Abstract
The discovery of a plethora of small non-coding RNAs (ncRNAs) has fundamentally changed our understanding of how genes are regulated. In this study, we employed the power of deep sequencing of RNA (RNA-seq) to examine the repertoire of ncRNAs present in small ribonucleoprotein particles (RNPs) of Trypanosoma brucei, an important protozoan parasite. We identified new C/D and H/ACA small nucleolar RNAs (snoRNAs), as well as tens of putative novel non-coding RNAs; several of these are processed from trans-spliced and polyadenylated transcripts. The RNA-seq analysis provided information on the relative abundance of the RNAs, and their 5′- and 3′-termini. The study demonstrated that three highly abundant snoRNAs are involved in rRNA processing and highlight the unique trypanosome-specific repertoire of these RNAs. Novel RNAs were studied using in situ hybridization, association in RNP complexes, and ‘RNA walk’ to detect interaction with their target RNAs. Finally, we showed that the abundance of certain ncRNAs varies between the two stages of the parasite, suggesting that ncRNAs may contribute to gene regulation during the complex parasite’s life cycle. This is the first study to provide a whole-genome analysis of the large repertoire of small RNPs in trypanosomes.
INTRODUCTION
In recent years, non-coding RNAs (ncRNAs) have become a major focus of research in species ranging from bacteria to man. In prokaryotes, the majority of ncRNAs function in regulation of gene expression with a minor proportion playing housekeeping roles, and recent and extensive RNA deep sequencing (RNA-seq) analysis has expanded their repertoire to include 80 family members (1). Functions assigned to ncRNAs include regulation of the stress response, metabolic regulation and pathogenesis. Most ncRNAs affect gene expression by base-pairing to their target and inhibiting translation, or by affecting mRNA stability (1). In eukaryotes, the most abundant ncRNAs are involved in translation (rRNA, tRNA), protein translocation (7SL RNA), splicing [U small nuclear RNAs (U snRNAs)], rRNA processing and RNA modification [small nucleolar RNAs (snoRNAs)] and RNA interference (RNAi) [microRNAs, and small interfering RNAs (siRNAs)] (2). The latter two classes are known regulators of translation and mRNA stability, respectively. More recent studies in humans described over 1000 ncRNAs, which do not belong to the known groups of small RNAs and are present in small ribonucleoprotein particles (RNPs) (3).
Protozoa of the family Trypanosomatidae include several medically and economically important parasites, such as Trypanosoma brucei, Trypanosoma cruzi and various Leishmania species. In addition to being human pathogens, trypanosomatids have attracted the attention of the scientific community because several previously unknown mechanisms of regulation of gene expression, such as trans-splicing (4,5) and mitochondrial RNA editing (6), were first described in these organisms. Moreover, their transcription is polycistronic, and gene regulation occurs mainly at the post-transcriptional level (7). Most recently, RNA-seq analysis and mapping of trans-splicing and poly (A) sites revealed 1114 new transcripts, including 103 ncRNAs. The data also lent support to the bidirectional nature of RNA polymerase II transcription initiation and indicated that transcription initiation is not restricted to regions at the beginning of gene clusters, but may occur at internal sites (8).
Our current knowledge of the complement of small ncRNAs in trypanosomes has been attained through a variety of approaches, including homology searches, functional studies and bioinformatic tools. However, no whole-genome analysis of small ncRNAs has been carried out to date. Nevertheless, trypanosome RNA research has led to some interesting and unique observations. For instance, most of the spliceosomal snRNAs are the shortest in nature and lack domains present in their higher eukaryotic counterparts (4), and the 7SL RNA Alu-like domain has been replaced by a unique tRNA-like molecule (9–11). However, several trypanosomal small RNAs with predicted essential functions remain to be identified, including a number of snoRNAs, telomerase, and RNase P RNA.
snoRNAs, which methylate (C/D class) or pseudouridylate (H/ACA class) rRNA residues, are a relatively well studied group of small RNAs in trypanosomes. The trypanosome snoRNA genes exist in clusters, and as shown in T. brucei (12) and L. major (13), most clusters carry interspersed H/ACA and C/D snoRNAs sequences. snoRNA genes are transcribed polycistronically (14) by RNA polymerase II, and individual RNAs are then processed from the primary transcript by an unknown RNase III enzyme, using information residing in the intergenic regions. Recently, it was demonstrated that the primary snoRNAs transcripts are trans-spliced and polyadenylated (8).
Whereas the trypanosome C/D snoRNAs structure conforms to the eukaryotic consensus, the H/ACA RNAs consist of a single hairpin, unlike the eukaryotic prototypical structure that contains two hairpins, and carry an AGA instead of the classical ACA box (12,13,15,16). The SLA1 (spliced leader associated RNA1) is a unique H/ACA RNA that directs pseudouridylation on the spliced leader RNA (SL RNA) (16). RNAi silencing of the pseudouridine synthase (CBF5) that binds H/ACA RNA, affects pseudouridylation on rRNAs and snRNAs, rRNA processing, and unexpectedly, trans-splicing as well (17). On the other hand, silencing of fibrillarin (NOP1), which binds to C/D snoRNAs, affects 2′-O-methylation of rRNAs and snRNAs, as well as rRNA processing (18).
In eukaryotes, processing of the primary rRNA transcripts to form mature 18, 28 and 5.8S RNAs involves several cleavage events (19). In trypanosomes, the rRNA processing pathway is fundamentally different from that of most higher eukaryotes; specifically, the large subunit rRNA is processed into two large fragments, LSUα and β, and five small fragments, termed srRNA 1–6 (20). Analysis of the rRNA processing defects that were observed in NOP1 and CBF5 silenced cells suggested the existence of trypanosome-specific guide RNAs to carry out the rRNA processing cleavages (17,18). In addition, systematic mapping of nucleotide modification sites (Nms) on T. brucei rRNAs identified 47 new Nms in addition to the previously identified 84, suggesting that further C/D snoRNAs remain to be identified (18). The majority of snoRNAs identified so far were implicated in rRNA modifications, and only homologues of higher eukaryotic U3 (21), snR30 and MRP RNA (17,18) were evident. More recently, the function of the TB11Cs2C2 snoRNA, present in a cluster together with SLA1 and snR30, was elucidated. We found that TB11Cs2C2 interacts by base-pairing with the small rRNAs srRNA-2 and srRNA-6, and that down regulation of TB11Cs2C2 by RNAi (snoRNAi) results in defects in the generation of srRNA-2 and LSUβ rRNA. This is the first snoRNA described so far to engage in trypanosome-specific processing events (22). On the other hand, bioinformatic searches failed to identify U8, U14 and U22 homologues that are involved in rRNA processing in other eukaryotes (19), suggesting that trypanosomes may possess unique snoRNAs to carry out ubiquitous as well as trypanosome-specific rRNA processing events.
In this study, we used deep-sequencing and bioinformatic analysis to assemble a library of potential ncRNAs purified from size-fractionated small RNPs of T. brucei procyclics. Although the majority of the newly predicted ncRNAs do not belong to known families, a few of them were found among the list of new trans-spliced and polyadenylated transcripts recently described in procyclics (8). We validated the existence of a number of small ncRNAs and predicted their potential function using a variety of molecular and cell biological approaches. The study confirmed the existence of 79 C/D and 63 H/ACA snoRNAs in the T. brucei genome, which were shown to be expressed in the procyclic form of the parasite. Functional evidence using snoRNAi (22,23) and ‘RNA walk’ (11) methodologies indicated that the abundant snoRNAs: TB10Cs4C4, TB6Cs1C3, TB9Cs2C1, are involved in trypanosome-specific rRNA processing steps, and that TBsRNA-4, a non-C/D and non-H/ACA snoRNA is a novel snoRNA. Lastly, our results suggest that differential ncRNA expression may contribute to gene regulation during the T. brucei complex life cycle.
MATERIALS AND METHODS
All the oligonucleotides used in this study are listed in Supplementary Figure S1.
Cell culture, constructs and transfection
Procyclic T. brucei strain 29–13 was grown in SDM-79 medium. The T7 RNA polymerase silencing constructs with opposing promoters were prepared as previously described (24) using oligonucleotides listed in Supplementary Figure S1. Cells were transfected as previously described, and a clonal population was selected (25).
Preparation of RNP complexes for RNA-Seq
Procyclic cells were grown and 109 cells (1 l of 106 cells/ml) were used for extract preparation. Extracts were prepared after washing the cells in PBS and re-suspending in 1 ml of 20 mM Tris–HCl (pH 7.7), 150 mM KCl, 3 mM MgCl2, 0.5 mM DTT and 0.1% Tween 20. Protease inhibitor cocktail (Roche Applied Science) and 0.5 µl of RNasin were added. Cells were homogenized with a dounce until complete lysis was microscopically observed. To the extract, KCl was added dropwise to 300 mM and the extract was gently stirred on ice for 30 min. The cell debris were removed by centrifugation at 13000 r.p.m. for 10 min, and the supernatant was centrifuged for 2 h at 33 000 r.p.m. in a Beckman 70.1 Ti rotor to pellet the ribosomes. A 500 µl aliquot was loaded on a Superdex 200 gel filtration column (Amersham Biosciences), equilibrated with (10 mM Tris pH 7.7, 150 mM KCl, 0.5 mM MgCl2, 10 mM DTT) at a flow rate of 0.5 ml/min. Fractions of 0.5 ml were collected. The elution of bovine serum albumin (66 kDa) and β-amylase (200 kDa) were used as markers to follow the fractionation.
Preparation of the RNA-seq library
RNA was extracted from FPLC fractions and 1/10 of the sample was labeled at the 5′-end with γ-ATP using T4 RNA ligase. The labeled and unlabeled material were mixed and fractionated on a 15% polyacrylamide/7M Urea gel. After exposure to a phosphorimager screen, RNAs between 20 and 30 nt were identified and excised from the gel, eluted and purified by phenol extraction. The RNA was processed for library construction for Illumina sequencing, essentially according to a protocol that was kindly provided to us by Prof Gregory Hannon (CSHL, NY, USA), with minor modifications (the protocol is available upon request).
Primer extension and northern analysis
RNA was extracted from T. brucei cells using the TRI-Reagent (Sigma). Primer extension analysis was performed as described (25) using 5′-end-labeled oligonucleotides specific to each target RNA. The extension products were analyzed on a 6% polyacrylamide/7 M urea gel and visualized by autoradiography. For northern analysis, total RNA was extracted, separated on a 10% polyacrylamide-denaturing gel and analyzed using RNA or DNA probes (26).
RT–PCR and RNase protection assay
The RNA was treated with the ‘DNase-free’ reagent (Ambion) according to the manufacturer’s protocol. Reverse transcription was performed by random priming (Reverse transcription system, Promega). The resulting cDNA was used for PCR amplification using primers as specified in Supplementary Figure S1. RNase protection was carried out as described in Ref. (25) using probes for the various internal transcribed spacer (ITS) prepared with primers described in Supplementary Figure S1.
‘RNA walk’ protocol
Cross-linking was performed essentially as described in (10). Briefly, T. brucei cells were harvested at 1 × 107 cells/ml and washed twice with PBS. Cells (∼109) were concentrated and incubated on ice. 4′-Aminomethyl-trioxsalen hydrochloride (AMT) (Sigma) was added to the cells at a concentration of 0.2 mg/ml. Cells treated with AMT were kept on ice and irradiated using a UV lamp at 365 nm at a light intensity of 10 µW/cm2 for 30 min. Next, the cells were washed once with PBS and deproteinized by digestion with proteinase K (Roche) (200 µg/ml in 1% SDS for 60 min). RNA was prepared using TRIzol reagent (Sigma). Around 250 μg of RNA extracted from UV treated cells (+UV) and untreated cells (−UV), was subjected to affinity selection essentially as described in (11,27). After affinity selection, the RNA was subjected to RT–PCR described above.
Fractionation of deproteinized extracts on sucrose gradients
Extracts were treated with 2% SDS and 2 mg/ml proteinase K and incubated at 14°C for 15 min. The lysate was cleared at 50 000g for 30 min, and the remaining RNA, with or without treatment at 65°C for 10 min, was centrifuged through a 10–30% (w/v) sucrose gradient at 4°C for 16 h at 22 000 r.p.m. in a Beckman SW41 rotor.
In situ hybridization
In situ hybridization with SL RNA was performed as recently described (28). The slides were incubated with 1:400 diluted primary anti-NHP2 antibodies, which were detected using IgG conjugated to Cy3. Nuclei were stained using 4′-6′-diamidino-2-phenylindole (DAPI) or propidium iodide (PI). The cells were visualized under a Zeiss LSM 510 META inverted microscope.
Bioinformatic analysis
The sequence reads obtained from the Illumina Genome Analyzer were first filtered for the presence of Illumina adapters, and reads having a consecutive match of over 11 bases to an adapter were discarded from subsequent analysis. The remaining reads were mapped to the T. brucei draft genome (V4; http://www.sanger.ac.uk/Projects/T_brucei/) using BLAST (Blastall v2.2.20) with the following parameters [-p blastn -e 0.0001 -b 20 -v 20 -m 0 –W 11 -F F], allowing up to six mismatches and requiring a minimal alignment length of 20 bp. An Excel spreadsheet was prepared that maps each read to its corresponding genomic location. Using PERL scripts, the reads were merged into single contigs based on the assumption that reads that mapped to coordinates less than 10 nt apart were likely to originate from the same transcript. The mapped reads were then compared to the T. brucei genome V4 annotation. Reads mapping to unannotated loci were chosen as potential novel ncRNAs.
In order to determine if the reads mapping to unannotated loci were derived from known annotated sequences, they were further analyzed by BLAST (29) against the T. brucei and L. major known coding sequences. Those reads that were not similar to any known coding sequence were then run as input to a variety of programs to identify putative snoRNAs. The programs used included snoScan version 0.9b (30), snoReport (31), snoGPS (32) and PsiScan (33), to test if the ncRNA candidates were likely to be snoRNAs. The snoscan program searches for features characteristic of C/D snoRNAs. The snoGPS program searches for features characteristic of H/ACA snoRNAs. The PsiScan program searches for features distinguishing H/ACA snoRNAs in trypanosomatids. The snoReport program scans sequences for both C/D and H/ACA snoRNAs, but does not require any target information. In addition, BLAST (29) was used to search for a complementary match to an RNA target. In the final stage, manual curation of the remaining sequences was done to look for motifs, followed by folding with MFOLD (34).
The topography of the distribution of reads was examined across a single contig by generating a histogram from the reads at each nucleotide. The data from the histogram were superimposed on the prototype or MFOLD-generated RNA structures (34).
The annotation of the newly identified non-coding RNAs, including the novel snoRNAs identified in this study, was submitted to GeneDB.
RESULTS
Fractionation of small RNPs and generation of an ncRNA library
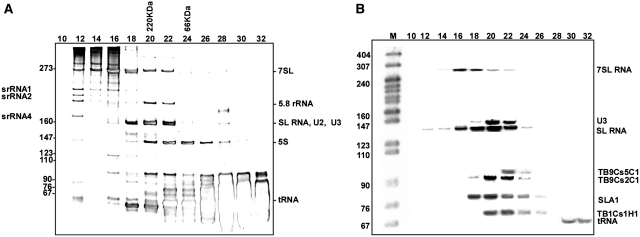
To identify novel ncRNAs, we decided to construct a library of RNAs co-purifying with small RNPs fractionated by size on an FPLC column. As starting material, we used a whole-cell extract from procyclic trypanosomes, which is largely depleted of ribosomal complexes. The RNA content of selected fractions was analyzed by electrophoresis on a denaturing gel followed by silver staining or northern hybridization (Figure 1A and B, respectively). The elution profile of several known small RNAs indicated that the fractionation procedure was successful in separating tRNAs from complexes carrying 7SL RNA, snRNAs or snoRNAs.
Figure 1.
Size fractionation of T. brucei RNPs by FPLC. Whole-cell extracts from 2 × 108 cells were fractionated on a Superdex-200 gel filtration column as described in ‘Materials and Methods’ section. The RNA was extracted from the fractions and separated on a 6% denaturing gel. (A) Silver staining of the gel. (B) The RNA was subjected to northern analysis with anti-sense RNA probes. The elution positions of marker proteins BSA (66 kDa) and β-amylase (200 kDa) are indicated. A pBR322 DNA-MspI digest was used as a marker.
To prepare the RNA-seq library, we selected fractions 20–24 because they appeared to be enriched in snoRNA-size molecules. Although the RNA pattern in Figure 1A did not show evidence of degradation, upon 3′-end labeling and analysis of the RNA on a high resolution sequencing gel, we observed the presence of a minor cluster of fragments of about 20–30 nt (data not shown). We reasoned that this material was likely to represent siRNAs, which in trypanosomes are 24–26 nt long (35). This possibility was tested by generating a library with Illumina adaptors, followed by cloning and Sanger sequencing of a dozen clones to verify the quality of the library. Much to our surprise, we discovered that the library contained sequences derived from U snRNAs, snoRNAs and rRNA, most likely because a low level of RNA degradation had occurred during the RNP preparation and/or manipulation of the sample. We decided to sequence this library and performed two technical replicas, which gave qualitatively similar results, but differed in the total number of sequence reads, with the second experiment providing ∼7-fold higher coverage than the first one, and a total of 14 × 106 reads that matched the T. brucei genome version 4.0. Comparison of the snoRNAs reads in the two replicas showed a Pearson coefficient of R = 0.872 (P < 0.01) (Supplementary Figure S5). The data presented below were derived from the analysis of the second experiment. We first clustered the reads into individual contigs and this analysis revealed 16 955 contigs, of which about one quarter (3674) was not annotated in GeneDB. We then discarded from subsequent analysis regions with low read representation (<50 reads) or when reads appeared as single pillars (i.e. <50 bp in length), because they were unlikely to be informative. The majority of known small RNAs were represented in our contig collection as summarized in Supplementary Figure S2. Specifically, we successfully identified 90% of C/D and 84% of H/ACA snoRNAs that were described previously, as well as all tRNAs and all the known siRNAs, suggesting that the library coverage was fairly comprehensive.
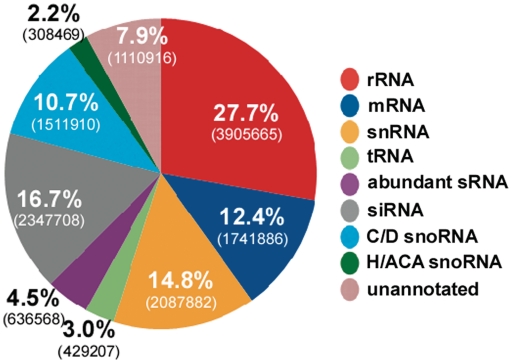
Next, to determine which RNA types were enriched in our library, we clustered the reads into different RNA classes, as illustrated in the pie diagram (Figure 2), and then compared their relative percentage to the known cellular abundance of each RNA class in eukaryotes (http://bionumbers.hms.harvard.edu/). The results indicated 66-fold enrichment of small RNAs (30% of the reads), which usually represent only 0.5% of total cellular RNA. On the other hand, tRNA-derived reads constituted only 3% of the total, as compared to the actual 10–15% abundance. This low representation of tRNA fragments in our library most likely resulted from the choice of fractions for library construction, which did not coincide with the tRNA peak (Figure 1A), and from the fact that tRNAs are highly modified and thus reverse transcribe inefficiently. Despite the enrichment of small RNAs, the sample contained 28% rRNA and 13% mRNA. Among the small RNAs, 1 820 000 reads (13%) represented snoRNAs, and 2 350 000 reads (17%) were derived from SLACS and INGI retroposons, and conceivably represent siRNAs (35) (Figure 2). The distribution of reads for the other known small RNAs not always reflects the actual abundance of these RNAs in trypanosomes. For instance, we found more reads for U1 snRNA than for 7SL RNA, although 7SL RNA is the most abundant trypanosome small RNA and U1 is less abundant compared to SL RNA (36). This discrepancy may stem from several causes, including: (i) the size of the RNP complexes, which determines their chromatographic behavior and thus their prevalence in the fractions we chose for library construction; and (ii) the accessibility of the RNA to nuclease attack, which is a major factor for inclusion in our library. For instance, 7SL RNA is part of the SRP which is a large complex, is highly structured and quite insensitive to micrococcal nuclease attack (37).
Figure 2.
Annotation of the reads obtained from RNA-seq of the RNP complexes. The percentage of the different RNA molecules among the reads and the number of reads from the second run are summarized in the pie chart. The abundant sRNA class includes reads from MRP, SL RNA and 7SL RNA.
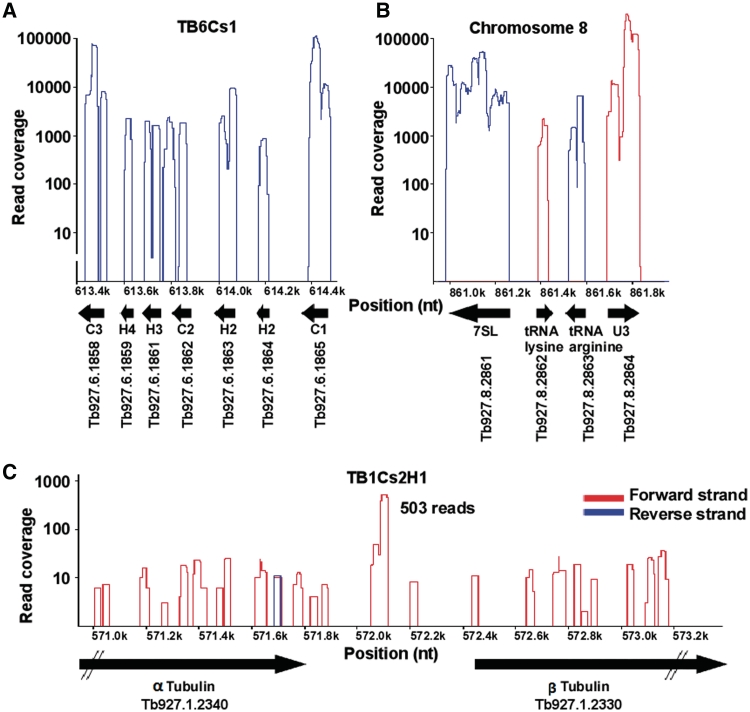
Next, the reads were imported and visualized in a customized genome browser. Figure 3 illustrates examples of three different genomic arrangements of small RNA genes and the read distribution along the respective coding regions. Most snoRNA genes are found in clusters, as exemplified in Figure 3A.
Figure 3.
Viewer presentation of selected small RNAs. The presentation demonstrates the number of individual sequence reads for each of the RNA species forming the pillars. The color of the pillars represents the direction of transcription. (A) The TB6Cs1 snoRNA cluster; (B) views of the 7SL RNA and U3 locus showing the upstream tRNA genes transcribed from the opposite strand; (C) location of TB1Cs2H1, a ‘solitary’ snoRNA gene situated between the α and β tubulin genes. The GeneDB identifier and their chromosomal positions are given.
Importantly, the RNA-seq analysis led to the discovery of a new class of snoRNA genes that are not part of clusters (termed ‘solitary’ snoRNA genes). Figure 3C shows one very interesting case: The coding region for snoRNA TB1Cs2H1 is located in the intergenic region between the α and β-tubulin genes. In addition, inspection of the loci coding for 7SL RNA, U3 snoRNA and the associated tRNA genes (38) revealed that the reads covered the entire RNA coding sequences and demonstrated that the upstream tRNA genes, which serve as extragenic promoter elements of the 7SL RNA and U3 RNA gene (38), are indeed expressed (Figure 3B).
RNA-seq expands the repertoire of snoRNAs
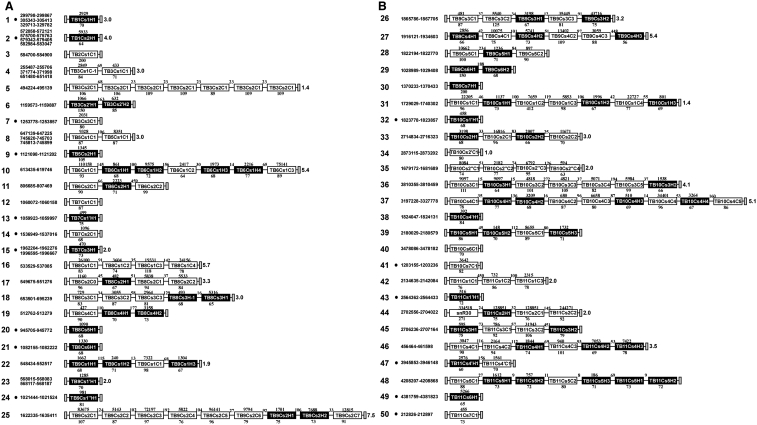
Previous insights into the rich repertoire of trypanosome snoRNAs were obtained mainly using in silico analysis. The first described set of snoRNAs consisted of 94 members (12). Subsequently, an algorithm used to identify AGA molecules in T. brucei revealed 16 additional snoRNA species, and other approaches, including comparative genomics between T. brucei and T. cruzi and L. major, identified an additional 12 snoRNA family members (18,33,39,40). Here, we identified 20 new snoRNAs (14 H/ACA and 6 C/D, Figure 4). Combining our past and present findings, we have so far identified 50 snoRNA loci in T. brucei, which are schematically depicted in Figure 4 alongside the number of reads for each snoRNA coding region (the new snoRNAs are highlighted by a bullet point). Supplementary Figure S4 shows a compilation of the sequences of all the snoRNAs described so far, and the number of reads obtained for each snoRNA in the two replicas. Remarkably, 14 out of the 20 new snoRNA genes were ‘solitary’. Although in earlier studies, we predicted that such snoRNAs were likely to exist (17,18), ‘solitary’ snoRNA genes were not previously identified, because the previous searches were biased toward snoRNAs that are located in repeated clusters (12).
Figure 4.
Schematic representation of snoRNA clusters in T. brucei. The C/D snoRNAs are shown as white boxes, whereas H/ACA-like snoRNAs appear in black boxes. Thinner lines indicate intergenic regions, and their sizes are indicated above the line. The length of the snoRNA genes are indicated below the box and the number of reads obtained in run II is shown above the box. The number on the right side of each cluster indicates the number of times the cluster is repeated in the genome. The small numbers before each cluster indicate the position of the cluster within the genome database, version 5 (http://www.genedb.org/genedb). The black dots indicate the clusters or loci identified in this study. (A) depicts clusters 1–25, and (B) shows clusters 26–50.
Among the 142 genes described in Figure 4, reads were obtained for 119 transcripts. The snoRNAs for which we did not detect expression are nevertheless localized in expressed clusters. No evidence could be obtained for the expression of cluster TB3Cs2C1. We have previously demonstrated differences in expression of snoRNAs between the two stages in the lifecycle of the parasite (18). The inability to detect the expression of a few putative snoRNAs may reflect their differential expression in the two life stages.
One striking feature of our analysis is the great variability in the number of reads for snoRNAs transcribed either from different or the same cluster(s) (Figure 4). Read values ranged from hundreds to even hundreds of thousands per RNA. Since snoRNAs have highly similar structures and are complexed with the same set of proteins we think it is safe to assume that the RNA-seq data reflect the relative abundance of this class of molecules and indicate that snoRNAs vary widely in abundance. We have previously noticed this phenomenon on a small scale in T. brucei (12) as well as in L. major (13). To verify the validity of the RNA-seq data, we performed primer extension as a means to measure the relative abundance of a number of snoRNAs and significant difference were detected (data not shown).
Next, to provide experimental evidence in support of our snoRNA identification, we chose eight candidates and examined their levels after silencing NOP58 for C/D snoRNAs (Supplementary Figure S6A-a) and CBF5 for H/ACA RNAs (Supplementary Figure S6A-b). The results demonstrated a significant reduction in the steady-state level of these snoRNAs in the silenced cells (quantified in Supplementary Figure S6A-c), as reported previously for the two classes of snoRNAs (17) and (18). The predicted targets of these new snoRNAs are illustrated in Supplementary Figure S6B-a and -b. Surprisingly, TB1Cs2H1 is predicted to guide pseudouridylation in the intergenic region of rRNA. Although such snoRNAs are rare, we have previously identified a C/D snoRNA whose target is located in the ITS2 of pre-rRNA (12).
Based on the recent genome-wide mapping of poly (A) addition sites (8), it was possible to examine the position of snoRNA genes with respect to mRNAs. We found that most snoRNA coding regions are located in intergenic regions, with the exception of a few located in the predicted 3′-UTR of certain genes. For instance, cluster TB10Cs5 was found in the UTR of Tb10.6k15.1510, and in other cases the 3′-UTR carried a solitary snoRNA (TB1Cs2H1 present in the 3′-UTR of β-tubulin and TB11Cs7C1 present in the UTR of Tb11.03.0310 mRNAs). These snoRNAs were likely processed since these were detected by RNA-seq. However, it is possible that if snoRNAs are not cleaved off the primary transcripts in the nucleus, the 3′-UTR of the corresponding mRNAs would encompass these unprocessed snoRNAs. The biological significance of a UTR carrying snoRNAs is currently unknown. Although snoRNAs were usually on the same transcription unit of their neighboring genes (+strand), one snoRNA cluster (TB9Cs1) was found in the opposite direction relative to a locus that codes for two hypothetical proteins (in the + strand). Such an unusual genomic organization was also detected for U5 snRNA and tRNA-sec. Surprisingly, snoRNAs were also part of loci annotated as hypothetical proteins, for instance the solitary TB9Cs1’H1 within Tb09.160.1270. In addition, Kolev et al. (8) detected 1114 ‘new transcripts’ that were not annotated previously, based on the fact that these carry both SL and poly(A) sites. Among these new transcripts, 103 were characterized as non-coding RNA, since they lack an ORF that is larger than 25 amino acids. Eleven RNAs detected in this study overlap with the ncRNA list of Kolev et al. (8), (Supplementary Figure S12). In addition, 13 new snoRNAs described in this study (Supplementary Figure S11), as well as 40 snoRNAs previously reported by our laboratory (Supplementary Figure S10) (12) were found among the ‘new transcripts’. The list in Supplementary Figure S11 shows both solitary as well as clustered snoRNAs.
Recently, the boundaries of ∼60 polycistronic transcription units were reported to be marked by distinct histone modifications and nucleosome organization (8,41), suggesting that transcription by RNA polymerase II occurs within the so called strand-switch regions (SSRs), which separate divergently transcribed polycistronic units. Additionally, Kolev et al. (8) provided evidence that polyadenylated RNA molecules carrying a 5′ triphosphate, the hallmark of primary transcripts, originated from within the SSRs. Thus, it was of interest to examine if snoRNA genes are located in these domains. Only two examples of snoRNAs present in a switch region were found. Both are solitary snoRNAs; TB7Cs3H1, a new H/ACA identified in this study, and TB10Cs6C1 (40), suggesting that snoRNAs are not over-represented in SSRs.
Functional analysis of snoRNAs implicated in rRNA processing
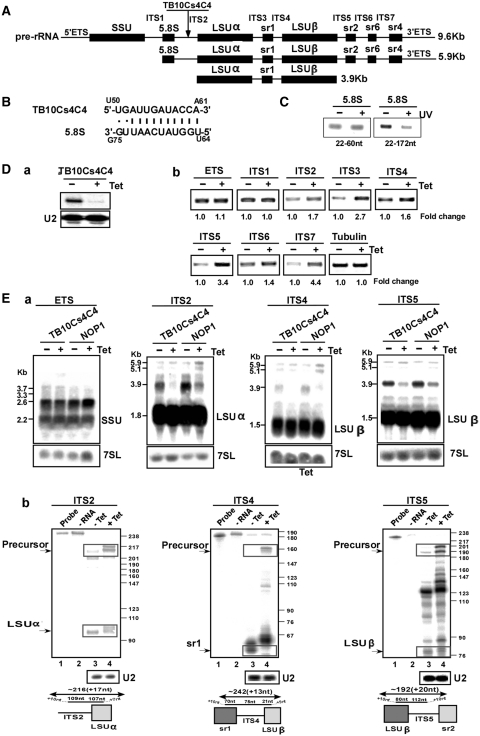
snoRNAs involved in rRNA processing, such as TB11Cs2C1/C2, are the most abundant snoRNAs expressed in T. brucei (22). To examine if the abundant snoRNAs identified here are involved in rRNA processing, we chose three candidates for functional analysis. The functional analyses included bioinformatic predictions of potential interactions between the snoRNA and rRNA (precursor and mature forms), and verification of the interactions predicted by bioinformatics using ‘RNA walk’ (11,22), and snoRNAi (22,23). The results for TB10Cs4C4 are presented in Figure 5. rRNA processing in trypanosomes involves several cleavages; trypanosome-specific cleavages generate the two LSU subunits, LSUα and LSUβ, releasing the srRNA fragments sr1, 2, 4 and 6. Cleavage at the internal spacer 1 (ITS1) releases a 5.9 kb precursor that is further cleaved at ITS2 and ITS5 to form the 3.9 kb precursor (depicted in Figure 5A). TB10Cs4C4 was shown to interact with SSU and to potentially guide the methylation on Gm1931. In addition, the RNA has the potential to interact with 5.8S rRNA (Figure 5B). To validate this interaction, we treated the cells with AMT followed by UV cross-linking and performed ‘RNA walk’ analysis (11,22). This method involves affinity selection of the small RNA and mapping the cross-linking adducts by RT–PCR. Domains that are cross-linked to the small RNA cannot be amplified by RT–PCR (11,22). Affinity selection using an anti-sense biotinylated oligonucleotide complementary to TB10Cs4C4 was used to enrich rRNA that was cross-linked to the snoRNA. Next, the adducts were mapped on the target. cDNA was prepared from the RNA before and after cross-linking, and the different domains were amplified by PCR. Marked reduction in the amplification of the fragment implicated in the interaction with the snoRNA was observed, in contrast to another adjacent domain that was efficiently amplified (Figure 5C). These results therefore validate the bioinformatics prediction.
Figure 5.
The role of TB10Cs4C4 in rRNA processing. (A) Schematic representation of the pre-rRNA. The coding sequences are shown in black. The identity of the intergenic regions is given. (B) Proposed interaction domain between the snoRNA and 5.8S rRNA. (C) Mapping the interaction domain of TB10Cs4C4 with 5.8S rRNA. RT–PCR along the 5.8S RNA. cDNA was prepared from the affinity selected RNA of untreated cells or after treatment with AMT and UV cross-linking. The cDNA was subjected to PCR using the set of primers indicated in the panel, and the PCR products were separated on a 1% agarose gel and stained with ethidium bromide. (D-a) Silencing of TB10Cs4C4. Total RNA (10 µg) before and after 3 days of induction was subjected to primer extension with primer specific to the snoRNA. The level of RNA was determined using U2-specific probe. (D-b) RT–PCR to examine rRNA processing defects under TB10Cs4C4 silencing. RNA from uninduced cells or cells after 3 days of induction was used to prepare cDNA. cDNA was amplified with primers situated in the intergenic regions specified in Supplementary Figure S1. The level of tubulin mRNA was used to control the level of RNA in each sample. The identity of the intergenic regions is indicated. (E) Monitoring the accumulation of rRNA precursors in TB10Cs4C4 silenced cells. (E-a) Northern analysis. Total RNA was extracted from cells carrying the TB10Cs4C4 RNAi construct without induction (−Tet), and after 3 days of induction (+Tet) and separated on a 1.2% agarose gel containing 2.2 M formaldehyde. The RNA was blotted and hybridized with indicated probe. The 7SL RNA probe was used to control the amount of RNA of each sample. The marker is indicated in Kb. (E-b) RNase protection to detect rRNA processing defects. RNA from cells expressing the TB10Cs4C4 silencing construct was used for RNase protection assays with the different probes, as indicated. The products were separated on a 6% denaturing gel. The structure of the antisense probe is schematically presented. The precursor and the mature snoRNAs are boxed. The sizes of the molecular weight marker, pBR322 DNA-MspI digest, are indicated. In each panel: Lane 1, probe; lane 2, protection assay in the absence of RNA; lane 3, RNA from uninduced cells; lane 4, RNA from cells after 3 days of induction.
Next, the snoRNA was silenced by RNAi and defects in rRNA processing were monitored using three approaches; RT–PCR, northern analysis and RNase protection. SnoRNAi was efficient, since a significant decrease in the steady-state level of this RNA was observed (Figure 5D-a).
To detect rRNA processing defects, the accumulation of pre-rRNA was examined in the silenced cells by RT–PCR, using probes covering the entire pre-rRNA. The results demonstrated accumulation of all pre-rRNA situated downstream to ITS1 (Figure 5D-b), suggesting a defect in the processing of LSU rRNA. We detected accumulation of the 5.9 kb precursor, and reduction of the 3.9 kb one (Figure 5E-a), a phenotype similar to that observed when fibrillarin (NOP1) is silenced (18). The defects suggest failure in the separation of LSU rRNA from the pre-rRNA. RNase protection analysis (Figure 5E-b) further supported the accumulation of ITS2, 4 and 5 which is in agreement with the accumulation of the 5.9 kb precursor. Given the bioinformatics prediction, TB10Cs4C4 is most probably involved in cleavage at ITS2, thus separating 5.8S from LSU rRNA. In mammalian cells, U8 mediates the cleavage at ITS2 (42). All our searches to identity the U8 homologue in trypanosomes failed. TB10CsC4 might therefore represent a functional homologue of U8 that cleaves within ITS2, since this RNA interacts with 5.8S rRNA, and the data suggest that ITS2 accumulates under its depletion.
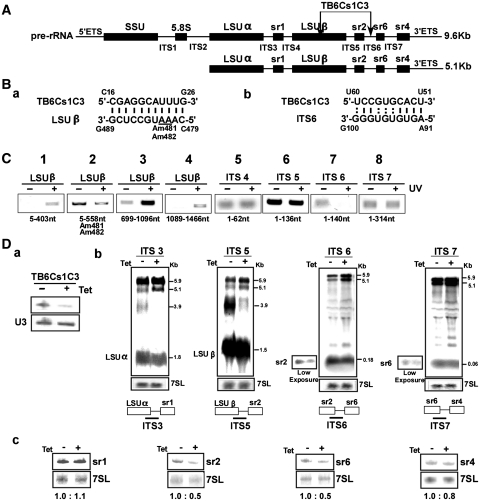
Next, we examined the role of TB6Cs1C3 in rRNA processing. The results are presented in Figure 6. Figure 6A illustrates the positions where TB6Cs1C3 is predicted to interact. The bioinformatics predictions suggest that in addition to potentially guiding two modifications on LSUβ, the snoRNA has potential to interact with ITS6 (Figure 6-B). The validity of the two possible interactions was examined by ‘RNA walk’. The ‘RNA walk’ was performed on different domains along the pre-rRNA, and the results show clear reduction in the amplification of domains predicted to interact with the snoRNA and that are located in LSUβ and ITS6 [Figure 6C, see lanes (2) and (7)].
Figure 6.
The role of TB6Cs1C3 in rRNA processing. (A) Schematic representation of the pre-rRNA. The coding sequences are in black. The identity of the intergenic regions is given, and the position of the interaction of TB6Cs1C3 with the target are indicated. (Ba- and b-), Proposed interaction domain between the snoRNA and its targets. (C) ‘RNA walk’ to validate the interaction of TB6Cs1C3 with its target. RT–PCR along the pre-mRNA. cDNA was prepared from the affinity selected RNA of untreated cells or after treatment with AMT and UV cross-linking. The cDNA was subjected to PCR using the set of primers indicated, and the PCR products were separated on a 1% agarose gel and stained with ethidium bromide. (D) Monitoring the accumulation of rRNA precursors in TB6Cs1C3 silenced cells. (D-a) Silencing of TB6Cs1C3. a-RNA was prepared from uninduced cells and cells 3 days after silencing, and subjected to primer extension with the snoRNA specific primer and U3 primer. (D-b) Northern analysis. Total RNA was extracted from cells carrying the TB6Cs1C3 RNAi construct without induction (−Tet), and after 3 days of induction (+Tet) and separated on a 1.2% agarose gel containing 2.2 M formaldehyde. The RNA was blotted and hybridized with the indicated probes. The 7SL RNA probe was used to control the amount of RNA of each sample. The marker sizes are indicated in kilobase pairs. (D-c) Monitoring defects in srRNA processing. Total RNA was extracted from cells carrying the TB6Cs1C3 RNAi construct without induction (−Tet), and after 3 days of induction (+Tet) and separated on a 10% acrylamide denaturing gel and subjected to northern analysis with srRNA probes. The 7SL RNA probe was used to control the amount of RNA in each sample.
Downregulation of TB6Cs1C3 by RNAi (Figure 6D-a) resulted in the accumulation of the 5.9 and 5.1 kb precursors and reduction in the level of the 3.9 kb species (Figure 6D-b), suggesting defects in processing downstream from ITS2. A defect characteristic of this silencing is the accumulation of the 5.1 kb RNA, suggesting that the decrease in the 3.9 kb precursor stems from the failure to cleave downstream to LSUβ. Based on the validated interaction of the snoRNA with ITS6, this snoRNA should direct cleavage at ITS5 and potentially at ITS6, both trypanosome-specific processing events. To localize the defects in srRNA processing, RNA was subjected to northern analysis monitoring changes in the level of srRNA-1, 2, 6 and 4. The results (Figure 6D-c) demonstrated decreased levels of srRNA-2 and 6, with no effect on the level of srRNA-1, and only a slight effect on srRNA-4, supporting the role of the snoRNA in cleavage at ITS6 for separating srRNA-2 and srRNA-6.
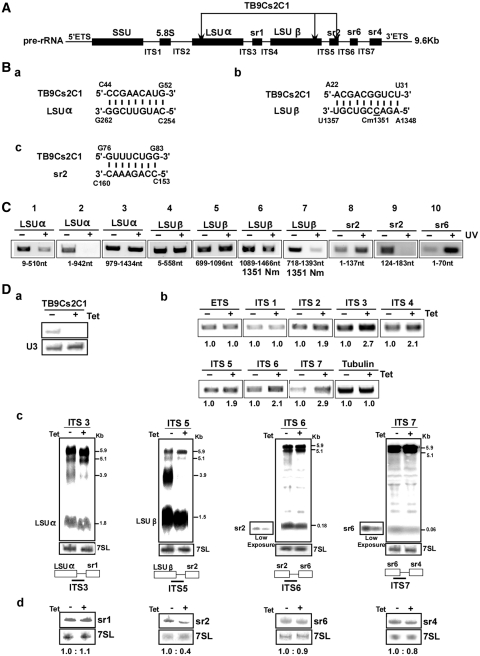
The third abundant snoRNA that was analyzed is TB9Cs2C1. According to the proposed interaction domains (Figure 7A) and the bioinformatic predictions (Figure 7B) there are three possible targets for TB9Cs2C1 on the rRNA; one of these has the potential to guide methylation at position C1351 of LSUβ. ‘RNA walk’ was performed to examine if all the three sites are used by the snoRNA to interact with rRNA. This analysis suggested the presence of adducts on LSUα, LSUβ and srRNA-2 (Figure7C, lanes 1, 2, 6, 7, 9). To examine the role of this snoRNA in rRNA processing, the snoRNA was silenced by RNAi (Figure 7D-a). rRNA defects observed by RT–PCR demonstrated accumulation of precursors from ITS2 and downstream (Figure 7D-b). Northern analysis also demonstrated defects in LSU processing showing accumulation of the 5.9 kb, but not the 5.1 kb precursor and reduction in 3.9 kb precursor, suggesting that this snoRNA is essential for processing on both sides of LSU (Figure 7D-c). Based on the observed rRNA defects and the bioinformatic predictions, we suggest that TB9Cs2C1 functions in cleavages situated at both the 5′- and 3′-ends of LSU (Figure 7D-c). To obtain further support for the specific role of this snoRNA in processing on srRNA-2, RNA from induced and uninduced cells was analyzed for the level of srRNAs. The results (Figure 7D-d) demonstrated significant reduction in the level of srRNA-2, further supporting the role of this snoRNA in srRNA-2 processing.
Figure 7.
The role of TB9Cs2C1 in rRNA processing. (A) Schematic representation of the pre-rRNA. The coding sequences are shown in black. The identity of the intergenic regions is given and the positions of the interaction with TB9Cs2C1 with the targets are indicated (B) Proposed interaction domains between the snoRNA and its target. (C). ‘RNA walk’ analysis. RT–PCR along the rRNA precursor. cDNA was prepared from the affinity selected RNA of untreated cells or after treatment with AMT and UV cross-linking. The cDNA was subjected to PCR using the set of primers indicated, and the PCR products were separated on a 1% agarose gel and stained with ethidium bromide. (D-a) Silencing of TB9Cs2C1. Total RNA (10 µg) before and after 3 days of induction was subjected to primer extension with primer specific to the snoRNA. The level of RNA was determined using a U3-specific probe. (D-b) RT–PCR to examine rRNA processing defects under TB9Cs2C1 silencing. RNA from uninduced cells or cells after 3 days of induction was used to prepare cDNA. cDNA was amplified with primers situated in the intergenic regions specified in Supplementary Figure S1. The level of Tubulin mRNA was used to control the level of RNA in each sample. The identity of the intergenic regions is indicated. (D-c) Monitoring the accumulation of rRNA precursors in TB9Cs2C1 silenced cells. Northern analysis. Total RNA was extracted from cells carrying the TB9Cs2C1 RNAi construct without induction (−Tet), and after 3 days of induction (+Tet) and separated on a 1.2% agarose gel containing 2.2 M formaldehyde. The RNA was blotted and hybridized with the indicated probes. The 7SL RNA probe was used to control the amount of RNA of each sample. Marker sizes are indicated in kilobase pairs. (D-d) Monitoring defects in srRNA processing. Total RNA was extracted from cells carrying the TB9Cs2C1 RNAi construct without induction (−Tet), and after 3 days of induction (+Tet), separated on a 10% acrylamide denaturing gel, and subjected to northern analysis with srRNA probes. The 7SL RNA probe was used to control the amount of RNA in each sample.
RNA-seq enables mapping the boundaries of small RNAs and helps predict their correct secondary structure in an RNP complex
In our previous genome-wide analysis, we predicted that the 5′-termini of C/D snoRNAs were 1–5 nt upstream from the C-box, and the 3′-termini 1–3 nt downstream of the D-box (12). Likewise, for the termini of H/ACA snoRNAs, we suggested that, in a manner similar to other eukaryotes, the 3′-end is located 3′-nt downstream from the AGA box (12). We also noticed that the 5′-end is always situated 3-nt upstream to the first stem–loop. Inspection of the snoRNAs termini derived from the RNA-seq data indicated that in 62% of the cases the predictions agreed with the boundaries previously suggested (12). In the case of C/D snoRNAs, we analyzed 54 molecules, and 25 molecules were found to differ from the consensus; 7 were longer by >5 nt at the 3′-end and 3 differed at the 5′-end by >5 nt. Only three predicted RNAs appeared shorter at both the 5′- and 3′-ends. A list of RNA-seq-derived boundaries for these RNAs is given in Supplementary Figure S7. Inspection of the H/ACA candidates indicated that 10 out of 39 predicted RNAs deviated from the consensus; in two cases, the RNAs were longer at the 5′-end, three had a gap in the middle of the RNA and five had a shorter 3′-end (Supplementary Figure S7).
To examine whether the 5′- and/or 3′-extensions represented the true ends of the molecules or instead were derived from potential pre-snoRNA processing intermediates, two cases were examined experimentally (Supplementary Figure S8). The distribution of the reads along the previously predicted coding regions (indicated in white) and the predictions derived from the RNA-seq data (indicated in gray) are depicted in Supplementary Figure S8A-a and B-a. We first used northern analysis (Supplementary Figure S8A-b) and primer extension (Supplementary Figure S8A-c and B-b) to determine whether the read’s distribution was consistent with the size of corresponding RNAs in steady-state. The results of both analyses showed that the sizes of the two RNAs and their corresponding 5′ termini were in agreement with our previous predictions. Specifically, the results of the primer extension analysis indicated that the sequences marked in gray are removed during processing to mature RNAs. Thus, it seems likely that the flanking sequences, predicted by the RNA-seq data, emerged from pre-snoRNA intermediates, suggesting that processing of certain pre-snoRNAs is less efficient than that of others.
Of special interest was the case of TB10Cs3C1 and TB10Cs3H1, shown in Supplementary Figure S8C. There was no interruption of reads between these RNA coding sequences, thus raising the possibility that if such a molecule existed it may be a homologue of scaRNAs, which are chimeric snoRNAs containing both C/D and H/ACA RNA sequences and activity (Supplementary Figure S8C-a) (43). However, primer extension demonstrated that the mature forms of TB10Cs3C1 and TB10Cs3H1 are individual snoRNAs (Supplementary Figure S8C-b). In addition, as determined by Northern analysis, the level of the ∼150 nt RNA decreased only under NOP58 silencing, whereas that of the ∼76 nt RNA changed only under CBF5 silencing (Supplementary Figure S8C-c), further suggesting that TB10Cs3C1 and TB10Cs3H1 are two separate snoRNAs.
Novel small ncRNAs
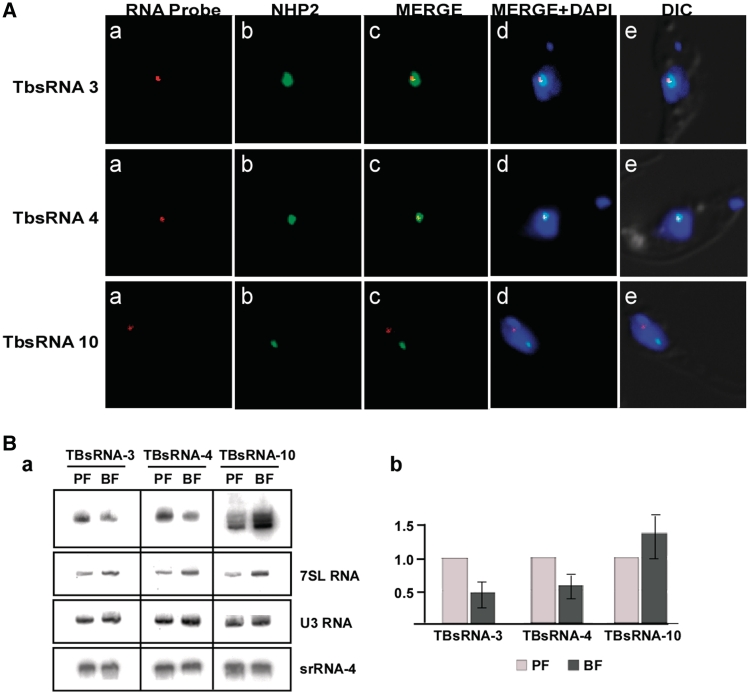
Initially, to discover new small ncRNAs we focused our analysis on the 3674 contigs that were not annotated in the genome. From these, we selected 274 contigs that were 50 nt or longer and had a minimum coverage of 50 reads. However, close inspection indicated that many of the contigs were derived from annotation ‘mistakes’ and included ribosomal RNAs, mRNA coding regions and UTRs. After elimination of these sequences, we were left with 52 contigs including 20 new snoRNAs. A list of the remaining putative RNAs is given in Supplementary Figure S9. Three of these RNAs (sRNA-1, sRNA-2 and sRNA-5) were not detected by primer extension. Thus, we turned our attention to the loci covered by high numbers of reads, as the corresponding RNAs were likely to be abundant and easy to detect. We chose three candidate molecules that we termed TBsRNA-3, -4 and -10. Figures 8 and 9 show the various approaches we took to elucidate the function of these RNAs. To investigate the localization of the RNAs we performed in situ hybridization in combination with immunofluorescence staining of NHP2, a nucleolus marker (Figure 8A). The results showed that both TBsRNA-3 and -4 co-localized with NHP2 at the nucleolus. In contrast, TBsRNA-10 was mostly found in the nucleus and was detected as a single fluorescent focus, clearly distinct from the nucleolus.
Figure 8.
Elucidating the function of novel ncRNAs, TBsRNA 3, 4 and 10. (A) In situ hybridization. In situ hybridization combined with immunofluorescence was performed as described in ‘Materials and Methods’ section. (a) in situ hybridization with TBsRNAs probe monitored with Alexa-Red; (b) Immunofluorescence with anti-NHP2 antibodies; (c) merge of (A-a) and (A-b); (A-d) DAPI-stained nuclei merged with (a) and (b); (e) DIC merged with (d). (B) Expression of the small RNAs during the life cycle of the parasite. RNA (20 µg) was prepared from the procyclic and bloodstream forms and separated on a 10% denaturing gel. (a) The blot was hybridized with the indicated probes. (b) Quantitation of the differential level of the RNA. The blot was subjected to Image J analysis.
Figure 9.
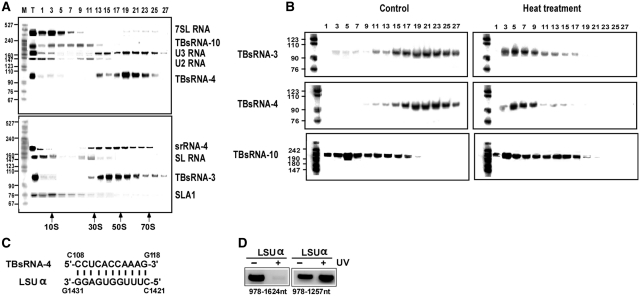
(A) Distribution of RNAs in RNP complexes. A whole-cell extract was prepared from 2 × 108 cells and fractionated on a 10–30% sucrose gradient at 35 000 r.p.m. for 3 h. The fractions were deproteinized, and the RNA was separated on a 10% polyacrylamide-denaturing gel and subjected to northern analysis with the indicated probes. A pBR322 MspI digest was used as MW marker. The S-value markers of E. coli ribosomes and catalase are indicated. (B) Association of TBsRNAs with other RNAs. RNA was prepared after gentle treatment with Proteinase K and fractionated on sucrose gradients before (control) and after heat treatment. The gradients were fractionated at 35 000 r.p.m. for 22 h, and RNA from the different fractions was separated on a 10% denaturing gel, and subjected to northern analysis with the indicated probes. (C) The potential base-pair interaction between TBsRNA-4 and its rRNA target. (D) ‘RNA walk’ analysis. RT–PCR of domains on LSUα. cDNA was prepared from the affinity-selected RNA of untreated cells, or after treatment with AMT and UV cross-linking. The cDNA was subjected to PCR using the oligonucleotides indicated. The PCR products were separated on a 1% agarose gel and stained with ethidium bromide.
Next, the levels of expression of several small RNAs were examined in procyclic and bloodstream form parasites (Figure 8B-a and -b). The results showed that there were minor changes in the level of U3 and rRNA in the two life-cycle stages, but that 7SL RNA was more abundant in the bloodstream as compared to procyclic forms. This observation is not surprising, since bloodstream trypanosomes most likely require high levels of SRP to support the intensive protein secretion demands that exist during this life-cycle stage. Interestingly, TBsRNA-3, -4 and -10 also seemed differentially expressed: TBsRNA-3 and -4 were more abundant in the procyclic stage, whereas, TBsRNA-10 was highly expressed in the bloodstream form.
To further gauge the potential function of TBsRNA-3, -4 and -10, we decided to examine the sedimentation behavior of the corresponding RNPs, as this analysis can be highly informative in suggesting the role of specific RNAs. To this end, whole-cell extracts were fractionated on sucrose gradients and the sedimentation of the RNAs was revealed by northern analysis. The results (Figure 9A) demonstrated that under the experimental conditions used we were able to separate ribosomal from spliceosomal complexes. The U2 and the SL RNP mono-particles (fractions 1–3) were found at the top of the gradient, but also in a distinct ∼40S complex (fractions 9–11), as described previously (44), whereas ribosomal complexes were detected in fractions 13–27. We found that TBsRNA-3 and -4 co-sedimented with ribosomal complexes. In contrast, the TBsRNA-10 ribonucleoprotein complexes sedimented in the lighter portions of the gradient and away from spliceosomes and ribosomes.
RNA-RNA association is another criterion that can help pinpoint the function of a novel RNA. Thus, we asked whether TBsRNA-3, -4 or -10 partnered with other cellular RNAs. To this end we extracted RNA from cells, using mild conditions that were expected to preserve RNA-RNA interactions, and then fractionated ‘native’ and ‘denatured’ (heat- treated) pairs of samples on parallel sucrose gradients. If the small RNAs associate with other RNAs, the heat-treatment should disrupt this interaction and result in fractionation of the RNAs at lower S values. In this analysis, the sedimentation behavior of TBsRNA-3 and -4 significantly changed upon heat treatment, most likely because these RNAs interact with rRNA. No shift in sedimentation was observed for TBsRNA-10, indicating that this RNA may not stably associate with another RNA under steady-state conditions (Figure 9B).
Due to the tight association of TBsRNA-4 with the ribosome, we examined if this RNA belongs to C/D or the H/ACA family by examining its level under depletion of CBF5 or NOP58. No change was observed in the level of this RNA in the silenced cells, suggesting that this is a novel type of snoRNA not belonging to either the C/D or the H/ACA families (data not shown). Indeed, this RNA can interact by perfect base-pairing with LSUα (Figure 9C). Future studies will investigate the role of this RNA in rRNA processing. In support of the predicted interaction, ‘RNA walk’ revealed a block in the region where the expected adduct should map, but not in a neighboring domain (Figure 9D).
The function of TBsRNA-10 is currently unknown, but BLAST analysis revealed a 19 nt sequence with perfect complementarity to the mRNA of Tb10.6k15.1430, a hypothetical protein coding gene.
DISCUSSION
We describe here the first whole-genome study by RNA-seq of small RNP complexes in procyclic trypanosomes. We identified new snoRNAs from the C/D and H/ACA families, as well as additional snoRNAs that do not belong to these families. Importantly, we provide evidence that a novel abundant snoRNA is most probably involved in trypanosome-specific rRNA processing. We assembled a library of potential ncRNAs and showed that this library provides a fertile ground for discovery of novel ncRNAs. To complement the bioinformatic approaches used, we also established a coherent experimental strategy, which is invaluable for analyzing the potential function of novel ncRNAs, and demonstrate its application to three novel RNAs.
Did this study complete the description of the snoRNA repertoire?
The present study, together with our previous analysis, indicates that trypanosomes have a rich repertoire of snoRNAs: 79 C/D and 63 H/ACA. Our previous mapping data predicted the presence of at least 131 Nms on rRNA (18), as compared to at least 100 Nms in Crithidia fasiculata (45) and 200 Nms in Euglena gracilis, which is phylogenetically related to trypanosome (46). The current repertoire of 79 C/D snoRNAs has the potential to guide ∼150 Nms, suggesting that we are close to cataloging the complete repertoire of C/D snoRNAs. However, about 40 Nms remain, for which we failed to find the corresponding snoRNAs, suggesting that additional snoRNAs are likely to exist. Since C/D snoRNAs appear to vary widely in abundance, we anticipate that there might be low abundance snoRNAs, which were not detected in our experiment. To complete the description of the repertoire, we are currently preparing a library using RNA affinity selected by the core C/D protein, SNU13. There are 63 H/ACAs reported in this study. However, since no genome wide mapping of pseudouridines on trypanosomes rRNA has been reported, we cannot predict the number of H/ACA RNAs in the genome. This will await the RNA-seq analysis of RNA selected via the H/ACA binding protein, NHP2.
One characteristic feature of trypanosome rRNA modifications and their guide RNAs is that, in contrast to yeast and humans, the number of predicted Nms exceeds the number of pseudouridines. The current study also identified more C/D (79) as compared to H/ACA (63) RNAs. It was previously proposed that the hypermethylation that exists in plants and thermophiles help sustain ribosome function at high temperatures (47). Similarly, the hypermethylation in the parasite may enable it to cope with the temperature shifts during the cycling between the procyclic stage (26°C) and the bloodstream form stage (37°C). Indeed, we demonstrated higher expression of C/D snoRNAs in the bloodstream form, possibly leading to elevated methylation during this life-cycle stage, and thereby supporting the notion that hypermethylation may be an important determinant of ribosome function at elevated temperature (18). However, it was noted that in several organisms not undergoing these temperature shifts, the number of C/D snoRNAs also exceeds that of H/ACA (48).
Interestingly, our screen did not detect expression of 23 known snoRNAs. Perhaps these RNAs are preferentially expressed in bloodstream forms. Since this study revealed new solitary C/D snoRNAs, additional molecules of this kind may be expressed in the bloodstream form. It will be therefore of great interest to compare the expression of snoRNAs between bloodstream and procyclic stages.
Highly abundant snoRNAs function in rRNA processing
This study sheds light on the complex processing mechanism of rRNA in trypanosomes by revealing 13 snoRNAs, which were represented by high number of reads. As described in the Introduction, rRNA processing in trypanosomes is unique in nature. Briefly, the first cleavage occurs between the SSU and 5.8S RNA, at position b1 (49), whereas in yeast and mammals the rRNA precursor is first cleaved at the 5′ ETS while later cleavages separate the SSU from 5.8S RNA (50). Most striking is the trypanosome LSU rRNA processing pathway through which 28S rRNA is cleaved into two large fragments, LSUα and LSUβ, and four small RNA fragments, sr1, sr2, 6 and 4, are released (20). Based on our previous studies that screened for rRNA processing defects under depletion of CBF5 or NOP1, we predicted the existence of trypanosome-specific snoRNAs (17,18). Indeed, we recently proposed that the TB11Cs2C1 snoRNA is a functional homologue of U14 (22). In addition, TB11Cs2C2, was found to be a trypanosome-specific snoRNA that directs cleavage upstream of srRNA2 and downstream of srRNA6 (22). The results presented in this study suggest that snoRNA TB10Cs4C4 is required for cleavage within ITS2, analogous to the function of U8 in mammals (42). Indeed, neither the U8 homologue nor its 29 kDa-specific protein (51) were identified in trypanosomes . Our results indicate that conventional snoRNAs present in other eukaryotes are replaced in trypanosomes by unique and specific trypanosome RNAs.
This study extends the repertoire of snoRNAs implicated to function in trypanosome-specific cleavage events. Such snoRNAs include: TB6Cs1C3, which functions in processing at the 3′ end of LSU, and TB9Cs2C1, which instead is involved in both 5′- and 3′-end processing of LSU. Our current results, together with our previous findings demonstrate that trypanosome-specific rRNA cleavages are indeed mediated by trypanosome-specific snoRNAs. The fine mapping of the defects observed under depletion of TB11Cs2C2 (22), or TB6Cs1C3 and TB9Cs2C1 (this study) suggest that these snoRNAs are all essential for proper processing of srRNA-2, whereas TB11Cs2C2 and TB6Cs1C3 are essential for processing of srRNA-6. However, each snoRNA functions independently, and thus results in specific defects when it is silenced. The many snoRNAs involved in cleavages at the 3′ end of LSU suggest that several snoRNA species function in these processing events. Indeed, recent data from our group based on RNA-seq of Leishmania snoRNAs suggest that homologues of TB10Cs4C4, TB9Cs2C1 and TB6Cs1C3 exist and are very abundant RNAs. These snoRNAs interact with the same intergenic domains albeit with different sequences.
Genome organization of snoRNAs, their differential expression and mode of processing and regulation of rRNA processing
The RNA-seq data revealed 14 ‘solitary’ snoRNAs among the 50 snoRNA loci. Some of the ‘solitary’ snoRNAs were found near abundant mRNAs, suggesting that another strategy to highly express a snoRNA is via its location near a highly expressed gene. The best example is the snoRNA located at the tubulin locus.
The RNA-seq data also provided insight into snoRNA processing and showed that although in 62% of the cases the snoRNA termini agree with our previous predictions, in 17% of the cases, additional snoRNA-flanking sequences were detected. Experimental validation (Supplementary Figure S8) showed that the extra flanking sequences are likely part of pre-snoRNAs, as they do not exist as part of the mature RNA. We suggest that pre-snoRNA processing, especially from the 3′ end, is not uniform among different molecules, most probably because of differential association with the exosome, which in other organisms carries out snoRNA 3′-end processing.
The differential abundance of snoRNAs observed in this study, was previously noted (12,13). A machine learning approach was used to predict the parameters that affect snoRNA abundance, and it predicted the abundance in 65% of the cases. However, the recent finding that the polycistronic pre-snoRNA transcripts from genomic clusters are polyadenylated (8) raises the possibility that 3′-end cleavage and polyadenylation also contribute to snoRNA stability and abundance. We propose that the level of snoRNA in steady-state emerges from the interplay between two processes that act to generate the snoRNP, namely pre-snoRNA trimming by exonucleolytic activities and snoRNP assembly. We have recently obtained evidence that the most abundant snoRNAs are polyadenylated in a complex manner by two different polymerases. We speculate that prolonging the process of 3′-end polyadenylation may slow down recruitment of the exosome and promote assembly of snoRNAs with the core proteins before exonucleolytic trimming begins. This type of processing takes place at the end of the clusters or next to abundant snoRNAs (our unpublished data). However, this peculiar mechanism of polyadenylation does not generate the majority of the snoRNAs, but only the most abundant ones. Most likely, the abundance of most of the snoRNAs is determined by the efficiency of cleavage of the individual snoRNAs from the polycistronic precursor by an as yet unknown endonuclease, as well as by the factors mentioned above.
In addition, little is known about how rRNA processing is regulated. In humans, an ncRNA was shown to associate with the rRNA promoter, thereby affecting heterochromatin formation, nucleosome positioning and rDNA silencing (52). However, TBsRNA-3, which is derived from the ETS, does not seem to belong to such a family of regulators, and we suggest that it is involved in regulating rRNA processing itself. One can envision that this RNA binds crucial factors essential for processing and thus may participate in regulation. The identification of its binding proteins should provide hints regarding its precise function. Likewise, TBsRNA-4 most probably has a role in rRNA processing as well, but further investigation is required since this is a novel RNA, has no obvious homologue in other organisms, and does not belong to known snoRNA families.
How does the number of trypanosome small ncRNAs compare to that of other eukaryotes?
Recent evidence suggest that the human genome codes for hundreds of thousands ncRNAs (53). However, many of these transcripts may represent transcriptional noise. Recently, RNPomics study defining the ncRNAs associated with human and mouse cells was analyzed and revealed several hundred miRNAs, snoRNAs and retroposon RNAs, but also ∼1000 new ncRNAs (3), which could not be grouped into these families. Furthermore, recent RNA-seq studies in bacteria suggest that their genome encodes for tens of such ncRNAs (80 in Escherichia coli, 55 in Salmonella Typhi and 50 in Listeria) (1).
Before we began this study, it was evident that trypanosomes express several hundred ncRNAs, many of which have counterparts in other eukaryotes. By sequencing RNAs derived from a small RNP preparation, we have provided strong support for the notion that trypanosomes express novel classes of ncRNA, which do not appear to fall into previously defined families. As the RNA-seq library was prepared from a selected size class of RNPs, we anticipate that in these parasites the full repertoire of ncRNAs may be much larger than anticipated. The next challenge will be to deepen the coverage of ncRNAs by employing affinity purification methods based on known or yet to be identified novel RNA binding proteins. Another important question to be addressed in future experiments is whether the repertoire of ncRNAs differs between trypanosomes in the bloodstream versus procyclic stages. Finally, the approaches presented in this study form the foundation for deciphering the role of any novel small RNA identified by RNA-seq.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure S1–S12.
FUNDING
Israel-US Binational Science Foundation (BSF); National Institutes of Health (RO1 AI056333 to E.U.); David and Inez Myers Chair in RNA silencing of diseases (to S.M.); ERC Starting Grant (to R.S.); the Minerva Foundation (to R.S.); the Azrieli Foundation (to O.W.). Funding for open access charge: National Institutes of Health (RO1 AI056333 to E.U.).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Sorek R, Cossart P. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat. Rev. Genet. 2010;11:9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- 2.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15(Spec No. 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 3.Rederstorff M, Bernhart SH, Tanzer A, Zywicki M, Perfler K, Lukasser M, Hofacker IL, Huttenhofer A. RNPomics: defining the ncRNA transcriptome by cDNA library generation from ribonucleo-protein particles. Nucleic Acids Res. 2010;38:e113. doi: 10.1093/nar/gkq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang XH, Haritan A, Uliel S, Michaeli S. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunzl A. The pre-mRNA splicing machinery of trypanosomes: complex or simplified? Eukaryot Cell. 2010;9:1159–1170. doi: 10.1128/EC.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6:e1001090. doi: 10.1371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beja O, Ullu E, Michaeli S. Identification of a tRNA-like molecule that copurifies with the 7SL RNA of Trypanosoma brucei. Mol. Biochem. Parasitol. 1993;57:223–229. doi: 10.1016/0166-6851(93)90198-7. [DOI] [PubMed] [Google Scholar]
- 10.Liu L, Ben-Shlomo H, Xu YX, Stern MZ, Goncharov I, Zhang Y, Michaeli S. The trypanosomatid signal recognition particle consists of two RNA molecules, a 7SL RNA homologue and a novel tRNA-like molecule. J. Biol. Chem. 2003;278:18271–18280. doi: 10.1074/jbc.M209215200. [DOI] [PubMed] [Google Scholar]
- 11.Lustig Y, Wachtel C, Safro M, Liu L, Michaeli S. ‘RNA walk’ a novel approach to study RNA-RNA interactions between a small RNA and its target. Nucleic Acids Res. 2010;38:e5. doi: 10.1093/nar/gkp872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang XH, Uliel S, Hury A, Barth S, Doniger T, Unger R, Michaeli S. A genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Trypanosoma brucei reveals a trypanosome-specific pattern of rRNA modification. RNA. 2005;11:619–645. doi: 10.1261/rna.7174805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang XH, Hury A, Hoze E, Uliel S, Myslyuk I, Apatoff A, Unger R, Michaeli S. Genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Leishmania major indicates conservation among trypanosomatids in the repertoire and in their rRNA targets. Eukaryot. Cell. 2007;6:361–377. doi: 10.1128/EC.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang XH, Ochaion A, Xu YX, Liu Q, Michaeli S. Small nucleolar RNA clusters in trypanosomatid Leptomonas collosoma. Genome organization, expression studies, and the potential role of sequences present upstream from the first repeated cluster. J. Biol. Chem. 2004;279:5100–5109. doi: 10.1074/jbc.M308264200. [DOI] [PubMed] [Google Scholar]
- 15.Liang XH, Liu L, Michaeli S. Identification of the first trypanosome H/ACA RNA that guides pseudouridine formation on rRNA. J. Biol. Chem. 2001;276:40313–40318. doi: 10.1074/jbc.M104488200. [DOI] [PubMed] [Google Scholar]
- 16.Liang XH, Xu YX, Michaeli S. The spliced leader-associated RNA is a trypanosome-specific sn(o) RNA that has the potential to guide pseudouridine formation on the SL RNA. RNA. 2002;8:237–246. doi: 10.1017/s1355838202018290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barth S, Hury A, Liang XH, Michaeli S. Elucidating the role of H/ACA-like RNAs in trans-splicing and rRNA processing via RNA interference silencing of the Trypanosoma brucei CBF5 pseudouridine synthase. J. Biol. Chem. 2005;280:34558–34568. doi: 10.1074/jbc.M503465200. [DOI] [PubMed] [Google Scholar]
- 18.Barth S, Shalem B, Hury A, Tkacz ID, Liang XH, Uliel S, Myslyuk I, Doniger T, Salmon-Divon M, Unger R, et al. Elucidating the role of C/D snoRNA in rRNA processing and modification in Trypanosoma brucei. Eukaryot. Cell. 2008;7:86–101. doi: 10.1128/EC.00215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 20.Campbell DA, Kubo K, Clark CG, Boothroyd JC. Precise identification of cleavage sites involved in the unusual processing of trypanosome ribosomal RNA. J. Mol. Biol. 1987;196:113–124. doi: 10.1016/0022-2836(87)90514-6. [DOI] [PubMed] [Google Scholar]
- 21.Hartshorne T, Agabian N. RNA B is the major nucleolar trimethylguanosine-capped small nuclear RNA associated with fibrillarin and pre-rRNAs in Trypanosoma brucei. Mol. Cell. Biol. 1993;13:144–154. doi: 10.1128/mcb.13.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta SK, Hury A, Ziporen Y, Shi H, Ullu E, Michaeli S. Small nucleolar RNA interference in Trypanosoma brucei: mechanism and utilization for elucidating the function of snoRNAs. Nucleic Acids Res. 2010;38:7236–7247. doi: 10.1093/nar/gkq599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang XH, Liu Q, Michaeli S. Small nucleolar RNA interference induced by antisense or double-stranded RNA in trypanosomatids. Proc. Natl Acad. Sci. USA. 2003;100:7521–7526. doi: 10.1073/pnas.1332001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- 25.Mandelboim M, Barth S, Biton M, Liang XH, Michaeli S. Silencing of Sm proteins in Trypanosoma brucei by RNA interference captured a novel cytoplasmic intermediate in spliced leader RNA biogenesis. J. Biol. Chem. 2003;278:51469–51478. doi: 10.1074/jbc.M308997200. [DOI] [PubMed] [Google Scholar]
- 26.Mandelboim M, Estrano CL, Tschudi C, Ullu E, Michaeli S. On the role of exon and intron sequences in trans-splicing utilization and cap 4 modification of the trypanosomatid Leptomonas collosoma SL RNA. J. Biol. Chem. 2002;277:35210–35218. doi: 10.1074/jbc.M201910200. [DOI] [PubMed] [Google Scholar]
- 27.Wachtel C, Michaeli S. Functional analysis of non-coding RNAs in trypanosomes: RNA walk, a novel approach to study RNA-RNA interactions between small RNA and its target. Methods Mol. Biol. 2011;718:245–257. doi: 10.1007/978-1-61779-018-8_15. [DOI] [PubMed] [Google Scholar]
- 28.Hury A, Goldshmidt H, Tkacz ID, Michaeli S. Trypanosome spliced-leader-associated RNA (SLA1) localization and implications for spliced-leader RNA biogenesis. Eukaryot. Cell. 2009;8:56–68. doi: 10.1128/EC.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 31.Hertel J, Hofacker IL, Stadler PF. SnoReport: computational identification of snoRNAs with unknown targets. Bioinformatics. 2008;24:158–164. doi: 10.1093/bioinformatics/btm464. [DOI] [PubMed] [Google Scholar]
- 32.Schattner P, Decatur WA, Davis CA, Ares M, Jr, Fournier MJ, Lowe TM. Genome-wide searching for pseudouridylation guide snoRNAs: analysis of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2004;32:4281–4296. doi: 10.1093/nar/gkh768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myslyuk I, Doniger T, Horesh Y, Hury A, Hoffer R, Ziporen Y, Michaeli S, Unger R. Psiscan: a computational approach to identify H/ACA-like and AGA-like non-coding RNA in trypanosomatid genomes. BMC Bioinformatics. 2008;9:471. doi: 10.1186/1471-2105-9-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 35.Djikeng A, Shi H, Tschudi C, Ullu E. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24-26-nucleotide RNAs. RNA. 2001;7:1522–1530. [PMC free article] [PubMed] [Google Scholar]
- 36.Tkacz ID, Gupta SK, Volkov V, Romano M, Haham T, Tulinski P, Lebenthal I, Michaeli S. Analysis of spliceosomal proteins in Trypanosomatids reveals novel functions in mRNA processing. J. Biol. Chem. 2010;285:27982–27999. doi: 10.1074/jbc.M109.095349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andreazzoli M, Gerbi SA. Changes in 7SL RNA conformation during the signal recognition particle cycle. EMBO J. 1991;10:767–777. doi: 10.1002/j.1460-2075.1991.tb08008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakaar V, Dare AO, Hong D, Ullu E, Tschudi C. Upstream tRNA genes are essential for expression of small nuclear and cytoplasmic RNA genes in trypanosomes. Mol. Cell. Biol. 1994;14:6736–6742. doi: 10.1128/mcb.14.10.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doniger T, Katz R, Wachtel C, Michaeli S, Unger R. A comparative genome-wide study of ncRNAs in trypanosomatids. BMC Genomics. 2010;11:615. doi: 10.1186/1471-2164-11-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doniger T, Michaeli S, Unger R. Families of H/ACA ncRNA molecules in trypanosomatids. RNA Biol. 2009;6:370–374. doi: 10.4161/rna.6.4.9270. [DOI] [PubMed] [Google Scholar]
- 41.Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010;38:4946–4957. doi: 10.1093/nar/gkq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrissey JP, Tollervey D. U14 small nucleolar RNA makes multiple contacts with the pre-ribosomal RNA. Chromosoma. 1997;105:515–522. doi: 10.1007/BF02510488. [DOI] [PubMed] [Google Scholar]
- 43.Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22:4283–4293. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang XH, Liu Q, Liu L, Tschudi C, Michaeli S. Analysis of spliceosomal complexes in Trypanosoma brucei and silencing of two splicing factors Prp31 and Prp43. Mol. Biochem. Parasitol. 2006;145:29–39. doi: 10.1016/j.molbiopara.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 45.Gray MW. The ribosomal RNA of the trypanosomatid protozoan Crithidia fasciculata: physical characteristics and methylated sequences. Can. J. Biochem. 1979;57:914–926. doi: 10.1139/o79-111. [DOI] [PubMed] [Google Scholar]
- 46.Russell AG, Schnare MN, Gray MW. Pseudouridine-guide RNAs and other Cbf5p-associated RNAs in Euglena gracilis. RNA. 2004;10:1034–1046. doi: 10.1261/rna.7300804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dennis PP, Omer A, Lowe T. A guided tour: small RNA function in Archaea. Mol. Microbiol. 2001;40:509–519. doi: 10.1046/j.1365-2958.2001.02381.x. [DOI] [PubMed] [Google Scholar]
- 48.Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94:83–88. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 49.Hartshorne T, Toyofuku W. Two 5′-ETS regions implicated in interactions with U3 snoRNA are required for small subunit rRNA maturation in Trypanosoma brucei. Nucleic Acids Res. 1999;27:3300–3309. doi: 10.1093/nar/27.16.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 51.Tomasevic N, Peculis B. Identification of a U8 snoRNA-specific binding protein. J. Biol. Chem. 1999;274:35914–35920. doi: 10.1074/jbc.274.50.35914. [DOI] [PubMed] [Google Scholar]
- 52.Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 53.Brosnan CA, Voinnet O. The long and the short of non-coding RNAs. Curr. Opin. Cell Biol. 2009;21:416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.