Abstract
What physical mechanism leads to organization of a highly condensed and confined circular chromosome? Computational modeling shows that confinement-induced organization is able to overcome the chromosome's propensity to mix by the formation of topological domains. The experimentally observed high precision of separate subcellular positioning of loci (located on different chromosomal domains) in Escherichia coli naturally emerges as a result of entropic demixing of such chromosomal loops. We propose one possible mechanism for organizing these domains: regulatory control defined by the underlying E. coli gene regulatory network requires the colocalization of transcription factor genes and target genes. Investigating this assumption, we find the DNA chain to self-organize into several topologically distinguishable domains where the interplay between the entropic repulsion of chromosomal loops and their compression due to the confining geometry induces an effective nucleoid filament-type of structure. Thus, we propose that the physical structure of the chromosome is a direct result of regulatory interactions. To reproduce the observed precise ordering of the chromosome, we estimate that the domain sizes are distributed between 10 and 700 kb, in agreement with the size of topological domains identified in the context of DNA supercoiling.
INTRODUCTION
Even the simplest organisms need to physically organize their chromosomes. Bacterial chromosomes form a compact DNA–protein complex called the nucleoid (1,2) where the interplay between compaction of the genetic material and its accessibility ensure vital cell functions such as DNA replication, segregation, gene expression and repair. In this work, we investigate an intriguing biological and physical problem: what physical mechanisms lead to organization of highly condensed and confined circular chromosomes?
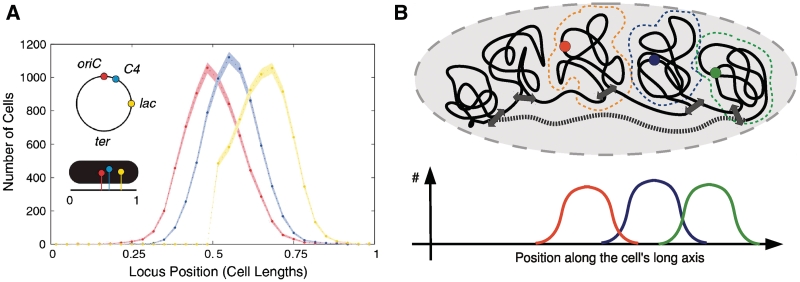
One of us previously measured and analyzed the position fluctuations of 15 single genetic loci (in G and early S phase), among which are the origin of replication OriC as well as two loci named ‘lac’ and ‘C4’ for convenience [for more details see (3)]. Loci in the body of the nucleoid were found to show a precision of positioning of better than 10% of the cell length and the precision of interlocus distance of genomically proximate loci was found to be better than 4%. Moreover, the linear relationship between positions of the genes on the chromosome and their spatial location within the cell was confirmed. While most studies of the localization of chromosomal loci in bacteria have focused on their position along the length of the cell, a recent study reports data concerning loci positioning across the width of the Escherichia coli cell finding ter-borne loci localized at the nucleoid periphery (4).
By physicochemical approaches, it has long been shown that the circular E. coli DNA molecule is organized into separate chromosomal loops or superhelical domains that are relaxed independently when DNA is cut (2,6,13–15). One purpose of these topological domains appears to be the prevention of chromosome unraveling as a result of DNA damage since the loss of chromosomal supercoiling leads to cell death (16). However, cross-links between random positions on the chromosome would also protect against unwinding (2). Instead of that genomic neighbors appear to be cross-linked suggesting that the so formed domains might serve structure–function relationships with respect to gene expression, too (7,17–21).
Experimental determinations of the average sizes of these domains differ between 10 kb (6) and 100 kb (23), report intermediate values (24), and even indicate an organization into a ring compacted by four macrodomains and two less structured regions (7,25). Thus, there already is evidence of a non-trivial nucleoid structure where smaller domains are organized within higher order ‘super’ structures.
Several drivers responsible for the observed reliable orientation and high level of organization of the E. coli chromosome have been suggested, including intranucleoid interactions such (i) macromolecular crowding (5), (ii) DNA supercoiling (6) or (iii) protein–DNA interactions (7–9) as well as explicit mechanisms of external positioning such as (iv) cellular confinement (3,10) or (v) tethering of the chromosome (11,12).
Increasing lines of evidence link the three-dimensional (3D) packaging of genes to the proper coordination of gene expression (7,17–21). Transcription factors (TFs) are the key controlling elements for appropriate gene expression in bacteria, where a functional network of regulatory interactions between TFs and target genes, which can themselves be TFs, is formed (20,22). Assuming the spatial nucleoid structure to influence transcription and vice versa, specific correlations are expected to arise in gene expression patterns (2). In fact, expression patterns correlate at short (<16 kb), medium (∼100 kb) and long (∼600–700 kb) distances in E. coli (2,26,27). The short-range correlations might result from small elementary domains of the nucleoid, while the long-range correlations could result from higher scales of organization, i.e. from long-range interactions between regions of the chromosome that are genomically distant, but spatial proximate when the chromosome is packaged within the nucleoid (2).
Underpinning the evidence for ‘regulatory domains’, a recent experimental study on the spatial organization of mRNA in E. coli shows that mRNAs display limited dispersion from their site of transcription (19). The high localization of mRNA implies that chromosome architecture might act as a spatial organizer, which compartmentalizes the cell interior such that dedicated (regulatory) proteins are produced within those subcellular regions, where their regulatory intervention is needed (8,19,28,29).
The assumption of functional domains is supported by another work on the role of transcriptional regulation in shaping the organization of genes on a chromosome (20). It was demonstrated that the more target genes a TF regulates, the higher is its need to be expressed in higher concentrations to regulate targets located dispersedly on the chromosome. In contrast, local or dedicated TFs were found to be expressed in much lower concentrations explaining the reasons for their proximity on the chromosome to their target genes (20). This aspect justifies an a posteriori conformational organization of DNA to produce colocalization phenomena since it is a natural way to make 3D targeting and assembly of complexes more efficient and error free.
Additionally, there is experimental evidence from other bacteria suggesting that there is a link between the final expression products and chromosome organization (30). Visualization of replicated DNA within living cells of Bacillus subtilis show that genes from distant chromosomal regions co-localize within a similar subcellular location for the purpose of coregulation (30).
How do bacterial cells operate such coordinated movement of specific sites within the compacted genome rapidly and faithfully? This is a place, in which physical modeling of polymers in confined space can help to relate experimental observations on E. coli nucleoid structure and to quantitatively test our models for chromosome organization.
First, we demonstrate that confinement and condensation are not sufficient to spontaneously organize the chromosome. Without dividing the chromosome into topological domains, confinement-induced organization cannot overcome propensity of the chromosome to mix. The high precision of separate subcellular positioning of loci located on different chromosomal domains naturally emerges as a result of entropic demixing of these structural subunits. This concept might have important implications for chromosome segregation since the increasing topological complexity of the stacked sequence of chromosomal loops implies a stronger repulsion between replicated DNA chains, and consequently more precise organization and faithful segregation (31,32).
In a subsequent step, we propose and investigate one possible mechanism for organizing these domains: the gene regulatory network. It was demonstrated that in the gene regulatory network in E. coli, regulatory genes need to be expressed in different concentrations in dependence of the genomic distance from their target genes and of the number of target genes regulated (20). Additionally, it was shown that expressed mRNAs largely display limited dispersion from their sites of transcription, which suggests that translation is spatially organized by using the chromosome layout as a template (19). In light of these recent findings, we suggest that regulatory control requires the co-localization of TF genes and target genes. We do not propose a detailed mechanism generating these attractive interactions [which might result from protein–protein or from protein–RNA interactions (33,34)], but rather explore its consequences in shaping the physical structure of the E. coli chromosome.
In fact, we find that the DNA chain self-organizes into several topologically distinguishable domains where the interplay between the entropic repulsion of chromosomal loops and their compression due to the confining geometry induces a formation into a stacked sequence of interlinked domains. These domains are sufficient to generate the observed precision of E. coli chromosome structure and we estimate the domain sizes to be distributed between 10 and 700 kb, in agreement with the size of topological domains identified in the context of DNA supercoiling.
MATERIALS AND METHODS
Ring polymers or polymer loops do not intermingle, but entropically repel each other both in free space and even more strongly in confinement (31,35,36). This can be understood by noting that two ring polymers suffer a loss of conformational entropy when being brought together within a distance smaller than their gyration radius (31,32,36). This tendency to segregate, which holds to a lesser extend for linear domains, too, leads to compartmentalization. The repulsive effect of loop formation is highlighted in Supplementary Figure S1 confirming that the density clouds of adjacent polymeric loops are indeed well separated.
These results suggest that the experimentally observed separate subcellular localization of the three genetic loci in (3) and displayed in Figure 1 could emerge due to their positioning on different chromosomal domains/loops. In fact, it is the mutual entropic repulsion between chromosomal domains that constitutes an elegant, self-organized mechanism of ordering in confinement due to compartmentalization. However, without dividing the chromosome into topological domains, confinement-induced organization cannot overcome the propensity of the chromosome to mix. Additionally, the experimental observation of equal variance of the position distributions displayed in Figure 1 suggests the absence of tethering interactions.
Figure 1.
(A) Histogram of long-axis locus position for IL01t C4 cells [adapted from (3)]. The genomic locus positions are shown schematically in the inset. Loci in the body of the nucleoid show a precision of positioning within the cell of better than 10% of the cell length. (B) A cartoon of the chromosomal domains being the ‘building blocks’ of the E. coli chromosome. The dark gray double arrows represent TF–gene interactions which ‘restrain’ the domains. The domain ‘walls’ indicated by the red, blue and green dashed lines are able to diffuse and thus subject to position fluctuations. The subcellular position distributions of the red, blue and green loci [fluorescently tagged in experiments (3,38,47,48)] examplify the precise separate positioning of genetic sites on different domains due to their tendency of demixing. The gray dashed line represents the ter-proximate region acting as a linker that connects the two polymer arms. A detailed description of the polymer model can be found in section ‘Domain Topology and Confinement Shape E. coli Chromosome Packaging’ in Supplementary Data.
In order to quantitatively test our assumptions, (i) we apply the Metropolis Monte Carlo method (37) to model the circular chromosome by a ring polymer being compacted due to fixed size loops within a rod-shaped geometry representing the nucleoid. The ter-region is represented by a stretched linker connecting the two polymer arms. This approach is based on a recent work finding the E. coli chromosome to be organized with a linker that connects the outer edges of the nucleoid (38). Figure 1B illustrates the employed polymer model and section ‘Domain Topology and Confinement Shape E. coli Chromosome Packaging’ in Supplementary Data explains further details on the polymer description. Comparing this approach to a ‘null model’, which consists of a simple ring polymer confined to the same geometry, allows for the investigation of the impact of domain formation on the packaging of highly confined chromosomes.
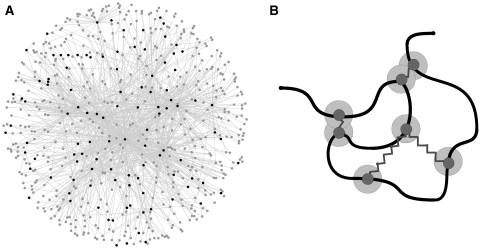
In a subsection step (ii), we propose that the E. coli gene regulatory network permits the genetic loci to identify and form domains with genetic neighbors. We investigate the consequences of this assumption by applying numerical simulations, where the chromosome is modeled as a self-avoiding polymer in confinement. TFs along the chromosome are associated with the respective sites along the polymer chain. These sites interact with their target sites according to an effective attractive potential, mimicking a regulatory interplay and driving TFs and their target genes to colocalize in space. Figure 2A illustrates the transciptional regulatory network as used in this work. Additionally, Figure 2 shows a 2D cartoon of the 3D self-avoiding DNA chain, which mimics the regulatory control between TF genes and target genes by assuming a harmonic interaction between these sites. Further details on the polymer description as well as on the E. coli gene regulatory network are shown in section ‘Gene Expression and Colocalization’ in Supplementary Data.
Figure 2.
(A) Graphical illustration of the transcriptional regulatory network describing the regulatory interplay between the TFs and their target genes as applied in our polymer model. Black nodes represent TF genes, dark gray nodes represent the target genes and links represent regulatory interactions between them. A detailed description of the network can be found in section ‘Gene Expression and Co-localization in Supplementary Data’. Global regulators are those TF genes that regulate lots of target genes, while other regulatory proteins are local, dedicated regulators. (B) 2D cartoon of the 3D self-avoiding DNA chain [adapted from (39)] mimicking the regulatory control between TF genes and target genes by assuming a harmonic interaction between these sites. Sites that can interact with other sites are represented by small blue filled circles connected by blue springs. The outer blue circles define the strength of the harmonic interaction potential. See section ‘Gene Expression and Co-localization’ in Supplementary Data for further details on the polymer description and the TF–gene network.
RESULTS
Division of the chromosome into domains is key to confinement-induced organization
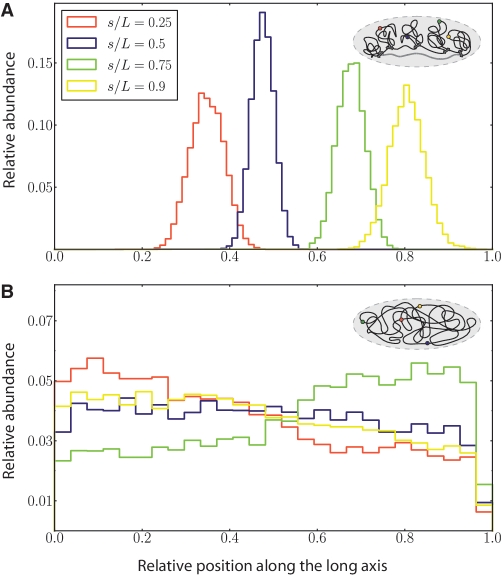
The position distribution of four sites along the polymer backbone with respect to the long axis of the confining cavity both for the E. coli polymer model as well as for the ‘null model’ are displayed in Figure 3. In case of the bare ring polymer, the absence of distinguished domains leads to a loss of spatial ordering that is reflected in the polymer's mobility throughout the confining geometry. In contrast, the specific topology of the E. coli polymer model leads to an interplay between the domain's tendency of de-mixing and the pressure exerted by confinement. As a result, the stacking of polymer loops inside the rod-shaped geometry induces a high level of spatial positioning illustrated by the well-defined and separated position distributions in Figure 3.
Figure 3.
The position distribution of four sites located at the relative positions 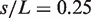 , 0.5, 0.75 and 0.9 along the polymer backbone L are shown with respect to the long axis of the confining cavity for (A) the domain model and (B) the ‘null model’. In case of the bare ring polymer, the absence of distinguished domains leads to a loss of spatial ordering which is reflected in the polymer's mobility throughout the confining geometry. In contrast, the specific topology of the E. coli polymer model induces a high level of spatial positioning. The interplay between the domain's tendency of de-mixing and the pressure exerted by confinement leads to a stacking of polymer loops inside the rod-shaped geometry.
, 0.5, 0.75 and 0.9 along the polymer backbone L are shown with respect to the long axis of the confining cavity for (A) the domain model and (B) the ‘null model’. In case of the bare ring polymer, the absence of distinguished domains leads to a loss of spatial ordering which is reflected in the polymer's mobility throughout the confining geometry. In contrast, the specific topology of the E. coli polymer model induces a high level of spatial positioning. The interplay between the domain's tendency of de-mixing and the pressure exerted by confinement leads to a stacking of polymer loops inside the rod-shaped geometry.
The specific spatial organization is able to explain the general linear correlation between positioning of genes on the chromosome and their location in the cellular volume: adjacent loops are displaced along the long axis of the rod-shaped confining volume in such a way to form a sequence of stacked ‘neighbor’ loops or an ‘effective nucleoid filament’ (3). The linear relationship between genomic and subcellular position is apparent in Figure 3, too.
An analysis of how the SMC-like protein MukBEF condenses DNA revealed its likely involvement in organizing the chromosome in a series of loops orthogonal to the cell axis, which was argued to account for the orderly arrangement of the chromosome (40). Here, we show that entropy might provide a purely physical driving force to support the dedicated action of specific proteins such as chromosome condensation proteins in order to create the right physical conditions for chromosome packaging. Notably, this result is confirmed by the experimental observation that cells with the MukBEF deletion are capable of almost wild-type chromosome structure.
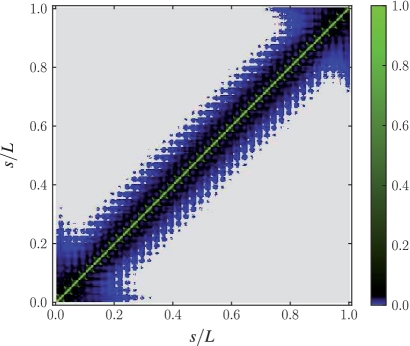
An established measure of compartmentalization are the contact probabilities between pairs of sites along the polymer as shown in Figure 4. The sites are numbered consecutively and the contact map displays their probabilities of establishing a contact, i.e. whenever they are spatially closer than a threshold distance dthreshold. In fact, the very low contact probability of non-diagonal contacts in Figure 4 shows that domain formation induces contacts among sites within the same and between sites of genomically neighboring domains. Thus, the experimentally observed high precision of subcellular locus positioning of better than 10% of the cell length (3) for genetic sites lying on different chromosomal domains has to be interpreted within the concept of structural units as the building blocks of the chromosome consisting of (supercoiled) DNA loops stabilized by DNA-binding proteins (41). The size of the structural units simultaneously influences the organization of the chromosome (segregation of chromosomal domains versus mixing) and its conformation (ordered versus random) in confined space (32). Consequently, the size of the structural unit is reflected in the decay width of the contact probability backbone shown in Figure 4. While Valens et al. (25) provide one measure of contact probabilities, Hi-C would provide another complementary approach with the opportunity to observe the interactions in a different biochemical context (cross-linking) and with a higher resolution in order to test our predictions (42).
Figure 4.
The contact probability map shows the probability for pairs of sites along the polymer backbone 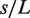 to form a contact, i.e. whenever both sites are spatially closer than a threshold distance
to form a contact, i.e. whenever both sites are spatially closer than a threshold distance 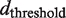 . Domain formation induces contacts among sites within the same domain and between sites of neighboring domains. The experimentally observed high precision of subcellular locus positioning can be explained by noting that the mobility of sites belonging to different chromosomal domains is restricted to the radius of the so-defined structural subunit.
. Domain formation induces contacts among sites within the same domain and between sites of neighboring domains. The experimentally observed high precision of subcellular locus positioning can be explained by noting that the mobility of sites belonging to different chromosomal domains is restricted to the radius of the so-defined structural subunit.
Moreover, while other domain organization models conclude that domain barriers are not placed stably at fixed sites on the chromosome, but are effectively randomly distributed hard walls (6), we have shown that their position need not be stochastic, since the domain walls are able to diffuse and thus subject to position fluctuations.
A recent study has evaluated the position of E. coli (43) chromosomal loci across the width of the cell by tagging loci with fluorescent proteins and comparing the measured distributions with simulated ones from different cell width models (4). The terminus region of the chromosome is found to be excluded from the body of the nucleoid and preferentially located at its periphery (4,44). Our model of the E. coli chromosome topology displays the same characteristic feature. The polymer chain region, which represents the ter-borne chromosome part, is preferentially located at the periphery of the confining cavity as is shown in Figure 5. Within our modeling approach, this result can be understood by noting that the (mostly linearly stretched) ter-region has less topological complexity compared with the looped ‘filament’-like structure of the remaining nucleoid. The topological complexity of the domains not only leads to an entropic repulsion between adjacent chromosomal domains, but also pushes the ter-region towards the envelope of the confining cavity. A donut like topology is observed if the topological complexity of chromosome is uniform (45).
Figure 5.
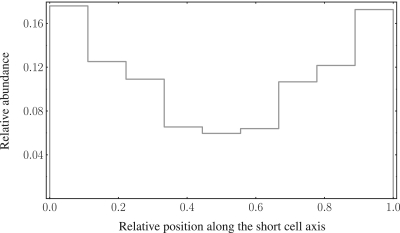
The relative abundance of ter-proximate sites as a function of their positioning with respect to the short axis of the confining geometry (cell's small axis). Meile and coworkers (4) have determined the position distributions of loci in the terminus region with respect to the short axis of the cell. They find these ter-borne loci localized at the nucleoid periphery. In our model, the polymer chain region representing the stretched ter-proximate region connecting the two nucleoid edges is preferentially located near the confining envelop, too, thus confirming the experimental observation. Within our modeling approach, this can be understood by noting that the (mostly linearly stretched) ter-region has less topological complexity compared with the looped ‘filament’-like structure of the remaining nucleoid. The topological complexity of the domains not only leads to an entropic repulsion between adjacent chromosomal domains, but also pushes the ter-region towards the envelope of the confining cavity in agreement with the experimental findings (4).
The gene regulatory network as a mechanism for domain formation
We have shown that the interplay between entropic repulsion of chromosomal domains and pressure exerted by the envelope of the confining cavity can be one driving force for nucleoid organization (and segregation) supporting the action of dedicated cellular machinery (46,47) or internal pushing forces (31,32,48). However, while the existence of chromosomal domains is widely accepted, their size and the mechanism that gives rise to them is still under debate. In this work, we investigate one possible mechanism for organizing chromosomal domains: the gene regulatory network.
The decision about gene expression or repression is controlled by TFs, which use metabolic or environmental signals to trigger a transcriptional response (22) within a functional network of regulatory interactions between TFs and target genes (21,49–51). Additionally, the proper genome-wide coordination of gene expression has been shown to be linked to the spatial organization of the chromosome within the nucleoid (8,22,52,53). In this respect, one can distinguish between ‘analog’ control, i.e. regulatory action by chromosome topology, and ‘digital’ control, i.e. regulation mediated by transcription factors (21,50,51,54).
Regarding the observed existence of chromosome domains in E. coli, we propose that one-dimensionally distant target genes, i.e. genes that are genomically far away from their regulative TF, colocalize with it in order to facilitate transcription. This assumption is supported by a recent work on the role of transcriptional regulation in shaping the organization of genes on a chromosome (20). It was demonstrated that the higher a TF is in the transcriptional hierarchy, the higher is its need to be expressed in higher concentrations to regulate target genes located dispersedly on the chromosome. In contrast, local or dedicated TFs, that are lower in the network hierarchy, were found to be expressed in much lower concentrations explaining the reasons for their proximity on the chromosome to their target genes. This aspect justifies an a posteriori conformational organization of DNA to produce colocalization phenomena since it is a natural way to make 3D targeting and assembly of complexes more efficient and error free. Thus, the formation of a specific chromosome topology (chromosomal domains) stabilized by nucleoid-associated proteins (NAPs) who are in charge of most chromosomal remodeling tasks could be seen as a feature of analog control.
Since a model for E. coli domain organization involves the recognition of a domain-specific pattern by a protein which would isolate it from other domains, various experimental groups have been looking for dedicated proteins that bind specifically to a single domain to organize the chromosome. A few examples of domain forming proteins have been identified. In B. subtilis, the DNA-binding ParB-like protein Spo0J appears to ensure proper arrangement and partitioning of chromosomal DNA by recruiting the condensin structural maintenance of chromosomes (SMCs) complex to the replication origin region (55,56). In E. coli, SMC-like complex MukBEF appears to colocalize with the origin of replication, but the mechanism is unknown. A recent study has identified a protein MatP that structures the terminus macrodomain. ChIP experiments have revealed that proteins SeqA and SlmA both appear to be excluded from the terminus region (7,57,58). However, the existence of these known examples is insufficient to explain the precision of structure observed on the chromosome (7). Here, we propose that structure proteins have been hiding in plain sight. They are the TFs that are already known to target small regions of the chromosome specifically.
We mimic the regulatory control between TF genes and target genes in the transciptional regulatory network by assuming a harmonic interaction between these sites as illustrated in Figure 2. Within this rather general framework, we find that the DNA chain indeed self-organizes into several topologically distinguishable domains. Figure 6 displays a snapshot of the genome organization as obtained after the equilibration of the self-interacting DNA chain, i.e. when the system fluctuates around its global energy minimum.
Figure 6.
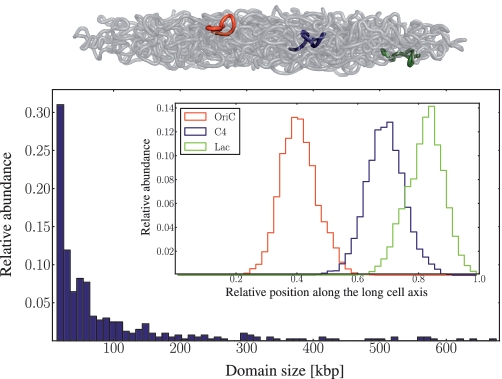
The 3D chromosome organization is obtained after the equilibration of the interacting self-avoiding polymer chain in a rod-shaped geometry. Modeling the regulatory interplay between TFs and their targets based on the E. coli transcriptional regulatory network, we find that genes on a DNA chain self-organize into several topologically distinguishable domains of different sizes. The chromosomal loop to which OriC is associated with is displayed in red, while the blue and green marked chromosomal regions refer to the chromosome domains that contain the genetic loci C4 and lac, respectively. It becomes clear that the three genetic loci are well separated with respect to projections on the long axis of the confining envelope. The formed domains are sufficient to generate the observed precision of E. coli chromosome structure and we estimate the domain sizes to be distributed between 10 and 700 kb.
Local TFs regulating a small number of target genes tend to localize peripherally, while global TFs which regulate a large number of genes assume more central localizations within the confining geometry. Several models have been proposed that lead to such a macro-arrangement of the nucleoid (59,60). In particular, the microarray experiments by Jeong et al. (27) show a high degree of correlation between the transcriptional signal of genes close together on the chromosome, where the observed stability and range of correlations extend far beyond the expected size of the average operon. Such dependence offers an intriguing hypothesis about the physical basis of the short-range transcriptional correlations: the transcription of the genes within a chromosomal domain is more similar to each other than to genes in other domains (27).
The folding of the nucleoid in domains due to gene colocalization puts into spatial contact distant chromosomal regions. The inset of Figure 6 shows the position distributions for the three genetic loci oriC, C4 and lac concurrently visualized in (3) as obtained by our model of the E. coli nucleoid. We find the three genetic loci to be located on different domains emerging due to TF–gene colocalization. In agreement with this finding, the genomic distances between the loci oriC and C4 as well as between the loci C4 and lac being ∼300 and 600 kb, respectively, formally confirm the sites' positioning on different domains even when an upper loop size limit of 700 kb (emerging from our numerical calculations) is assumed. Additionally, we find the terminus region to self-organize at the mid-cell position connecting the two chromosome arms in agreement with Ref. (3)
Moreover, the inset of Figure 6 also shows that the assumption of TF–gene interactions as drivers of domain formation (among other possible mechanisms) are able to reproduce the linear correlation between the position of a gene on the chromosome (i.e. site along the polymer) and its subcellular position inside the nucleoid (i.e. the long axis of the confining cavity). In fact, the strong linear correlation is a direct consequence of the fact that DNA is compacted and confined. Notably, there is only a linear correlation for sufficiently large domain sizes since the absence of domains leads to a loss of precision of positioning and consequently linear ordering (61).
Thus, we study the distribution of loop sizes in Figure 6 finding them to range between 10 and 700 kb in agreement with the size of topological domains identified in the context of DNA supercoiling (27,62). We find a mean domain size of 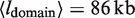 . The long tail of large loop sizes (>150 kb) can be explained by noting that the larger domains are themselves built up by smaller subdomains in agreement with the observed long-range correlations in gene expression patterns (2,26,27).
. The long tail of large loop sizes (>150 kb) can be explained by noting that the larger domains are themselves built up by smaller subdomains in agreement with the observed long-range correlations in gene expression patterns (2,26,27).
DISCUSSION
In this work, we have (i) proposed a mechanism by which chromosomal domains are formed and (ii) quantitatively investigated this model applying numerical simulations. We assume the E. coli transciptional regulatory network to give rise to colocalization of TFs and their target sites due to attractive interactions between genetic loci. Under these conditions, we find that the DNA chain self-organizes into several topologically distinguishable domains where the interplay between the entropic repulsion of chromosomal loops and their compression due to the confining geometry induces a formation into a stacked sequence of interlinked domains or ‘rosettes’ (63). Thus, the experimentally observed high precision of separate subcellular positioning of genetic loci located on different chromosomal domains naturally emerges as a result of entropic de-mixing of loops where the precision of localization is related to the position fluctuations of each chromosomal domain. To recover the precision of organization observed in E. coli, we estimate the domain sizes to be distributed between 10 and 700 kb, in agreement with the size of topological domains identified in the context of DNA supercoiling.
However, the question has to be raised whether the circular chromosome behaves as an equilibrated polymer (64). Calculation of the rate of uncoiling of the DNA molecule can be computed by taking into account the increase of entropy on unwinding as well as the viscous resistance of the surrounding medium (65). Thus, at ∼300 K the bidirectional replication of a 4-Mb chromosome would require ∼20 min (65), which might leave a substantial amount of time for the non-replicating phase where chromosome domains could be rearranged. Notably, the study of E. coli chromosome organization is influenced by the experimental conditions, which might not be judicious due to different genetic background and growth conditions. Due to the high packing density of the nucleoid, the bacterial chromosome might not be able to explore the whole configuration space of possible conformations but rather a restricted subspace starting from similar initial configurations emerging due to the progressive segregation and DNA compaction after each replication cycle. In fact, numerical calculations on time scales that are small compared with the relaxation time of our polymer system indicate even more precise locus positioning due to ‘frozen-in’ configurations.
Our concept could have important implications for chromosome segregation, too. It was previously shown that compaction of the bacterial chromosome and conformational entropy alone could direct and facilitate the segregation of newly replicated daughter strands of DNA (31,32). In this work, the sequence of stacked domains has higher internal topological complexity compared with linear or circular chains leading to even stronger repulsive interactions. Thus, segregation by entropic forces induced by chain topology in strong confinement might constitute a reliable mechanism, where no additional drivers such as a mitotic spindle-like machinery or dedicated proteins may be needed (32).
We have not taken into account the effect of the chromosomal domains being negatively supercoiled by either plectonemic or toroidal supercoils (62) since we have focused on the global physical properties of the chromosome on a more coarse-grained level. However, DNA gyrases, which cause branched supercoils and thus increase domains topological complexity, further promote entropic repulsion between the supercoiled chromosomal domains as pointed out above.
Summarizing, our approach offers a robust framework for understanding the basic physical principles underlying E. coli chromosome organization. Its advantage is that it does not depend on the microscopic details of the DNA chain or on specific DNA–protein interactions. In light of the difference in length scales between proteins and chromosomes, the question has to be raised whether local actions of specific proteins alone are able to globally shape chromosome organization (7,32). Thus, our model is based on the idea that nature exploits entropy, excluded volume, specific chromosome topologies and confinement as a driving force to create the right physical conditions for chromosome packaging eventually fine-tuned by the dedicated action of proteins such as NAPs that bridge and bend DNA (9) or chromosome condensation proteins [SMC (51) and MukBEF (66)]. Future experiments investigating the spatial distribution of TF genes and target genes would be insightful to test our predictions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figure 1 and Supplementary References [1–6].
FUNDING
Heidelberg Graduate School of Mathematical and Computational Methods for the Sciences (HGS MathComp to M.F.); Heinz-Goetze-Foundation (to S.L.); National Science Foundation (Grant PHY-084845 to P.A.W.). Funding for open access charge: National Science Foundation (Grant PHY-084845 to P.A.W).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Jané Kondev, Suckjoon Jun, Jonathan Machta, Lindsay Shopland and Benót Knecht for their comments and suggestions. For this work, the bwGRiD parallel computing facilities (http://www.bw-grid.de), member of the German D-Grid initiative, funded by the Ministry for Education and Research and the Ministry for Science, Research and Arts Baden-Wuerttemberg, as well as the high performance PC cluster HELICS II at the Interdisciplinary Center for Scientific Computing (IWR) were used.
REFERENCES
- 1.Toro E, Shapiro L. Bacterial chromosome organization and segregation. Cold Spring Harb. Perspect. Biol. 2010;2:a000349. doi: 10.1101/cshperspect.a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rocha EPC. The organization of the bacterial genome. Annu. Rev. Genet. 2008;42:211–233. doi: 10.1146/annurev.genet.42.110807.091653. [DOI] [PubMed] [Google Scholar]
- 3.Wiggins PA, Cheveralls KC, Martin JS, Lintner R, Kondev J. Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc. Natl Acad. Sci. USA. 2010;107:4991–4995. doi: 10.1073/pnas.0912062107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meile J-C, Mercier R, Stouf M, Pages C, Bouet J-Y, Cornet F. The terminal region of the E. coli chromosome localises at the periphery of the nucleoid. BMC Microbiol. 2011;11:1–10. doi: 10.1186/1471-2180-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marenduzzo D, Micheletti C, Cook PR. Entropy-driven genome organization. Biophys. J. 2008;90:3712–3721. doi: 10.1529/biophysj.105.077685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes. Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dame RT, Kalmykowa OJ, Grainger DC. Chromosomal macrodomains and associated proteins: implications for DNA organization and replication in Gram negative Bacteria. PLoS Genet. 2011;7:e1002123. doi: 10.1371/journal.pgen.1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vora T, Hottes AK, Tavazoie S. Protein Occupancy Landscape of a Bacterial Genome. Mol. Cell. 2009;35:247–253. doi: 10.1016/j.molcel.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luijsterburg MS, Noom MC, Wuite GJL, Dame RT. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 2006;156:262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Fritsche M, Heermann DW. Confinement driven spatial organization of semiflexible ring polymers: implications for biopolymer packaging. Soft Matter. 2011;7:6906–6913. [Google Scholar]
- 11.Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffith JD. Visualization of prokaryotic DNA in a regularly condensed chromatin-like fiber. Proc. Natl. Acad. Sci. USA. 1976;73:563–7. doi: 10.1073/pnas.73.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer M, De Jong MA, Woldringh CL, Nanninga N. Factors affecting the release of folded chromosomes from Escherichia coli. Eur. J. Biochem. 1976;63:469–75. doi: 10.1111/j.1432-1033.1976.tb10249.x. [DOI] [PubMed] [Google Scholar]
- 15.Kavenoff R, Ryder OA. Electron microscopy of membrane-associated folded chromosomes of Escherichia coli. Chromosoma. 1976;55:13–25. doi: 10.1007/BF00288323. [DOI] [PubMed] [Google Scholar]
- 16.Deng S, Stein RA, Higgins NP. Organization of supercoil domains and their reorganization by transcription. Mol. Microbiol. 2005;57:1511–1521. doi: 10.1111/j.1365-2958.2005.04796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frenster JH. Correlation of the binding to DNA loops or to DNA helices with the effect on RNA synthesis. Nature. 1965;208:1093. doi: 10.1038/2081093a0. [DOI] [PubMed] [Google Scholar]
- 18.Danchin A, Guerdoux-Jamet P, Moszer I, Nitschké P. Mapping the bacterial cell architecture into the chromosome. Philos. Trans. Roy. Soc. Lond. B Biol. Sci. 2000;355:179–190. doi: 10.1098/rstb.2000.0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–82. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janga SC, Salgado H, Martínez-Antonio A. Transcriptional regulation shapes the organization of genes on bacterial chromosomes. Nucleic Acids Res. 2009;37:3680–3688. doi: 10.1093/nar/gkp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marr C, Geertz M, Htt M-T, Muskhelishvili G. Dissecting the logical types of network control in gene expression profiles. BMC Syst. Biol. 2008;2:18. doi: 10.1186/1752-0509-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martínez-Antonio A, Medina-Rivera A, Collado-Vides J. Structural and functional map of a bacterial nucleoid. Genome Biol. 2009;10:247–250. doi: 10.1186/gb-2009-10-12-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Worcel A, Burgi E. On the structure of the folded chromosome of Escherichia coli. J. Mol. Biol. 1972;71:127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- 24.Romantsov T, Fishov I, Krichevsky O. Internal structure and dynamics of isolated Escherichia coli nucleoids assessed by fluorescence correlation spectroscopy. Biophys. J. 2007;92:2875–2884. doi: 10.1529/biophysj.106.095729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 2004;23:4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carpentier AS, Torresani B, Grossmann A, Henaut A. Decoding the nucleoid organization of Bacillus subtilis and Escherichia coli through gene expression data. BMC Genomics. 2005;6:84. doi: 10.1186/1471-2164-6-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong KS, Ahn J, Khodursky AB. Spatial patterns of transcriptional activity in the chromosome of Escherichia coli. Genome Biol. 2004;5:R86. doi: 10.1186/gb-2004-5-11-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailly-Bechet M, Danchin A, Iqbal M, Marsili M, Vergassola M. Codon usage domains over bacterial chromosomes. PLoS Comput. Biol. 2006;2:e37. doi: 10.1371/journal.pcbi.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho B-K, Palsson BØO. Can the protein occupancy landscape show the topologically isolated chromosomal domains in the E. coli genome?: an exciting prospect. Mol. Cell. 2009;35:255–256. doi: 10.1016/j.molcel.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 30.Berlatzky IA, Rouvinski A, Ben-Yehuda S. Spatial organization of a replicating bacterial chromosome. Proc. Natl Acad. Sci. USA. 2008;105:14136–14140. doi: 10.1073/pnas.0804982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: Lessons for the bacterial chromosome. Proc. Natl Acad. Sci. USA. 2006;103:12388. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jun S, Wright A. Entropy-driven chromosome segregation. Nat. Rev. Microbiol. 2010;8:600–607. doi: 10.1038/nrmicro2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohniwa RL, Morikawa K, Takeshita SL, Kim J, Ohta T, Wada C, Takeyasu K. Transcription-coupled nucleoid architecture in bacteria. Genes Cells. 2007;12:1141–1152. doi: 10.1111/j.1365-2443.2007.01125.x. [DOI] [PubMed] [Google Scholar]
- 34.Foleya PL, Wilsonb DB, Shuler ML. Macromolecular crowding can account for RNase-sensitive constraint of bacterial nucleoid structure. Biochem. Biophys. Res. Commun. 2010;395:42–47. doi: 10.1016/j.bbrc.2010.03.128. [DOI] [PubMed] [Google Scholar]
- 35.Bohn M, Heermann DW, Loureno̧ O, Cordeiro C. On the influence of topological catenation and bonding constraints on ring polymers. Macromolecules. 2010;43:2564–2573. [Google Scholar]
- 36.Bohn M, Heermann DW. Topological interactions between ring polymers: implications for chromatin loops. J. Chem. Phys. 2010;132:044904–044914. doi: 10.1063/1.3302812. [DOI] [PubMed] [Google Scholar]
- 37.Binder K, Heermann DW. Monte Carlo Simulation in Statistical Physics: An Introduction. Berlin: Springer; 2010. [Google Scholar]
- 38.Liu X, Wang X, Reyes-Lamothe R, Sherratt D. Replication-directed sister chromosome alignment in Escherichia coli. Mol. Microbiol. 2010;75:1090–1097. doi: 10.1111/j.1365-2958.2009.06791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Junier I, Martin O, Képès F. Spatial and topological organization of DNA chains induced by gene co-localization. PLoS Comput. Biol. 2010;6:e1000678. doi: 10.1371/journal.pcbi.1000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breier AM, Cozzarelli NR. Linear ordering and dynamic segregation of the bacterial chromosome. Proc. Natl Acad. Sci. USA. 2004;101:9175–9176. doi: 10.1073/pnas.0403722101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stavans J, Oppenheim A. DNA-protein interactions and bacterial chromosome architecture. Phys. Biol. 2006;3:R1. doi: 10.1088/1478-3975/3/4/R01. [DOI] [PubMed] [Google Scholar]
- 42.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit J, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Espeli O, Mercier R, Boccard F. DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol. Microbiol. 2008;68:1418–1427. doi: 10.1111/j.1365-2958.2008.06239.x. [DOI] [PubMed] [Google Scholar]
- 44.Mercier R, Petit M, Schbath S, Robin S, El Karoui M, Boccard F, Espéli O. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell. 2008;135:475–485. doi: 10.1016/j.cell.2008.08.031. [DOI] [PubMed] [Google Scholar]
- 45.Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14:212–223. [PMC free article] [PubMed] [Google Scholar]
- 46.Reyes-Lamothe R, Wang X, Sherratt D. Escherichia coli and its chromosome. Trends Microbiol. 2008;16:238–245. doi: 10.1016/j.tim.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 47.Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joshi MC, Bourniquel A, Fisher J, Brian TH, Kleckner N, Bates D. Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proc. Natl Acad. Sci. USA. 2011;108:2765–2770. doi: 10.1073/pnas.1019593108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hershberg R, Yeger-Lotem E, Margalit H. Chromosomal organization is shaped by the transcription regulatory network. Trends Genetic. 2005;21:138–142. doi: 10.1016/j.tig.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 50.Babu MM, Luscombe and NM, Aravind and L, Gerstein and M, Teichmann SA. Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 2004;14:283–291. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Janga SC, Collado-Vides J. Structure and evolution of gene regulatory networks in microbial genomes. Res. Microbiol. 2007;158:787–794. doi: 10.1016/j.resmic.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sonnenschein N, Geertz M, Muskhelishvili G. Analog regulation of metabolic demand. BMC Microbiol. 2011;5:1–13. doi: 10.1186/1752-0509-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sexton T, Schober H, Fraser P, Gasser S. Gene regulation through nuclear organization. Natl. Struct. Mol. Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 54.Marr C, Geertz M, Hütt MT, Muskhelishvili G. Dissecting the logical types of network control in gene expression profiles. BMC Syst. Biol. 2008;2:18. doi: 10.1186/1752-0509-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sullivan NL, Marquis KA, Rudner DZ. Recruitment of condensin to replication origin regions by ParB-parS organizes the origin and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 57.Sánchez-Romero MA, Busby SJ, Dyer NP, Ott S, Millard AD, Grainger DC. Dynamic distribution of SeqA protein across the chromosome of Escherichia coli K-12. mBio. 2010;1:e00012–e00010. doi: 10.1128/mBio.00012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tonthat NK, Arold ST, Pickering BF, Van Dyke MW, Liang S, Lu Y, Beuria TK, Margolin W, Schumacher MA. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 2011;30:154–164. doi: 10.1038/emboj.2010.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright MA, Kharchenko P, Chruch GM, Segre D. Chromosomal periodicity of evolutionary conserved gene pairs. Proc. Natl Acad. Sci. USA. 2007;104:10559–10564. doi: 10.1073/pnas.0610776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kepes F. Periodic transcriptional organization of the E. coli genome. J. Mol. Biol. 2004;340:957–964. doi: 10.1016/j.jmb.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 61.Buenemann M, Lenz P. A geometrical model for DNA organization in bacteria. PLoS ONE. 2010;5:e13806. doi: 10.1371/journal.pone.0013806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willenbrock H, Ussery DW. Chromatin architecture and gene expression in Escherichia coli. Genome Biol. 2004;5:252. doi: 10.1186/gb-2004-5-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bates D. The bacterial replisome: back on track? Mol. Macrobiol. 2008;69:1341–1348. doi: 10.1111/j.1365-2958.2008.06378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosa A, Everaers R. Structure and dynamics of interphase chromosomes. PLoS Comput. Biol. 2008;4:e1000153. doi: 10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Longuet-Higgins HC, Zimm BH. Calculation of the Rate of Uncoiling of the DNA Molecule. J. Mol. Biol. 1960;2:1–4. [Google Scholar]
- 66.Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol. Microbiol. 2007;65:1485–1492. doi: 10.1111/j.1365-2958.2007.05881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.