Abstract
It is generally accepted that functionally important RNA structure is more conserved than sequence due to compensatory mutations that may alter the sequence without disrupting the structure. For small RNA molecules sequence–structure relationships are relatively well understood. However, structural bioinformatics of mRNAs is still in its infancy due to a virtual absence of experimental data. This report presents the first quantitative assessment of sequence–structure divergence in the coding regions of mRNA molecules based on recently published transcriptome-wide experimental determination of their base paring patterns. Structural resemblance in paralogous mRNA pairs quickly drops as sequence identity decreases from 100% to 85–90%. Structures of mRNAs sharing sequence identity below roughly 85% are essentially uncorrelated. This outcome is in dramatic contrast to small functional non-coding RNAs where sequence and structure divergence are correlated at very low levels of sequence similarity. The fact that very similar mRNA sequences can have vastly different secondary structures may imply that the particular global shape of base paired elements in coding regions does not play a major role in modulating gene expression and translation efficiency. Apparently, the need to maintain stable three-dimensional structures of encoded proteins places a much higher evolutionary pressure on mRNA sequences than on their RNA structures.
INTRODUCTION
Secondary structure elements both in the untranslated (UTR) and coding (CDS) regions of mRNAs have been implicated in a variety of regulatory functions (1). For example, riboswitches modulate gene expression through conformational changes in response to various stimuli (2). In addition, translation initiation, elongation, termination and translation efficiency all depend on higher order mRNA secondary structures in non-coding regions (3,4). Coding region hairpins have also been suggested to play a role in the regulation of translation (5). The relationship between RNA structure and gene expression in the coding regions of mRNAs has been demonstrated both computationally and experimentally (6–10). In particular, reduced mRNA stability near the start codon has been observed in a wide range of species, probably as a mechanism to facilitate ribosome binding or start codon recognition by initiator tRNA (11). Computational studies show that native mRNA sequences have lower folding energies and hence more stable structure than codon-randomized ones (5). The three mRNA functional domains—5′-UTR, CDS and 3′-UTR—form largely independent folding units, with base pairing across domain borders being rare (12). Evolutionary conserved local secondary structures have been identified in the CDS regions (13,14) and shown to be functional (15).
There is a selective pressure toward maintaining both stable RNA structures of coding regions and the three-dimensional folds of their encoded proteins (16). It has been argued that the redundancy of the genetic code plays an important role in satisfying these selection requirements (12). In general, however, sequence–structure relationships in mRNA-coding regions remain elusive; and, their spatial structure is unknown. While hundreds of atomic resolution structures have been determined for smaller RNA molecules, most notably tRNAs, experimental structures of large RNAs are still rare (17). Until recently, direct experimental determination of mRNA structure has been impossible on a large scale. Furthermore, most insights into the evolutionary constraints acting on them arose from correlating predicted base paring patterns with the effects of site-directed mutagenesis on mRNA expression and degradation, as well as on the expression levels and activity of encoded protein products.
Significant progress has been made in predicting RNA secondary structure from sequence based on free-energy minimization (18), probabilistic models (19) and evolutionary information (20). However, the accuracy of current algorithms is still insufficient to model large molecules, primarily because the number of theoretically possible RNA secondary structures grows exponentially with the length of the sequence (21). Also, the free folding energy of millions of suboptimal structures is very close to the most stable structure. Lowest energy structures may not necessarily reflect folding in vivo (22) due to kinetic processes and protein–RNA interactions. Additionally, it is hard to model pseudoknots and unstructured regions (23).
More accurate prediction of RNA secondary structure can be achieved by using experimental constraints obtained from oligonucleotide data to guide free-energy minimization (24). Moreover, experimental methods have been developed that allow comprehensive monitoring of RNA structure at single nucleotide resolution. One such method, fragmentation sequencing, allows for reconstructing RNA structures by sequencing fragments of single-stranded RNA resulting from nuclease digestion. Another method, known as selective 2′-hydroxyl acylation and primer extension (SHAPE) (25), exploits the sensitivity of selective acetylation of the ribose 2′-hydroxyl position to local nucleotide flexibility, thereby allowing identification of those nucleotides that are conformationally constrained by base pairing. Accurate SHAPE-directed RNA structure determination has been reported for several types of RNA molecules, including Escherichia coli 16S RNA and yeast tRNAasp (26), as well as for the entire HIV-1 genome (27). This latter work highlighted the intricate relationship between RNA sequences and protein structure of the encoded proteins. In particular, it was found that flexible loops in protein structures correspond to highly structured RNA elements, implying a functional role of mRNA structure in the modulation of ribosome processivity at domain boundaries.
In recent work, Kertesz and colleagues (28) reported the first transcriptome-wide experimental analysis of mRNA structures using the novel technology called parallel analysis of RNA structure (PARS). PARS enables the determination of base pairing probabilities at single nucleotide resolution by refolding RNAs in vivo, treating them with structure-specific enzymes and then sequencing the resulting fragments. Structural profiles were obtained for more than 3000 transcripts from the budding yeast Saccharomyces cerevisiae. The work of Kertesz et al. revealed higher degree of structuredness in the mRNA-coding regions compared with the 3′- and 5′-untranslated regions, implying a functional role of RNA structure in coding regions in regulating gene expression. The global data set of PARS profiles represents a true treasure trove for investigating sequence–structure and structure–function relationships in mRNAs.
This report provides the first comprehensive analysis of sequence–structure relationships in the coding regions of yeast mRNAs based on base pairing propensities measured by the PARS technology. It was found that PARS profiles of paralogous mRNAs show very strong, essentially linear, correlation sequence for identity levels upwards of 85–90%. Yet, pairs of more distantly related yeast transcripts secondary structure appear to be unrelated. Interestingly, predicted secondary structures of yeast paralogs display a similar behavior with respect to sequence identity; and, there is a significant correlation between experimental and theoretical structures, as noted previously (28). Theoretical structures of orthologous mRNA pairs from yeast and Candida glabrata are also uncorrelated for low sequence identity levels while for highly similar sequences no conclusion could be made due to lack of data.
MATERIALS AND METHODS
Experimental data on yeast mRNA secondary structure
Secondary structure profiles of 3000 transcripts from the budding yeast S. cerevisiae have recently been determined using a novel experimental strategy called PARS (28). For each individual nucleotide position of mRNAs, a PARS score reflects its likelihood to be in a double-stranded conformation. PARS scores for yeast transcripts were downloaded from http://genie.weizmann.ac.il/pubs/PARS10. 5′- and 3′-UTR regions were identified by sequence comparison with yeast amino acid sequences, and then excluded from consideration. In the following, a vector of PARS scores for a given transcript is referred to as its experimental structure.
Yeast paralogs
Data on paralogous yeast proteins were kindly provided by Martin Münsterkötter and Ulrich Güldner from the fungal genomics group at the Institute for Bioinformatics and Systems Biology (German Research Center for Environmental Health, Munich). A list of protein pairs sharing significant similarity (identity at the amino acid level >50%) was extracted from the SIMAP database (29). Additionally, the putative paralogs were required to have not >10% difference in sequence length. In total, 243 paralog pairs involving 409 different yeast genes satisfied these conditions.
Amino acid sequences of paralogous yeast proteins were globally aligned using the ggsearch program from the FASTA software suite (30). Amino acid sequence alignments were subsequently converted into mRNA sequence alignments; and, the percent identity between each pair of coding regions was calculated by dividing the number of identical nucleotides by the length of the alignment.
Orthologs from C. glabrata
Sequence data for C. glabrata were downloaded from the PEDANT genome database (31). A list of orthologous protein pairs between S. cerevisiae and C. glabrata was extracted from the eggNOG database (32). In total, we obtained 2327 ortholog pairs. The alignment procedure was the same as for paralogs, see above.
PARS score distances between yeast paralogs
To assess global structural similarity between pairs of aligned mRNA sequences, root mean square deviations (RMSDs) between vectors of PARS scores were calculated for all alignment positions that did not contain gaps. Additionally, for each transcript pair, profiles of local structural similarity were obtained by calculating RMSDs between PARS scores in non-gapped alignment positions within a sliding window of varying length, typically between 100 and 1000 nt.
Prediction of mRNA secondary structures
For each nucleotide position of transcript sequences, the theoretical probability to be in double-stranded conformation was calculated using the RNAfold method from the Vienna RNA package (33). As done similarly for experimental PARS scores (see above), RNAfold probability values were used to calculate global and local measures of structural similarity between aligned coding regions of mRNAs based on RMSD. For brevity, a vector of predicted probabilities of RNA bases in double-stranded conformation for a given transcript is further referred to as its theoretical structure.
Data availability
All sequence alignments together with experimentally determined and predicted structures are available in Supplementary Data.
RESULTS
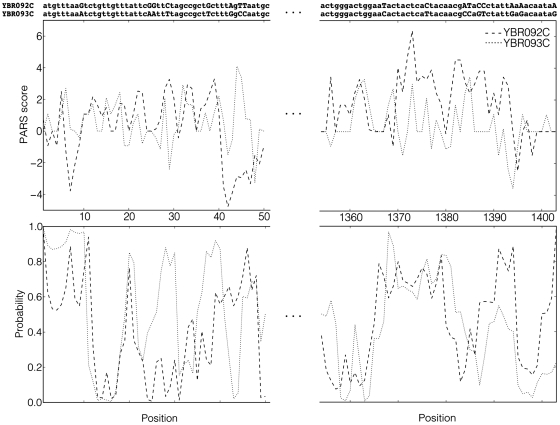
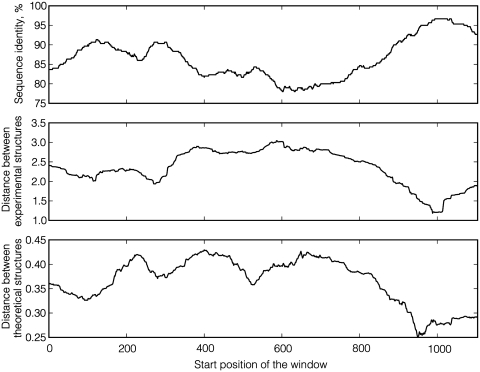
By illustrating the data used in this study on a concrete example, the research results can be readily presented. Two yeast mRNA sequences, YBR092C and YBR093C, share 86.5% sequence identity, and their partial alignment is depicted in the top part of Figure 1. The position-dependent PARS scores for both sequences are shown in the middle part of Figure 1. Both graphs display a rather high degree or correlation, albeit not perfect. In the bottom part of Figure 1, theoretical structures (probabilities for individual bases to be paired) are drawn along the sequence. Figure 2 shows how distances between experimental and theoretical structures of YBR092C and YBR093C vary along the mRNA sequence dependent on sequence identity in a local sequence window. As expected, highly similar regions generally correspond to more similar structures.
Figure 1.
Sequence alignment, experimental and theoretical structures of the first and last 50 nt for the pair of yeast mRNA sequences YBR092C (dashed lines) and YBR093C (dotted lines).
Figure 2.
The profile of local structural similarity versus local sequence identity for the pair of yeast mRNA sequences YBR092C and YBR093C. The length of the sliding window is 300. The global sequence identity between these two sequences is 86.5%.
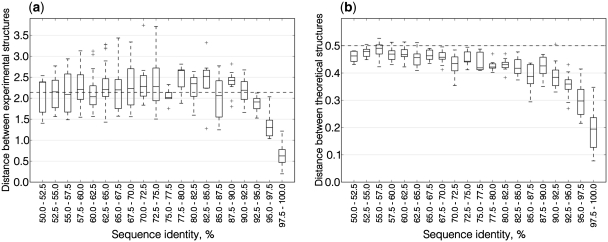
Calculations exemplified in Figures 1 and 2 were performed for all pairs of paralogous mRNA sequences in our data set. Table 1 summarizes pair-wise correlations between the three evolutionary measures considered in this work for different ranges of sequence identities. Figure 3a shows how the difference between experimental structures depends on sequence similarity. PARS scores appear to be entirely uncorrelated for identity levels of up to ∼85–90%. In this sequence identity range, the median RMSD between PARS score vectors does not differ from the median calculated for randomly selected mRNA pairs (dashed horizontal line in Figure 3a). For sequence identity levels over 85–90%, the distance between experimental structures shows essentially a linear dependence from sequence similarity (Supplementary Figure S1).
Table 1.
Correlation coefficients and P-values for different ranges of sequence identity
| Sequence identity range (%) | Sequence identity versus RMSD between experimental structures |
Sequence identity versus RMSD between theoretical structures |
RMSD between experimental structures versus RMSD between theoretical structures |
|||
|---|---|---|---|---|---|---|
| Correlation coefficient | P-value | Correlation coefficient | P-value | Correlation coefficient | P-value | |
| 50–60 | 0.12 | 0.39 | −0.07 | 0.62 | 0.14 | 0.31 |
| 60–70 | 0.14 | 0.22 | −0.10 | 0.37 | −0.02 | 0.87 |
| 70–80 | −0.08 | 0.67 | −0.08 | 0.67 | −0.24 | 0.21 |
| 80–90 | 0.01 | 0.91 | −0.14 | 0.40 | 0.04 | 0.79 |
| 90–100 | −0.92 | 5.66e−27 | −0.75 | 1.24e−12 | 0.69 | 3.56e−10 |
Figure 3.
Boxplots of distances between structures of aligned paralogous mRNAs in different ranges of sequence similarity. Each box corresponds to the range of similarity 2.5%. The box extends from the lower to the upper quartile values, with a horizontal line at the median value. Whiskers demonstrate the entire range of the data. Crosses show outliers. (a) Distances between experimental structures. The average level of PARS score distances for alignments of random sequence pairs is 2.14 (dashed line). (b) Distances between theoretical structures. The average level of probability distance for alignments of random sequence pairs is 0.5 (dashed line).
Upon conducting the same experiment with pairs of theoretical structures of yeast mRNAs, it was found that the distance between the structures also begins to depend on sequence similarity upward of roughly 85–90% identity (Figure 3b). For pairs with identity between sequences within the range from 97.5% to 100%, the median distance between theoretical structures constitutes 38% of the random level. Yet, for experimental structures, it is lower at 29%. The link between sequence and structure is thus stronger when experimental structures are considered. The distance between theoretical structures also shows a linear dependence from sequence similarity for sequence identity levels over 85–90% (Supplementary Figure S2).
Therefore, what is the significance of the sequence–structure dependence shown in Figure 3; and, how would it appear for codon-randomized mRNA sequences? Since experimental PARS scores are not available for randomly generated sequences, this issue could only be assessed for theoretical structures. For each pair of paralogs, one sequence was kept unchanged. In the second mRNA, however, mutations were randomly distributed along the sequence, keeping the encoded amino acid sequence, the codon usage and the total number of mutations between the paralogs unchanged. Overall, the divergence of structures between codon-randomized paralogs displays virtually the same dependence on sequence similarity as for native sequences (Supplementary Figure S3).
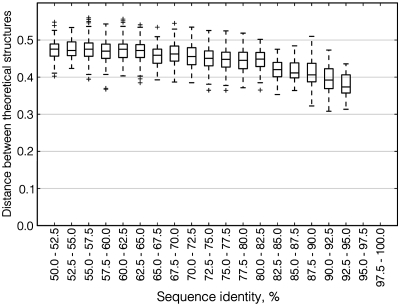
We also compared predicted structures between orthologous mRNAs from S. cerevisiae and the pathogenic yeast C. glabrata (Figure 4). Although C. glabrata is the most closely related organism to S. cerevisiae with a completely sequenced genome (34), no pair of orthologous mRNAs between these two organisms shares sequence identity >95% and thus no conclusion about structure divergence for very similar sequences could be made. However, for lower identity levels theoretical structures of orthologs are uncorrelated and thus behave the same way as paralogous structures.
Figure 4.
Boxplot of distances between theoretical structures of aligned orthologous mRNAs in different ranges of sequence similarity. Notation as in Figure 3.
DISCUSSION
In some sense, the current situation in RNA bioinformatics is reminiscent of the early days of structural bioinformatics of proteins, when the availability of a sufficiently large data set of X-ray structures allowed for the first comprehensive analysis of the relation between the divergence of sequence and structure in proteins (35). Until recently, studies of the evolutionary conservation of RNA structures were based on in silico predictions and largely limited to non-coding RNA. In the first large-scale study, Schudoma et al. (36) determined that in short RNA loops with known three-dimensional structures sequence identity >75% implies significant structural similarity. The most comprehensive investigation of sequence–structure relationships in RNA molecules to date is based on all-against-all pair-wise structural comparison of non-coding RNAs (tRNAs, rRNAs, riboswitches and riboswitches) with known spatial architectures (37). Assessment of evolutionary divergence revealed that the correlation between sequence and secondary structure conservation is highly significant for sequence identity levels in the range between just a few percentage points up to roughly 60% where this relationship saturates. Further increase of sequence similarity (60–100%) does not lead to an appreciable growth of secondary structure similarity. None of the studies mentioned above considered mRNAs because no mRNA structures are currently known at atomic resolution.
The principal finding of this research is that the correlation between sequence and structure in the coding regions of yeast mRNAs is much weaker than in small non-coding RNAs. Up to ∼85–90% sequence identity, the similarity of both experimental and theoretical base pairing propensities between paralogous yeast mRNAs is at random level; while, for more similar sequence pairs, sequence and structure are strongly correlated. This may imply that mRNAs do not experience a strong selective pressure to preserve a certain degree of structuredness. The fact that codon-randomized sequences display a similar behavior also indicates that there is no appreciable evolutionary pressure to preserve a particular RNA structure as long as the encoded protein remains unchanged. Taken together, these results underscore a high degree of evolutionary neutrality in yeast mRNA molecules, both at the level of primary (third codon position) and secondary (extent of base paring) structure.
On one hand, our findings are in strong contrast to many non-coding RNAs and cis-acting regulatory elements of mRNAs whose biological function is primarily mediated by their spatial architecture (38) stabilized by tertiary interactions, modified bases and interactions with proteins and small ligands. On the other hand, sequence–structure relationships observed in this work are compatible with the notion that, in general, RNA molecules do not have a single global structure. Instead, they exist as a highly dynamic ensemble of alternative conformations (39,40) that are often capable of performing different functions (41). The extent of base pairing may play a role in the regulation of pre-mRNA splicing, translation and mRNA degradation. Both experimentally determined PARS scores and computationally derived partition functions analyzed in this work are statistical measures that reflect the propensity of each nucleotide to form a base pair across a large number of metastable structures.
This analysis has several important limitations. First, PARS probes RNA structures in vitro rather than in the living cell and may not always reproduce functional RNA structures (42). Second, even if the base paring information obtained by the PARS technology were perfectly correct, it still merely represents a one-dimensional profile of structural propensities, a far cry from knowing the actual RNA secondary structure, let alone spatial architecture, for each individual molecule at any moment of time. Third, the findings do not rule out much stronger sequence–structure correlations in certain local structural elements of coding regions, such as reprogrammed genetic-decoding signals (43) or mRNA localization signals. We also cannot rule out the possibility that the degree of mRNA structuredness does have an important functional role in spite of quick erosion of structural similarity between paralogs with diminishing sequence similarity, and that this erosion reflects functional differentiation. However, we consider such explanation unlikely because the same behavior is observed between orthologous mRNAs. Finally, only a small subset of the PARS data constituted by pairs of sequence similar yeast mRNAs (paralogs) was explored. As a next step, it will be exciting to conduct comparative analyses of mRNA structuromes [the term coined by Westhof and Romby (44)], focusing on orthologous sequences from multiple organisms and taking into account important genomic variables, such as expression level and evolutionary rate. Given the current pace of high-throughput RNA analysis technologies there is no doubt that such data will become available in the near future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary figures S1–S3.
FUNDING
The DFG International Research Training Group ‘Regulation and Evolution of Cellular Systems’ (GRK 1563) and by the Russian Foundation for Basic Research (RFBR 09-04-92742). Funding for open access charge: German Research Center for Environmental Health, Munich.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dmitry Ivankov and Natalya Bogatyreva for helpful discussions and Janusz Bujnicki for illuminating comments on the article.
REFERENCES
- 1.Bevilacqua PC, Blose JM. Structures, kinetics, thermodynamics, and biological functions of RNA hairpins. Annu. Rev. Phys. Chem. 2008;59:79–103. doi: 10.1146/annurev.physchem.59.032607.093743. [DOI] [PubMed] [Google Scholar]
- 2.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray NK, Hentze MW. Regulation of protein synthesis by mRNA structure. Mol. Biol. Rep. 1994;19:195–200. doi: 10.1007/BF00986961. [DOI] [PubMed] [Google Scholar]
- 4.Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 5.Katz L, Burge CB. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 2003;13:2042–2051. doi: 10.1101/gr.1257503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum. Mol. Genet. 2003;12:205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 8.Ilyinskii PO, Schmidt T, Lukashev D, Meriin AB, Thoidis G, Frishman D, Shneider AM. Importance of mRNA secondary structural elements for the expression of influenza virus genes. OMICS. 2009;13:421–430. doi: 10.1089/omi.2009.0036. [DOI] [PubMed] [Google Scholar]
- 9.Carlini DB, Chen Y, Stephan W. The relationship between third-codon position nucleotide content, codon bias, mRNA secondary structure and gene expression in the drosophilid alcohol dehydrogenase genes Adh and Adhr. Genetics. 2001;159:623–633. doi: 10.1093/genetics/159.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, Maixner W, Diatchenko L. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 11.Gu W, Zhou T, Wilke CO. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput. Biol. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shabalina SA, Ogurtsov AY, Spiridonov NA. A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res. 2006;34:2428–2437. doi: 10.1093/nar/gkl287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer IM, Miklós I. Statistical evidence for conserved, local secondary structure in the coding regions of eukaryotic mRNAs and pre-mRNAs. Nucleic Acids Res. 2005;33:6338–6348. doi: 10.1093/nar/gki923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steigele S, Huber W, Stocsits C, Stadler PF, Nieselt K. Comparative analysis of structured RNAs in S. cerevisiae indicates a multitude of different functions. BMC Biol. 2007;5:25. doi: 10.1186/1741-7007-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol. Cell. Biol. 2005;25:4752–4766. doi: 10.1128/MCB.25.11.4752-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White HB, III, Laux BE, Dennis D. Messenger RNA structure: compatibility of hairpin loops with protein sequence. Science. 1972;175:1264–1266. doi: 10.1126/science.175.4027.1264. [DOI] [PubMed] [Google Scholar]
- 17.Holbrook SR. Structural principles from large RNAs. Annu. Rev. Biophys. 2008;37:445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]
- 18.Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Dowell RD, Eddy SR. Efficient pairwise RNA structure prediction and alignment using sequence alignment constraints. BMC Bioinformatics. 2006;7:400. doi: 10.1186/1471-2105-7-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernhart SH, Hofacker IL. From consensus structure prediction to RNA gene finding. Brief. Funct. Genomic Proteomic. 2009;8:461–471. doi: 10.1093/bfgp/elp043. [DOI] [PubMed] [Google Scholar]
- 21.Meyer IM, Miklós I. SimulFold: simultaneously inferring RNA structures including pseudoknots, alignments, and trees using a Bayesian MCMC framework. PLoS Comput. Biol. 2007;3:e149. doi: 10.1371/journal.pcbi.0030149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahen EM, Watson PY, Cottrell JW, Fedor MJ. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol. 2010;8:e1000307. doi: 10.1371/journal.pbio.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reeder J, Giegerich R. Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics. BMC Bioinformatics. 2004;5:104. doi: 10.1186/1471-2105-5-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan S, Mathews DH, Turner DH. Interpreting oligonucleotide microarray data to determine RNA secondary structure: application to the 3′ end of Bombyx mori R2 RNA. Biochemistry. 2006;45:9819–9832. doi: 10.1021/bi052618x. [DOI] [PubMed] [Google Scholar]
- 25.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 26.Low JT, Weeks KM. SHAPE-directed RNA secondary structure prediction. Methods. 2010;52:150–158. doi: 10.1016/j.ymeth.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rattei T, Tischler P, Götz S, Jehl MA, Hoser J, Arnold R, Conesa A, Mewes HW. SIMAP–a comprehensive database of pre-calculated protein sequence similarities, domains, annotations and clusters. Nucleic Acids Res. 2010;38:D223–D226. doi: 10.1093/nar/gkp949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson WR. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 31.Walter MC, Rattei T, Arnold R, Güldener U, Münsterkötter M, Nenova K, Kastenmüller G, Tischler P, Wölling A, Volz A, et al. PEDANT covers all complete RefSeq genomes. Nucleic Acids Res. 2009;37:D408–D411. doi: 10.1093/nar/gkn749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller J, Szklarczyk D, Julien P, Letunic I, Roth A, Kuhn M, Powell S, von Mering C, Doerks T, Jensen LJ, et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 2010;38:D190–D195. doi: 10.1093/nar/gkp951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dujon B. Yeast evolutionary genomics. Nat. Rev. Genet. 2010;11:512–524. doi: 10.1038/nrg2811. [DOI] [PubMed] [Google Scholar]
- 35.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schudoma C, May P, Nikiforova V, Walther D. Sequence-structure relationships in RNA loops: establishing the basis for loop homology modeling. Nucleic Acids Res. 2010;38:970–980. doi: 10.1093/nar/gkp1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capriotti E, Marti-Renom MA. Quantifying the relationship between sequence and three-dimensional structure conservation in RNA. BMC Bioinformatics. 2010;11:322. doi: 10.1186/1471-2105-11-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gruber AR, Bernhart SH, Hofacker IL, Washietl S. Strategies for measuring evolutionary conservation of RNA secondary structures. BMC Bioinformatics. 2008;9:122. doi: 10.1186/1471-2105-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mironov AA, Dyakonova LP, Kister AE. A kinetic approach to the prediction of RNA secondary structures. J. Biomol. Struct. Dyn. 1985;2:953–962. doi: 10.1080/07391102.1985.10507611. [DOI] [PubMed] [Google Scholar]
- 40.Danilova LV, Pervouchine DD, Favorov AV, Mironov AA. RNAKinetics: a web server that models secondary structure kinetics of an elongating RNA. J. Bioinform. Comput. Biol. 2006;4:589–596. doi: 10.1142/s0219720006001904. [DOI] [PubMed] [Google Scholar]
- 41.Zhao P, Zhang WB, Chen SJ. Predicting secondary structural folding kinetics for nucleic acids. Biophys. J. 2010;98:1617–1625. doi: 10.1016/j.bpj.2009.12.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mauger DM, Weeks KM. Toward global RNA structure analysis. Nat. Biotechnol. 2010;28:1178–1179. doi: 10.1038/nbt1110-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Namy O, Rousset JP, Napthine S, Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 44.Westhof E, Romby P. The RNA structurome: high-throughput probing. Nat. Methods. 2010;7:965–967. doi: 10.1038/nmeth1210-965. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence alignments together with experimentally determined and predicted structures are available in Supplementary Data.