacons to probe molecular interactions between lactate dehydrogenase and single-stranded DNA. Anal. Chem. 2000;72:3280–3285. doi: 10.1021/ac991434j. [DOI] [PubMed] [Google Scholar]
- 30.Heyduk T, Heyduk E. Molecular beacons for detecting DNA binding proteins. Nat. Biotechnol. 2002;20:171–176. doi: 10.1038/nbt0202-171. [DOI] [PubMed] [Google Scholar]
- 31.Heyduk E. Molecular beacons for detecting DNA binding proteins: mechanism of action. Anal. Biochem. 2003;316:1–10. doi: 10.1016/s0003-2697(03)00004-6. [DOI] [PubMed] [Google Scholar]
- 32.Knoll E, Heyduk T. Unimolecular beacons for the detection of DNA-binding proteins. Anal. Chem. 2004;76:1156–1164. doi: 10.1021/ac034985p. [DOI] [PubMed] [Google Scholar]
- 33.Famulok M. Oligonucleotide aptamers that recognize small molecules. Curr. Opin. Struct. Biol. 1999;9:324–329. doi: 10.1016/S0959-440X(99)80043-8. [DOI] [PubMed] [Google Scholar]
- 34.Famulok M, Mayer G, Blind M. Nucleic acid aptamers from selection in vitro to applications in vivo. Accounts Chem. Res. 2000;33:591–599. doi: 10.1021/ar960167q. [DOI] [PubMed] [Google Scholar]
- 35.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 36.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 37.Osborne SE, Ellington AD. Nucleic acid selection and the challenge of combinatorial chemistry. Chem. Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 38.Jhaveri S, Rajendran M, Ellington AD. In vitro selection of signaling aptamers. Nat. Biotechnol. 2000;18:1293–1297. doi: 10.1038/82414. [DOI] [PubMed] [Google Scholar]
- 39.Strehlitz B, Nikolaus N, Stoltenburg R. Protein detection with aptamer biosensors. Sensors. 2008;8:4296–4307. doi: 10.3390/s8074296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song S, Wang L, Li J, Fan C, Zhao J. Aptamer-based biosensors. TrAC Trend. Anal. Chem. 2008;27:108–117. [Google Scholar]
- 41.Balamurugan S, Obubuafo A, Soper S, Spivak D. Surface immobilization methods for aptamer diagnostic applications. Anal. Bioanal. Chem. 2008;390:1009–1021. doi: 10.1007/s00216-007-1587-2. [DOI] [PubMed] [Google Scholar]
- 42.Famulok M, Hartig JS, Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 43.Tombelli S, Minunni M, Mascini M. Analytical applications of aptamers. Biosens. Bioelecton. 2005;20:2424–2434. doi: 10.1016/j.bios.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Willner I, Zayats M. Electronic aptamer-based sensors. Angew. Chem. Int. Ed. 2007;46:6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto R, Baba T, Kumar PK. Molecular beacon aptamer fluoresces in the presence of Tat protein of HIV-1. Genes Cells. 2000;5:389–396. doi: 10.1046/j.1365-2443.2000.00331.x. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto R, Katahira M, Nishikawa S, Baba T, Taira K, Kumar PKR. A novel RNA motif that binds efficiently and specifically to the Tat protein of HIV and inhibits the trans-activation by Tat of transcription in vitro and in vivo. Genes Cells. 2000;5:371–388. doi: 10.1046/j.1365-2443.2000.00330.x. [DOI] [PubMed] [Google Scholar]
- 47.Hamaguchi N, Ellington A, Stanton M. Aptamer beacons for the direct detection of proteins. Anal. Biochem. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]
- 48.Bailey K, Bettelheim FR, Lorand L, Middlebrook WR. Action of thrombin in the clotting of fibrinogen. Nature. 1951;167:233–234. doi: 10.1038/167233a0. [DOI] [PubMed] [Google Scholar]
- 49.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 50.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li JJ, Fang X, Tan W. Molecular aptamer beacons for real-time protein recognition. Biochem. Biophys. Res. Commun. 2002;292:31–40. doi: 10.1006/bbrc.2002.6581. [DOI] [PubMed] [Google Scholar]
- 52.Nutiu R, Li Y. Structure-switching signaling aptamers. J. Am. Chem. Soc. 2003;125:4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 53.Heyduk E, Heyduk T. Nucleic acid-based fluorescence sensors for detecting proteins. Anal. Chem. 2005;77:1147–1156. doi: 10.1021/ac0487449. [DOI] [PubMed] [Google Scholar]
- 54.Yang CJ, Jockusch S, Vicens M, Turro NJ, Tan W. Light-switching excimer probes for rapid protein monitoring in complex biological fluids. Proc. Natl Acad. Sci. USA. 2005;102:17278–17283. doi: 10.1073/pnas.0508821102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonner JC. Regulation of PDGF and its receptors in fibrotic diseases. Cytokine Growth F. R. 2004;15:255–273. doi: 10.1016/j.cytogfr.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 56.Cao Y, Cao R, Hedlund E-M. R Regulation of tumor angiogenesis and metastasis by FGF and PDGF signaling pathways. J. Mol. Med. 2008;86:785–789. doi: 10.1007/s00109-008-0337-z. [DOI] [PubMed] [Google Scholar]
- 57.Katilius E, Katiliene Z, Woodbury NW. Signaling aptamers created using fluorescent nucleotide analogues. Anal. Chem. 2006;78:6484–6489. doi: 10.1021/ac060859k. [DOI] [PubMed] [Google Scholar]
- 58.Tang Z, Mallikaratchy P, Yang R, Kim Y, Zhu Z, Wang H, Tan W. Aptamer switch probe based on intramolecular displacement. J. Am. Chem. Soc. 2008;130:11268–11269. doi: 10.1021/ja804119s. [DOI] [PubMed] [Google Scholar]
- 59.He JL, Wu ZS, Zhang SB, Shen GL, Yu RQ. Fluorescence aptasensor based on competitive-binding for human neutrophil elastase detection. Talanta. 2010;80:1264–1268. doi: 10.1016/j.talanta.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 60.Lin Y, Padmapriya A, Morden KM, Jayasena SD. Peptide conjugation to an in vitro-selected DNA ligand improves enzyme inhibition. Proc. Natl Acad. Sci. USA. 1995;92:11044–11048. doi: 10.1073/pnas.92.24.11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang Y, Fang X, Bai C. Signaling aptamer/protein binding by a molecular light switch complex. Anal. Chem. 2004;76:5230–5235. doi: 10.1021/ac049565u. [DOI] [PubMed] [Google Scholar]
- 62.Friedman AE, Chambron JC, Sauvage JP, Turro NJ, Barton JK. A molecular light switch for DNA: Ru(bpy)2(dppz)2+ J. Am. Chem. Soc. 1990;112:4960–4962. [Google Scholar]
- 63.Rye HS, Glazer AN. Interaction of dimeric intercalating dyes with single-stranded DNA. Nucleic Acids Res. 1995;23:1215–1222. doi: 10.1093/nar/23.7.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou C, Jiang Y, Hou S, Ma B, Fang X, Li M. Detection of oncoprotein platelet-derived growth factor using a fluorescent signaling complex of an aptamer and TOTO. Anal. Bioanal. Chem. 2006;384:1175–1180. doi: 10.1007/s00216-005-0276-2. [DOI] [PubMed] [Google Scholar]
- 65.Li B, Wei H, Dong S. Sensitive detection of protein by an aptamer-based label-free fluorescing molecular switch. Chem. Commun. 2007:73–75. doi: 10.1039/b612080f. [DOI] [PubMed] [Google Scholar]
- 66.LePecq JB, Paoletti C. A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J. Mol. Biol. 1967;27:87–106. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- 67.Wang J, Li T, Guo X, Lu Z. Exonuclease III protection assay with FRET probe for detecting DNA-binding proteins. Nucleic Acids Res. 2005;33:e23. doi: 10.1093/nar/gni021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tak PP, Firestein GS. NF-κB: a key role in inflammatory diseases. J. Clin. Invest. 2001;107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Q, Verma IM. NF-[kappa]B regulation in the immune system. Nat. Rev. Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 70.Kumar A, Takada Y, Boriek AM, Aggarwal BB. Nuclear factor-kB: its role in health and disease. J. Mol. Med. 2004;82:434–448. doi: 10.1007/s00109-004-0555-y. [DOI] [PubMed] [Google Scholar]
- 71.Xu X, Zhao Z, Qin L, Wei W, Levine JE, Mirkin CA. Fluorescence recovery assay for the detection of protein-DNA binding. Anal. Chem. 2008;80:5616–5621. doi: 10.1021/ac8007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma D-L, Xu T, Chan DS-H, Man BY-W, Fong W-F, Leung C-H. A highly selective, label-free, homogenous luminescent switch-on probe for the detection of nanomolar transcription factor NF-κβ. Nucleic Acids Res. 2011;39:e67. doi: 10.1093/nar/gkr106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ikezoe T, Yang Y, Bandobashi K, Saito T, Takemoto S, Machida H, Togitani K, Koeffler HP, Taguchi H. Oridonin, a diterpenoid purified from Rabdosia rubescens, inhibits the proliferation of cells from lymphoid malignancies in association with blockade of the NF-κB signal pathways. Mol. Cancer Ther. 2005;4:578–586. doi: 10.1158/1535-7163.MCT-04-0277. [DOI] [PubMed] [Google Scholar]
- 74.Leung C-H, Grill SP, Lam W, Han Q-B, Sun H-D, Cheng Y-C. Novel mechanism of inhibition of nuclear factor-κB DNA-binding activity by diterpenoids isolated from isodon rubescens. Mol. Pharmacol. 2005;68:286–297. doi: 10.1124/mol.105.012765. [DOI] [PubMed] [Google Scholar]
- 75.Dai N, Kool ET. Fluorescent DNA-based enzyme sensors. Chem. Soc. Rev. 2011 doi: 10.1039/c0cs00162g. in press, doi:10.1039/C0CS00162G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leung C-H, Chan DS-H, Man BY-W, Wang C-J, Lam W, Cheng Y-C, Fong W-F, Hsiao W-LW, Ma D-L. Simple and Convenient G-Quadruplex-Based Turn-On Fluorescence Assay for 3′ → 5′ Exonuclease Activity. Anal. Chem. 2011;83:463–466. doi: 10.1021/ac1025896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gonçalves MST. Fluorescent labeling of biomolecules with organic probes. Chem. Rev. 2008;109:190–212. doi: 10.1021/cr0783840. [DOI] [PubMed] [Google Scholar]
- 78.O'Hare HM, Johnsson K, Gautier A. Chemical probes shed light on protein function. Curr. Opin. Struct. Biol. 2007;17:488–494. doi: 10.1016/j.sbi.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 79.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 80.Lu JQ, Xu MB, Zhou XW, Xu JG, Tao Q. A novel detection of single-stranded DNA binding protein based on ss-DNA modified chip using surface plasmon resonance microscopy. Chin. Chem. Lett. 2007;18:441–444. [Google Scholar]
- 81.Hao D, Ohme-Takagi M, Yamasaki K. A modified sensor chip for surface plasmon resonance enables a rapid determination of sequence specificity of DNA-binding proteins. FEBS Lett. 2003;536:151–156. doi: 10.1016/s0014-5793(03)00045-0. [DOI] [PubMed] [Google Scholar]
- 82.Pan S, Xu J, Shu Y, Wang F, Xia W, Ding Q, Xu T, Zhao C, Zhang M, Huang P, et al. Double recognition of oligonucleotide and protein in the detection of DNA methylation with surface plasmon resonance biosensors. Biosens. Bioelectron. 2010;26:850–853. doi: 10.1016/j.bios.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 83.Teh HF, Peh WYX, Su X, Thomsen JS. Characterization of protein-DNA interactions using surface plasmon resonance spectroscopy with various assay schemes. Biochemistry. 2007;46:2127–2135. doi: 10.1021/bi061903t. [DOI] [PubMed] [Google Scholar]
- 84.Wu L, Zhang Q, Su L, Huang M, Zhao J, Yang M. Effects of small molecular inhibitors on the binding between HIV-1 reverse transcriptase and DNA as revealed by SPR biosensor. Sens. Actuators, B. 2007;B122:243–252. [Google Scholar]
- 85.Yang N, Su X, Tjong V, Knoll W. Evaluation of two- and three-dimensional streptavidin binding platforms for surface plasmon resonance spectroscopy studies of DNA hybridization and protein-DNA binding. Biosens. Bioelectron. 2007;22:2700–2706. doi: 10.1016/j.bios.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 86.Pollet J, Delport F, Janssen KPF, Jans K, Maes G, Pfeiffer H, Wevers M, Lammertyn J. Fiber optic SPR biosensing of DNA hybridization and DNA-protein interactions. Biosens. Bioelectron. 2009;25:864–869. doi: 10.1016/j.bios.2009.08.045. [DOI] [PubMed] [Google Scholar]
- 87.Shumaker-Parry JS, Aebersold R, Campbell CT. Parallel, quantitative measurement of protein binding to a 120-element double-stranded DNA array in real time using surface plasmon resonance microscopy. Anal. Chem. 2004;76:2071–2082. doi: 10.1021/ac035159j. [DOI] [PubMed] [Google Scholar]
- 88.Zhang S, Metelev V, Tabatadze D, Zamecnik PC, Bogdanov A., Jr Fluorescence resonance energy transfer in near-infrared fluorescent oligonucleotide probes for detecting protein-DNA interactions. Proc. Natl Acad. Sci. USA. 2008;105:4156–4161. doi: 10.1073/pnas.0800162105. [DOI] [PMC free article] [PubMed] [Google Scholar]