Abstract
Nonsense-mediated mRNA decay (NMD) is a surveillance mechanism that detects and degrades mRNAs containing premature termination codons (PTCs). SMG-1-mediated Upf1 phosphorylation takes place in the decay inducing complex (DECID), which contains a ribosome, release factors, Upf1, SMG-1, an exon junction complex (EJC) and a PTC-mRNA. However, the significance and the consequence of Upf1 phosphorylation remain to be clarified. Here, we demonstrate that SMG-6 binds to a newly identified phosphorylation site in Upf1 at N-terminal threonine 28, whereas the SMG-5:SMG-7 complex binds to phosphorylated serine 1096 of Upf1. In addition, the binding of the SMG-5:SMG-7 complex to Upf1 resulted in the dissociation of the ribosome and release factors from the DECID complex. Importantly, the simultaneous binding of both the SMG-5:SMG-7 complex and SMG-6 to phospho-Upf1 are required for both NMD and Upf1 dissociation from mRNA. Thus, the SMG-1-mediated phosphorylation of Upf1 creates a binding platforms for the SMG-5:SMG-7 complex and for SMG-6, and triggers sequential remodeling of the mRNA surveillance complex for NMD induction and recycling of the ribosome, release factors and NMD factors.
INTRODUCTION
Eukaryotes have a conserved RNA surveillance mechanism to help maintain correct gene expression. Nonsense-mediated mRNA decay (NMD) is an mRNA surveillance mechanism that detects and degrades mRNAs containing premature termination codons (PTCs) to eliminate potentially harmful C-terminally truncated proteins (1–3). NMD also targets many physiological mRNAs to regulate abundance, including mRNAs encoding selenocysteine-containing proteins and mRNA-like non-coding RNAs (4–6). If C-terminally truncated proteins retain some of their function and/or PTC-read through produces functional proteins, NMD suppression leads to the phenotypic rescue of certain PTC-related mutations (7–9). In addition, NMD suppression can augment un-natural polypeptides, which are putative tumor-specific antigens encoded by frame-shift mutations on PTC-mRNAs (10). Thus, clarification of the mechanism of NMD is critical for the development of pharmacological reagents for genetic diseases and cancer (11,12).
The current model of mammalian PTC recognition posit a splicing-dependent deposition of the exon junction complex (EJC) components, 20–24 nt upstream of an exon–exon junction (13) and deposition of nine UPF/smg gene products, which are evolutionally conserved trans-acting factors of NMD (1,14,15). In the initial round of translation (16,17), if a translation termination codon is located upstream of an EJC, the SMG-1 kinase complex (SMG1C; SMG-1:SMG-8:SMG-9), Upf1 RNA helicase and translation termination factors form the SMG-1:Upf1:eRF1:eRF3 complex (SURF), which associates with the ribosome on the messenger ribonucleoprotein (mRNP) (18–21). The association of ribosome:SURF with the EJC forms the decay inducing complex (DECID), which can distinguish a PTC from a normal termination codon. The DECID induces SMG-1-mediated Upf1 phosphorylation, a rate-limiting step of NMD (19–25). Recognition of PTCs, independently of the downstream splicing junction, has also been demonstrated in mammals, and the molecular mechanism for this process is under debate (26–29). In contrast to the events before PTC recognition, those after PTC recognition are not well understood. For instance, whereas a ribosome is present in the DECID complex during PTC discrimination and PTC mRNA decay is expected to occur after dissociation of the mRNA from ribosomes, the timing and mechanism of ribosome dissociation from mRNA is unclear.
As noted above, a critical consequence of PTC recognition is the phosphorylation of Upf1 by SMG-1 (22,25). SMG-1 phosphorylates Upf1 in vitro at several serine/threonine–glutamine (S/TQ) motifs in the N- and C-terminal regions (22). Among them, S1078, S1096 and S1116 are phosphorylated in vivo in mammals (22,30,31). However, the functional importance of these phosphorylation sites remains to be clarified. In addition to phosphorylation, dephosphorylation is also necessary for NMD (30,32,33). SMG-5, SMG-6 and SMG-7 are involved in the dephosphorylation of Upf1, probably through the recruitment of protein phosphatase 2A (PP2A) (19,30,32–35). SMG-5, SMG-6 and SMG-7 are evolutionally conserved related proteins, but each is required for NMD (32,36). The majority of SMG-5 and SMG-7 forms a complex (the SMG-5:SMG-7 complex) that preferentially binds to phosphorylated Upf1 (phospho-Upf1) in vivo, whereas phospho-independent binding of the SMG-5:SMG-7 complex with the N-terminal region of Upf1 is observed during NMD (30). The binding of the SMG-5:SMG-7 complex to phospho-Upf1 induces Upf2 dissociation from Upf1, and this step is involved in NMD (30). SMG-5 and SMG-7 share the 14-3-3-like domain. The SMG-7 14-3-3-like domain can directly bind in vitro to a Upf1 phosphopeptide containing phospho-S1078 (37). However, the binding site of the SMG-5:SMG-7 complex on Upf1 in vivo remain to be clarified. SMG-7 is considered as mRNA decay mediator since it is tethering at either 3′- or 5′-UTR of mRNA induce Dcp2 (decapping enzyme) and Xrn1 (5′-3′-exonuclease) dependent mRNA decay (38). SMG-6 also shares the 14-3-3-like domain, which has been proposed to compete with the SMG-5:SMG-7 complex for binding to phospho-Upf1 (2,3), but association of SMG-6 with phospho-Upf1 has not been determined (39). SMG-5 and SMG-6 have a C-terminal PilT N-terminus (PIN) domain. The PIN domain of SMG-6 has endonuclease activity in vitro and catalytically inactive SMG-6 fails to support NMD in mammalian cells (40,41). While SMG-5, SMG-6 and SMG-7 are required for NMD, their mechanisms of action remain to be clarified.
Here, we demonstrate that the SMG-1-mediated phosphorylation of T28 and S1096 of Upf1 create binding platforms for SMG-6 and the SMG-5:SMG-7 complex, respectively. SMG-6 associates in vivo with phosphorylated Upf1 through its 14-3-3-like domain. We also show that the phospho-specific binding of SMG-6 and the SMG-5:SMG-7 complex to Upf1 is required for NMD. Furthermore, we provide evidence supporting the involvement of the SMG-5:SMG-7 complex in the dissociation of the ribosome from DECID after Upf1 phosphorylation. In addition, we suggest that the phospho-specific binding of SMG-6 is required for Upf1 dissociation from mRNA.
MATERIALS AND METHODS
Plasmids, antibodies and siRNAs
Expression vectors for wild-type Flag-HA-streptavidin binding peptide (SBP)-SMG-6, SMG-6 mutants (-mt1433, -mtPIN, -dCT), wild-type Upf1 and Upf1-mutants [-dCT (amino acids 6–1027), -dNCT (amino acids 64–1027), -S1078A, -S1096A, -S1116A, -T28A, -2SA (SS1078/1096AA), -4SA (SSSS1073/1078/1096/1116AAAA), -5T/SA (TSSSS28/1073/1078/1096/1116AAAAA)] were constructed in the mammalian expression vectors pcDNA5/FRT/TO/Flag-HA-SBP, pEF_Flag-HA-SBP or pSR-HA, following standard procedures. The wild-type Flag-HA-SBP-SMG-6 and SMG-6 mutants were mutated at coding sequence nucleotides to confer siRNA SMG-6 resistance by site-directed mutagenesis. HA-SMG-5, HA-SMG-5dCT and HA-Upf1-4SA plasmids were previously described (19,22,30).
The following siRNA target sequences were used: SMG-5, GAAGGAAATTGGTTGATAC; SMG-6, GGGTCACAGTGCTGAAGTA; SMG-7, CAGCACAGTCTACAAGCCA; non-silencing (NS), All Star Negative Control siRNA (Qiagen).
Anti-eIF4A3, anti-SMG-5 and anti-SMG-6 antibodies were generated against recombinant human eIF4A3 (amino acids 1–48), SMG-5 (amino acids 416–541) or SMG-6 (amino acids 58–181) peptides fused to glutathione S-transferase (GST) (anti-eIF4A3 and -SMG-5) or maltose-binding protein (MBP) (anti-SMG-6), respectively. Affinity purification of the antibody was performed following standard procedures. An anti-P-T28-Upf1 antibody was generated as described previously (30) using a keyhole limpet hemocyanin (KLH)-conjugated phosphopeptide [C-LGAD(P)TQGSEF, T28-P] as antigen. The anti-SMG-1, Upf1 (clone 5C3), Upf2, SMG-7, SMG-9 and P-1078/1096-Upf1 (clone 7H1) antibodies were generated as described previously (19,20,22,30). Antibodies to eRF1 (Abcam), rpL7a (Cell signaling technology), rpS16 (Abcam), Magoh (Abcam), Y14 (Abcam), eIF4E (Santa Cruz), PABPC1 (Abcam), PABPC4 (Bethyl), CBP80 (Bethyl), β-actin (Sigma), GAPDH (Abcam) and HA (3F10) (Roche) were used.
Transfection with plasmids and siRNAs
HeLa TetOff cells (Clontech) and HEK 293 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin. Plasmid transfections were performed in six-well plates, 10 or 15 cm dishes using Polyfect (Qiagen), Transfectin (BioRad) or LipofectaminLTX (Invitrogen) according to the manufacturer's recommendations. For affinity purification of SBP-tagged proteins, cells were harvested 48 h after transfection. siRNA transfections were performed in six-well plates or in 15 cm dishes, using siLentFect (BIORAD) or LipofectaminRNAiMAX (Invitrogen) according to the manufacturer's protocol, and cells were harvested 46–72 h later.
Immunoprecipitation, SBP purification and western blot analysis
HeLa TetOff cells were transfected using Lipofectamine LTX (Invitrogen) or Polyfect (Qiagen) and incubated for 30 min at 4°C in a lysis buffer containing 50 mM Tris–HCl at pH 7.4, 50 mM NaCl, 0.05% Tween-20, 100 nM okadaic acid (Calbiochem), phosphatase inhibitor cocktail, protease inhibitor cocktail (Roche) and cycloheximide (100 μg/ml, Sigma). The pre-cleared lysates were added to 200 μg/ml RNaseA (Qiagen), and then incubated with appropriate antibodies for 1 h with gentle rotation. Subsequently, lysates with antibodies were incubated with 30 μl of protein G-Sepharose (GE Biotech) for an additional 1 h at 4°C with gentle rotation. To immunoprecipitate HA-tagged proteins, 25 μl of anti-HA-affinity matrix (Roche), pre-adsorbed with antibody, was used instead of protein G-sepharose. For SBP purification, 30 μl of streptavidin-coated Sepharose (GE Biotech) was used and lyastes were incubated for 2 h at 4°C. The immunocomplexes (or SBP–streptavidin complex) were washed with washing buffer containing 50 mM Tris–HCl at pH 7.4, 50 mM NaCl, 0.05% Tween-20 and 100 nM okadaic acid, boiled in 100 μl of standard 1× SDS sample buffer and then analyzed by western blotting. In experiments, where HA-affinity matrix, Upf1-monoclonal antibody or streptavidin-coated sepharose was used, proteins recovered on the matrix were eluted by incubation at 37°C for 15 min (for HA-affinity matrix and Upf1-monoclonal antibody) or at 4°C for 30 min (for streptavidin-coated sepharose) in the presence of 50 μl washing buffer containing 1 mg/ml HA-peptide, 1 mg/ml Upf1 peptide or 2 mM biotin, boiled with 50 μl of 2 × sample buffer and then analyzed by western blotting. Except for Figure 3, anti-Upf1, P-T28 and P-S1078/1096 antibodies were used as probes after 8% polyacrylamide gel electrophoresis. In Figure 3, samples were separated using a 8–16% gradient polyacrylamide gel, which did not provide the resolution required for the Upf1 mobility shift.
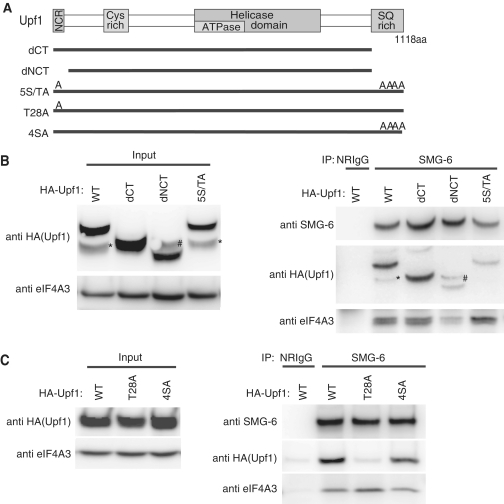
Figure 3.
Threonine residue 28 of Upf1 is necessary for the binding of SMG-6. (A) Schematic representation of Upf1 mutants. The N-terminal conserved region (NCR), cystein rich region, helicase domains and SQ-rich region of Upf1 are depicted as gray boxes. In Upf1-dCT, the C-terminal 90 amino acids are deleted. In Upf1-dNCT, the N-terminal 63 amino acids and the C-terminal 90 amino acids are deleted. In Upf1-5S/TA, T28, S1073, S1078, S1096 and S1116 are replaced by A. In Upf1-T28A, T28 is substituted for A. In Upf1-4SA, S1073, S1078, S1096 and S1116 are replaced by A. (B and C) HeLa TetOff cells were transfected with the plasmids shown above. The cell extracts were immunoprecipitated with anti-SMG-6 antibody or with normal rabbit IgG (NRIgG) in the presence of RNaseA. Immunoprecipitated fractions were analyzed by western blotting with the indicated antibody probes. ‘Asterisks’ indicates unexpected degradation product of HA-tagged-Upf1. ‘Hash’ indicates an uncharacterized band detected by the anti-HA antibody in Upf1-dNCT mutant-expressed extract.
All western blot experiments were detected with the ECL western blotting detection kit (GE Biotech) or with Lumi-Light western blot substrate (Roche) and quantified with a LuminoImager, LAS-3000 and Science Lab 2001 Image Gauge software (Fuji Photo Film). All experiments were performed at least three times, and typical results are shown.
NMD analysis in mammalian cells
NMD analysis was performed in essentially the same manner as described elsewhere (42), except that HeLa TetOff cells (1.8 × 106) were transfected with siRNAs and plasmid vectors, which are indicated in figures, in combination with 1.5 μg of pTRE-BGG-WT or pTRE-BGG-PTC. Briefly, 44 h after transfection, doxycyclin was added at time zero to inhibit de novo transcription. Cells were then harvested at the indicated time points, and total RNAs were analyzed by northern blotting using either a β-globin or GAPDH (control) probe. The quantities of BGG mRNA were normalized to GAPDH signals and then graphically plotted. Values in figures represented the mean ± SE for three to four independent experiments.
RESULTS
SMG-6 binds to phosphorylated Upf1 through its 14-3-3-like domain
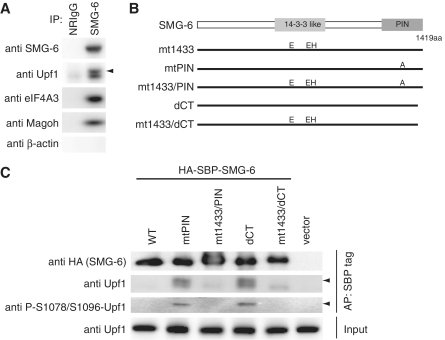
To investigate the processes of NMD subsequent to Upf1 phosphorylation, we analyzed whether endogenous SMG-6, involved in the dephosphorylation of Upf1, binds to phospho-Upf1. Immunoprecipitation experiments using HeLa TetOff cell lysates and an anti-SMG-6 antibody revealed that SMG-6 coprecipitated with Upf1 in the presence of RNaseA (Figure 1A). Note that the coprecipitated Upf1 produced a doublet band, suggesting the presence of phospho-Upf1 in the SMG-6 complex (Figure 1A). To confirm this notion, we transfected cells with HA-SBP-tagged SMG-6 and affinity purified the SMG-6 complex using streptavidin-conjugated sepharose beads. The binding between endogenous Upf1 and exogenous wild-type SMG-6 is not strong; therefore, we also analyzed the nuclease-inactive SMG-6-mtPIN and -dCT mutants (Figure 1B). These mutants were designed based on observations of the corresponding SMG-5 mutants, SMG-5-D860A and -dCT, which accumulate the phospho-Upf1:SMG-5 complex in cells (30). As shown in Figure 1C, the amount of endogenous Upf1 coprecipitated with SBP-tagged SMG-6 mutants was greater than that with wild-type SMG-6. Intriguingly, both mutants coprecipitated a doublet band of Upf1 similar to the endogenous SMG-6 precipitate. Highly mobility shifted band, indicated by arrowhead, was phosphorylated at least at serine (S) 1078 and 1096 residues, as detected by our phospho-Upf1 antibody, which recognizes phosphorylated S1078/S1096 (anti-P-S1078/S1096-Upf1) (Supplementary Figure S1) (20,30). Note that coprecipitated phospho-Upf1 with SMG-6 is mostly in the mobility shifted (hyper-phosphorylated) form, although the anti-P-S1078/S1096-Upf1 can detect a doublet in total cell extract (Supplementary Figure S1). These results indicate that, in vivo, SMG-6 preferentially binds to hyper-phosphorylated-Upf1, as the SMG-5:SMG-7 complex does (30).
Figure 1.
SMG-6 interacts with phosphorylated Upf1 via its 14-3-3-like domain. (A) Interactions between SMG-6 and components of the NMD machinery. HeLa TetOff cell lysates were immunoprecipitated with anti-SMG-6 antibody in the presence of RNaseA. Immunoprecipitates (IP) were analyzed by western blotting with the indicated antibody probes. (B) Schematic representation of SMG-6 mutants. The 14-3-3-like domain and PIN domain are depicted as gray boxes. In SMG-6-mt1433, two arginines and one tyrosine in the 14-3-3-like domain were replaced by glutamic acid and histidine, respectively (R656E, R737E and Y738H). In SMG-6-mtPIN, an aspartic acid in the PIN domain is replaced by alanine (D1251A). SMG-6-mt1433/PIN has both mt1433 and mtPIN mutations. In SMG-6-dCT, the C-terminal 11 amino acids are deleted. SMG-6-mt1433/dCT has both mt1433 and dCT mutations. (C) HeLa TetOff cell lysates were affinity purified with streptavidin sepharose in the presence of RNaseA. The cell lysates (input) and affinity-purified (AP) fractions were analyzed by western blotting with the indicated antibody probes. Arrow head indicates the hyper-phosphorylated isoform of Upf1 detected by our Upf1 and phospho-Upf1 antibodies.
The 14-3-3-like domain of SMG-7 can bind directly with a phosphorylated amino acid residue (37). The presence of a 14-3-3-like domain in SMG-6 suggests that SMG-6 can also bind to phospho-Upf1 through this domain. To evaluate this possibility, we made a mutant of SMG-6 containing the amino acid changes, R656E, R737E and Y738H, which are all located in the 14-3-3-like domain. Note that the corresponding amino acid residues of the SMG-7 14-3-3-like domain are required for direct contact with the phosphorylated peptide (37). As expected, both SMG-6-mt1433/PIN and -mt1433/dCT double mutants showed greatly reduced coprecipitation of phospho-Upf1 (Figure 1C). These results support the notion that, in vivo, SMG-6 binds to phospho-Upf1 via its 14-3-3-like domain. These mutations abolished the coprecipitation of phospho-Upf1, but non-phospho-Upf1 was still precipitated at similar levels as wild-type, suggesting the existence of additional phospho-independent binding of SMG-6 to Upf1 (Figure 1C).
The coprecipitation of endogenous EJC components, eIF4A3 and Magoh were also observed in endogenous SMG-6 immunoprecipitates (Figure 1A). The N-terminal region of SMG-6 contains two sequences homologous to the EJC binding region of Upf3. To test the involvement of the SMG-6 Upf3-homology region in EJC binding, we tagged SMG-6-mtEJC with SBP and performed affinity purification from cell extracts transfected with SBP-tagged wild-type SMG-6 or SMG-6-mtEJC (Supplementary Figure S2A and S2B). As expected, SMG-6-mtEJC failed to coprecipitate eIF4A3 and Magoh, whereas wild-type SMG-6 did coprecipitate eIF4A3 and Magoh (Supplementary Figure S2C). These results suggest that SMG-6 binds to EJC components via the Upf3-homology region, which is consistent with a recent report (39).
Binding of SMG-6 to phospho-Upf1 is required for NMD and dephosphorylation of Upf1
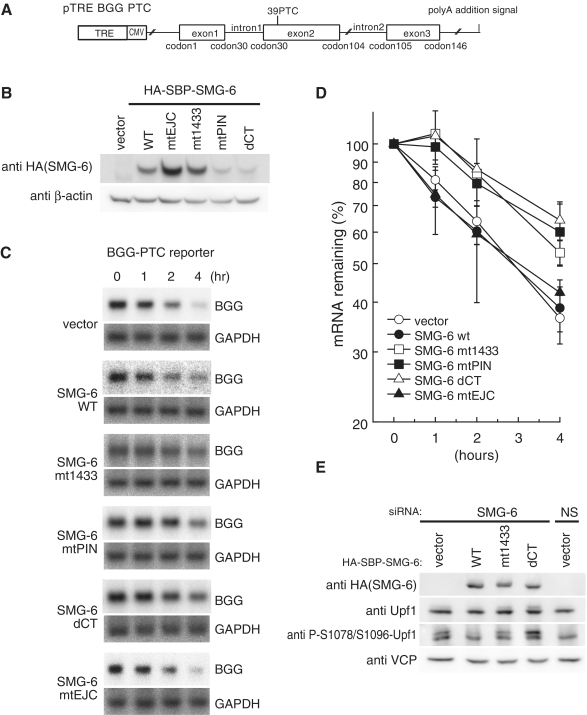
We next investigated the significance of the binding of SMG-6 to phosho-Upf1. For this purpose, we expressed wild-type SMG-6 or SMG-6-mt1433 and a PTC-containing β-globin mRNA reporter in HeLa TetOff cells (Figure 2A) and analyzed the dominant-negative effect of SMG-6-mt1433 on NMD. The cells were treated with doxycycline to repress the transcription of the reporter gene, and mRNAs extracted at indicated time points were subjected to northern blot analysis to estimate the half-life of the β-globin reporter mRNA. Overexpression of wild-type SMG-6 had no effect on the half-life of PTC-containing β-globin mRNA. In contrast, overexpressed SMG-6-mt1433 prolonged the half-life of PTC-containing β-globin mRNA without affecting that of wild-type β-globin mRNA (Figure 2C and D, data not shown). These results support the notion that binding of SMG-6 to phospho-Upf1 through its 14-3-3-like domain is required for PTC-containing β-globin mRNA decay.
Figure 2.
Phospho-dependent binding of SMG-6 to Upf1 is required for Upf1 dephosphorylation and PTC-containing β-globin mRNA decay. (A) Schematic representation of the Tet-inducible human β-globin gene (BGG) reporter construct containing a PTC in exon 2. The open reading frame (ORF) is represented by boxes and introns and UTRs by lines. (B and C) NMD is inhibited by overexpression of SMG-6 mutants. HeLa TetOff cells were transfected with the plasmids shown above (B) or to the left of the panel (C). Total cell lysates were analyzed by western blotting with indicated antibody probes (B). HeLa TetOff cells were co-transfected with the indicated plasmids and the Tet-inducible BGG-PTC plasmid. After the addition of doxycycline to repress the transcription of the reporter plasmids, total RNAs, prepared at the times indicated, were analyzed by northern blotting (C). (D) The quantities of BGG mRNA, normalized to GAPDH signals, were plotted. The means ± SD from three independent experiments are shown. (E) The 14-3-3-like domain of SMG-6 is required for Upf1 dephosphorylation. HeLa TetOff cells were transfected with siRNA targeting SMG-6, together with the indicated siRNA-resistant SMG-6 plasmid vectors. The total cell lysates were analyzed by western blotting with the indicated antibody probes. Valosin-containing protein (VCP) was used as a loading control.
Genetic analysis in Caenorbabditis elegans revealed that SMG-6 is required for the dephosphorylation of Upf1 (32). Therefore, we next investigated the physiological significance of the binding of SMG-6 to phosho-Upf1 for dephosphorylation of Upf1. The transfection of SMG-6-targeted siRNA induced the accumulation of phospho-Upf1, as detected by the anti-P-S1078/S1096-Upf1 antibody in total cell extracts (Figure 2E). This phenotype was rescued by the expression of siRNA-insensitive wild-type SMG-6 but not by the expression of SMG-6-mt1433 (Figure 2E). These results establish that the binding of SMG-6 to phospho-Upf1 through its 14-3-3-like domain is required for the dephosphorylation of Upf1.
We also assessed the effect of SMG-6-mtPIN, -dCT and -mtEJC mutants on NMD. The results indicate that overexpression of mutants, SMG-6-mtPIN and -dCT but not of -mtEJC, suppressed the decay of PTC-containing β-globin mRNA (Figure 2C and D). Similarly, rescue experiments showed that the -dCT mutant failed to rescue the SMG-6 knockdown-induced accumulation of phospho-Upf1, supporting the above notion (Figure 2E).
SMG-1-mediated phosphorylation of Upf1 at T28 is required for association with SMG-6
The preferential binding of SMG-6 to phospho-Upf1 prompted us to identify the phosphorylated amino acid residue(s) of Upf1 responsible for SMG-6 binding. Both the N- and C-terminal regions of Upf1 have evolutionally conserved S/TQ residues, which are phosphorylated by SMG-1 in vitro (Supplementary Figure S3) (22). Based on these observations, we created phospho-resistant mutants of SBP-tagged Upf1, which do not contain the putative phosphorylation sites. The first mutant, Upf1-dCT, lacked the C-terminal 90 amino acids, residues 1028–1118. The second mutant, Upf1-dNCT, lacked the N-terminal 63 amino acids and the C-terminal 90 amino acids, and the third mutant, Upf1-5S/TA, harbored alanine (A) substitutions at T28, S1073, S1078, S1096 and S1116 (Figure 3A). Immunoprecipitation analysis of endogenous SMG-6 in the presence of RNaseA from HeLa TetOff cell extracts transfected with HA-tagged wild-type Upf1, or the Upf1 mutants, -dCT, -dNCT or -5S/TA showed that endogenous SMG-6 precipitated exogenously expressed wild-type Upf1 and the Upf1-dCT mutant. On the other hand, SMG-6 precipitated only small amounts of Upf1-dNCT or -5S/TA mutants compared with wild-type (Figure 3B). Similar experiments using two additional mutants of Upf1, Upf1-T28A, in which T28 was substituted by A, and Upf1-4SA, in which S1073, S1078, S1096 and S1116 were substituted by A, revealed that endogenous SMG-6 precipitated only a faint amount of Upf1-T28A, whereas it precipitated similar amounts of Upf1-4SA and wild-type Upf1 (Figure 3C). These results demonstrate that T28 of Upf1 is needed for SMG-6 binding.
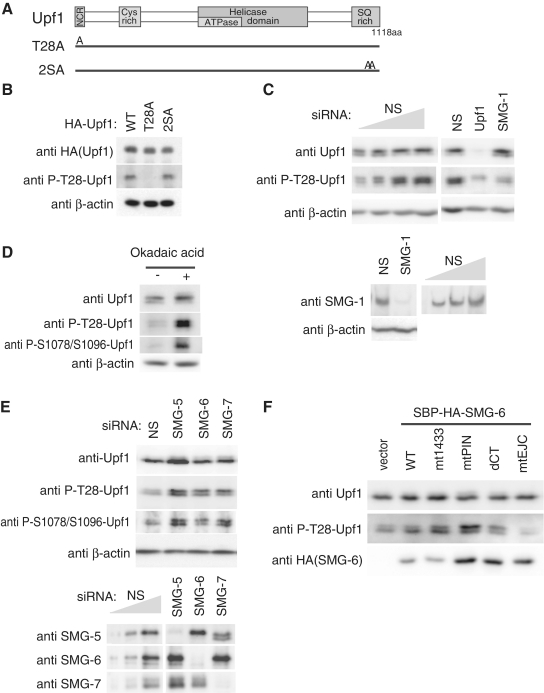
Preferential binding of SMG-6 to phospho-Upf1 (Figure 1) and the requirement of Upf1 T28 for this binding (Figure 3), in addition to the observation that a GST-fused peptide derived from the region of Upf1 T28 is efficiently phosphorylated by SMG-1 in vitro (22), suggest that, in vivo, SMG-1 phosphorylates T28 of Upf1 to create an SMG-6 binding site. To confirm this, we generated a phospho-specific polyclonal antibody against phosphorylated-T28 (P-T28-Upf1) and analyzed the reactivity to wild-type Upf1 and to Upf1 mutants -T28A and -2SA, in which S1078 and S1096 were substituted by A (Figure 4A). Anti-P-T28-Upf1 antibody recognized HA-SBP-tagged wild-type Upf1 and the mutant, Upf1-2SA, whereas it failed to detect the mutant, Upf1-T28A (Figure 4B). In addition, anti-P-T28-Upf1 detected an endogenous Upf1 phosphorylation signal that was strikingly reduced by Upf1 knockdown (Figure 4C). Furthermore, SMG-1-targeted siRNA treatment resulted in a decrease in anti-P-T28-Upf1 signal (Figure 4C). These results indicate that SMG-1 phosphorylates T28 of Upf1 in vivo, which was recognized by our anti-P-T28-Upf1 antibody. SMG-5, SMG-6 and SMG-7 are involved in the PP2A-mediated dephosphorylation of Upf1 at S1078 and S1096 in mammals (22,30,32). To assess the requirement for these factors in the dephosphorylation of the newly identified Upf1 phosphorylation site, T28, we analyzed P-T28-Upf1 signals in response to treatment with okadaic acid (OA), a PP2A inhibitor, knockdown of SMG-5, SMG-6 or SMG-7, or the overexpression of SBP-tagged SMG-6 mutants. We observed an increase in accumulation of the anti-P-T28-Upf1 signals after OA treatment (Figure 4D) and knockdown of SMG-5, SMG-6 or SMG-7 (Figure 4E), similar to the P-S1078/S1096 signals. Note that the amount of SMG-5 is slightly decreased in SMG-7 knocked-down and showed an unexpected mobility shift to the lower molecular weight side (Figure 4E). The overproduction of SMG-6-mt1433, -mtPIN and -dCT, but not of wild-type or -mtEJC, resulted in the accumulation of P-T28-Upf1 signals (Figure 4F). These results suggest that, similar to residues S1078/S1096, T28 of Upf1 is dephosphorylated by PP2A in an SMG-5, SMG-6 and SMG-7-dependent manner.
Figure 4.
SMG-1-mediated phosphorylation of Upf1 threonine residue 28. (A) Schematic representation of Upf1 mutants. The NCR, cystein rich region, helicase domains and SQ-rich region are depicted by gray boxes. In Upf1-T28A, T28 is substituted for A. In Upf1-2SA, S1078 and S1096 are replaced by A. (B) The anti-P-T28-Upf1 antibody recognizes T28 phosphorylation of Upf1 in vivo. HeLa TetOff cells were transfected with the plasmids indicated above the blot. Total cell extracts were probed with the antibodies indicated. (C) The anti-P-T28-Upf1 antibody detects SMG-1-mediated endogenous-Upf1 phosphorylation. Cells were transfected with the siRNAs shown above. Total cell extracts were probed with the antibodies indicated. (D) OA-sensitive phosphatase-mediated dephosphorylation of Upf1 P-T28. Total extracts of cells treated for 4 h with 50 nM OA were analyzed as above. (E and F) Knockdown of SMG-5, SMG-6 or SMG-7, or overexpression of SMG-6 mutants accumulates T28 phosphorylated Upf1. Cells were transfected with the siRNAs or SMG-6 plasmids indicated above the blot. Total cell extracts were probed with the antibodies indicated. To estimate the knockdown efficiency, 100, 50, 25 and 12.5% (B) or 100, 33 and 11% (E) of NS control samples were loaded.
SMG-1-mediated phosphorylation of Upf1 at S1096 is required for binding to SMG-5 and SMG-7
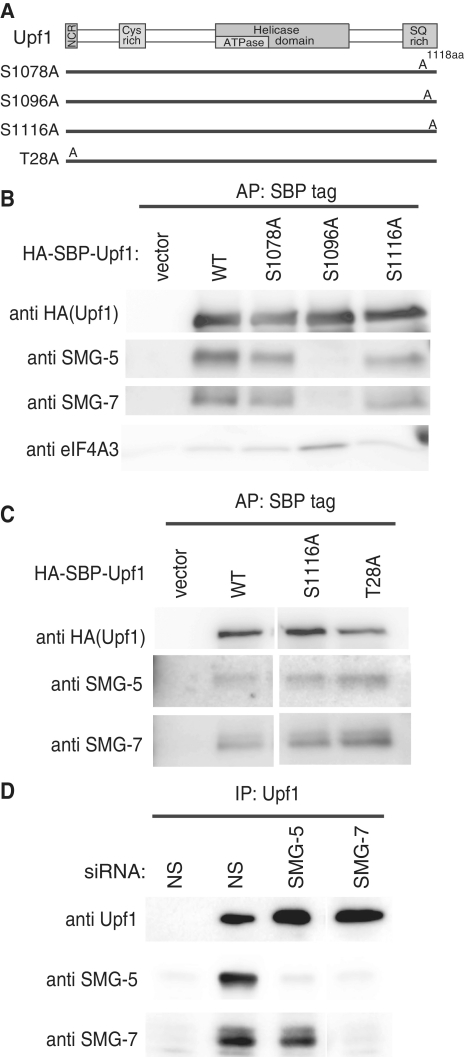
Although the SMG-5:SMG-7 complex also preferentially binds to phospho-Upf1, the exact binding site(s) of them are unknown (30). To identify the Upf1 residue(s) responsible for SMG-5:SMG-7 complex binding, we made three SBP-tagged Upf1 mutants, Upf1-S1078A, Upf1-S1096A and Upf1-S1116A, which harbored alanine substitutions at S1078, S1096 or S1116, respectively (Figure 5A). These mutants were chosen for the following two reasons: (i) SMG-7 binding to phospho-Upf1 is strikingly decreased in the Upf1-4SA mutant (19) and (ii) S1078, S1096 and S1116 residues of Upf1 are phosphorylated in vivo (22,30,31). Affinity purification of SBP-tagged Upf1 in the presence of RNaseA from cell extracts revealed that the S1096A mutant failed to coprecipitate endogenous SMG-5 or SMG-7, whereas mutants S1078 and S1116 coprecipitated equivalent amounts of SMG-5 and SMG-7 compared with wild-type Upf1 (Figure 5B). A similar experiment preformed using an SBP-tagged Upf1-T28A mutant showed that no apparent effect on SMG-5 or SMG-7 coprecipitation was observed for the Upf1-T28A mutant (Figure 5C). These results indicate that phosphorylated S1096 of Upf1 is integral to the binding site of the SMG-5:SMG-7 complex. Although both of SMG-5 and SMG-7 have a 14-3-3-like domain, only one serine residue, S1096, is responsible for the SMG-5:SMG-7 complex binding. To investigate which molecule is mainly involved in this binding, we immunoprecipitated endogenous Upf1 from cell extracts transfected with siRNA targeted against SMG-5 or SMG-7 in the presence of RNaseA. As shown in Figure 5D, depletion of SMG-7 decreased the SMG-5 coprecipitation compared with a non-silencing control. On the other hand, SMG-5 knockdown do not significantly decrease the coprecipitation of SMG-7. This suggests that SMG-7 links the SMG-5:SMG-7 complex to phospho-S1096 of Upf1.
Figure 5.
Upf1 serine residue 1096 is necessary for the binding of the SMG-5:SMG-7 complex. (A) Schematic representation of Upf1 mutants. The NCR, cystein rich region, helicase domains and SQ-rich region are depicted by gray boxes. In Upf1-S1078A, S1078 is substituted for A. In Upf1-S1096, S1096 is replaced by A. In Upf1-S1116A, S1116 is substituted for A. In, Upf1-T28A, T28 is replaced by A. (B and C) HeLa TetOff cells were transfected with the indicated plasmids or an empty vector (‘vector’). The cell extracts were purified with streptavidin sephorose in the presence of RNaseA. Purified fractions were analyzed by western blotting with the antibodies indicated. (D) HeLa TetOff cells were transfected with the indicated siRNAs. The cell extracts were immunoprecipitated with anti-Upf1 (5C3) antibody in the presence of RNaseA. The immunoprecipitates (IP) were analyzed by western blotting with the antibodies shown on the left of the panels.
Phosphorylation at both T28 and S1096 of Upf1 is required for NMD
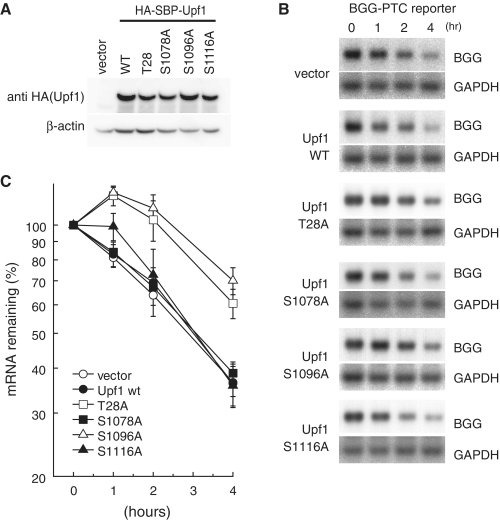
As described above, T28 and S1096 of Upf1 are required for SMG-6 and the SMG-5:SMG-7 complex, respectively, to bind to Upf1. If the binding of either of these factors is necessary for NMD to proceed in vivo, overproduction of mutant Upf1-T28A or -S1096A might inhibit NMD in a dominant-negative manner. To analyze this, we expressed wild-type Upf1 or the Upf1 mutants, -T28A, -S1078A, -S1096A or -S1116A in HeLa TetOff cells, and subsequently measured the half-lives of PTC-containing β-globin mRNA by northern blot analysis, as described in Figure 2. Overexpression of wild-type Upf1 or Upf1 mutants -S1078A or -S1116A had no affect on the half-lives of PTC-containing β-globin mRNA. In contrast, overexpression of Upf1-T28A or S1096A stabilized PTC-containing β-globin mRNA (Figure 6B and C). This result indicates that phosphorylation of both T28 and S1096 of Upf1 is required for PTC-dependent degradation of β-globin mRNA.
Figure 6.
Phosphorylation of Upf1 at both T28 and S1096 is required for PTC-containing β-globin mRNA decay. (A and B) HeLa TetOff cells were transfected with the plasmids shown above (A) or to the left of the panel (B). Total cell lysates were analyzed by western blotting with the indicated antibody probes (A). NMD is inhibited by overexpression of Upf1-T28A and -S1096 mutants. HeLa TetOff cells were co-transfected with the Tet-inducible β-globin (BGG)-PTC plasmid shown in Figure 2A and with Upf1 plasmids indicated on the left of the panel. After the addition of doxycycline to repress the transcription of the BGG reporter plasmid, total RNAs prepared at the times indicated were analyzed by northern blotting (B). (C) The quantities of BGG mRNA, normalized to GAPDH signals, were plotted. The means ± SD from three independent experiments are shown.
Distinct phospho-Upf1 complexes are formed in SMG-5 or SMG-6 inactivated cell extracts
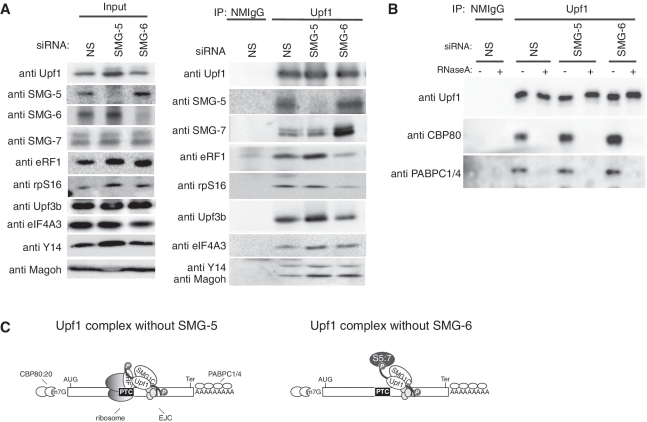
Depletion of NMD components or expression of specific NMD component mutants enabled us to ‘freeze’ the transient protein complex formed during the process of PTC recognition and allowed us to suggest the sequential mRNP remodeling of the mRNA surveillance complex (19,20,43,44). The preferential binding of the SMG-5:SMG-7 complex and SMG-6 to phospho-Upf1 and their essential role for NMD prompted us to capture the post-Upf1 phosphorylation process of NMD by the inactivation of these proteins. For this, we immunoprecipitated endogenous Upf1 from cell extracts transfected with siRNA targeted against SMG-5 or SMG-6 in the presence of RNaseA and cycloheximide, which stabilizes 80S ribosomes on mRNAs in vitro and prevents their dissociation during immunoprecipitation (45) during cell lysis. The Upf1 immune complexes eluted with the Upf1-peptide antigen contained eRF1, EJC components (Upf3b, Y14 and Magoh), rpS16 and SMG-7. Compared with a non-silencing control, depletion of either SMG-5 or SMG-6 caused an accumulation of phospho-Upf1 in the total cell extracts and in the Upf1 immune complex (Figure 4E; data not shown). Depletion of SMG-5 enhanced the amounts of eRF1, Upf3b and EJC components (eIF4A3, Y14 and Magoh) coprecipitated with Upf1, whereas the amounts of coprecipitated rpS16 and SMG-7 were not apparently altered. In contrast, SMG-6 depletion decreased the coprecipitation of rpS16 and eRF1 with Upf1, although it increased coprecipitation of SMG-5, SMG-7 and EJC components (eIF4A3, Y14 and Magoh) (Figure 7A). We failed to detect SMG-6 coprecipitation in this experimental condition, most likely because of the low binding affinity of the anti-SMG-6 antibody.
Figure 7.
SMG-5 or SMG-6 knockdown causes the accumulation on mRNP of phospho-Upf1 complexes containing different components. (A and B) HeLa TetOff cells were transfected with the indicated siRNAs. The cell extracts were immunoprecipitated with anti-Upf1 (5C3) antibody in the presence of RNaseA (A), or in the absence or presence of RNaseA (B). The cell extracts (Input) and immunoprecipitates (IP) were analyzed by western blotting with the antibodies shown on the left of the panels. (C) Schematic presentation of Upf1 complexes following SMG-5 or SMG-6 siRNA treatment.
Next, we tested whether the phospho-Upf1 complexes enriched in SMG-5 or SMG-6 depleted cell extracts are formed on mRNPs. For this, we immunopurified endogenous Upf1 from cells transfected with siRNAs targeting SMG-5 or SMG-6 in the presence or absence of RNaseA. RNaseA sensitive coprecipitation of CBP80, a nuclear cap binding protein and of PABPC1/4, cytoplasmic poly(A) binding proteins, was observed in the Upf1 immunoprecipitates from SMG-5- or SMG-6-depleted cell extracts, suggesting that the phospho-Upf1 complexes are formed on mRNPs (Figure 7B). Our model to explain these observations is depicted in Figure 7C.
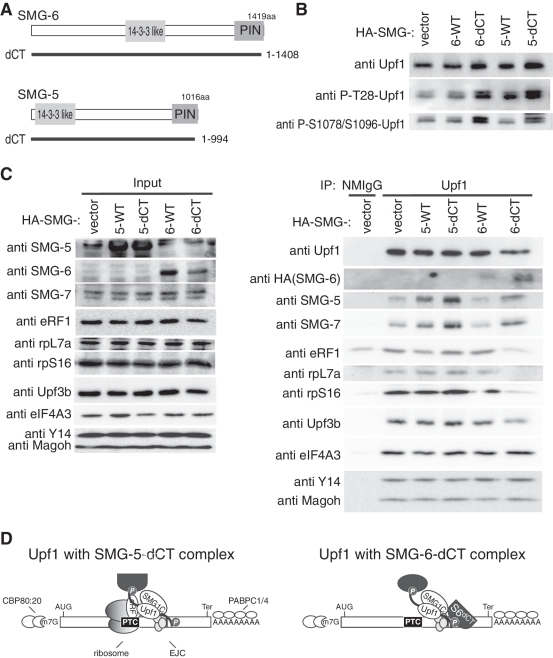
To further evaluate the distinct processes regulated by SMG-5 or SMG-6, we took advantage of the dominant-negative mutants of SMG-5, SMG-5-dCT and of SMG-6, SMG-6-dCT (Figure 8A). SMG-5-dCT preferentially binds to phospho-Upf1 and inhibits Upf1 dephosphorylation and NMD (30). SMG-6-dCT also showed a similar phenotype (Figures 1 and 2). We immunoprecipitated endogenous Upf1 from cell extracts transfected with wild-type HA-tagged-SMG-5, HA-tagged-SMG-5-dCT, wild-type SMG-6 or HA-tagged-SMG-6-dCT in the presence of RNaseA and cycloheximide. Compared with the wild-type, expression of either SMG-5-dCT or SMG-6-dCT caused a significant accumulation of phospho-Upf1 in cell extracts and in the Upf1 immune complex and showed greater levels of SMG-5dCT and SMG-6-dCT coprecipitation with Upf1 (Figure 8B; data not shown). Overexpression of the SMG-6-dCT mutant increased the coprecipitation of endogenous SMG-5 and SMG-7, and reduced the coprecipitation of ribosomal proteins, rpL7a and rpS16, eRF1 and Upf3b with Upf1 compared with wild-type SMG-6 (Figure 8C). On the other hand, overexpression of the SMG-5dCT mutant did not alter the coprecipitation of these proteins except for the increased coprecipitation of endogenous SMG-7 compared with wild-type SMG-5 (Figure 8C). No apparent alteration was observed for the coprecipitation of EJC components, Y14, Magoh and eIF4A3 in these precipitates (Figure 8C). In addition, CBP80 was coprecipitated with Upf1 only in the absence of RNaseA, suggesting that the phospho-Upf1 complexes with SMG-5dCT or SMG-6-dCT are formed on mRNPs (Supplementary Figure S4). Taken together with the knockdown experiment presented above, these results indicate that the phospho-Upf1 complex containing ribosomal proteins and eRF1 only appears with the inactivated SMG-5, but not with the inactivated SMG-6. The decrease of Upf3b in the Upf1 complex accumulated with SMG-6-dCT might reflect competitive binding of SMG-6 and Upf3b to phospho-Upf1. Our model is depicted in Figure 8D.
Figure 8.
Overexpression of SMG-5-dCT or SMG-6-dCT causes the accumulation of phospho-Upf1 complexes containing different components. (A) Schematic representation of SMG-5dCT and SMG-6dCT mutants. The 14-3-3-like domain and PIN domain are depicted in gray boxes. SMG-5dCT and SMG-6dCT have deletions of the C-terminal 23 or 11 amino acids, respectively. (B) Overexpression of SMG-5 or SMG-6 mutants results in the accumulation of Upf1 phosphorylated at T28 and S1078/S1096. Cells were transfected with the SMG-5 or SMG-6 plasmids indicated above the blot. Total cell extracts were probed with the antibodies indicated. (C) HeLa TetOff cells were transfected with the indicated plasmids. The cell extracts were immunoprecipitated with anti-Upf1 (5C3) antibody in the presence of RNaseA. (D) Schematic presentation of Upf1 complexes following overexpression of SMG-5-dCT or SMG-6-dCT.
DISCUSSION
Here, we demonstrated that the SMG-1-mediated phosphorylation of Upf1 creates at least two distinct phospho-specific binding platforms for SMG-6 and the SMG-5:SMG-7 complex. Both bindings are required for the mRNP remodeling to dissociate Upf1 from mRNA and NMD. Further, the binding of SMG-5:SMG-7 complex, but not SMG-6, to pohospho-Upf1 leads the dissociation of ribosome and release factors from DECID complex. Figure 9 illustrates our view of the simultaneous recruitments of the SMG-6 and the SMG-5:SMG-7 complex for phospho-Upf1 that lead to the remodeling of post-PTC-recognized complex.
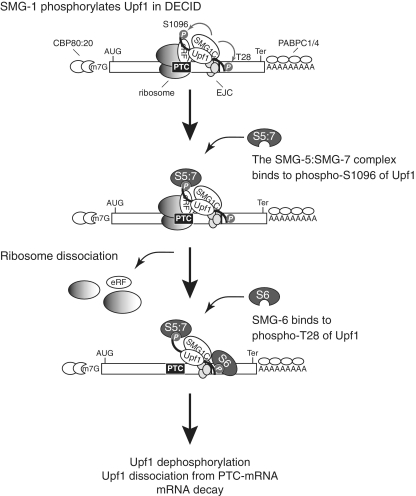
Figure 9.
Model depicting NMD from post-Upf1 phosphorylation to mRNA decay. PTC recognition is established by the formation of mRNA surveillance complexes called ‘DECIDs’ that contain a PTC-recognizing ribosome, eRF, Upf1, SMG1C (SMG-1:SMG-8; SMG-9 complex) and the EJC during the initial round of translation. DECID formation induces SMG-1–mediated Upf1 phosphorylation. The SMG-5:SMG-7 complex binds to phospho-S1096 of Upf1 to dissociate the ribosome and release factor from Upf1. SMG-6 binds to phospho-T28 of Upf1 to induce Upf1 dissociation from the mRNA. AUG, start codon; Ter, termination codon; eRF, eRF1:eRF3 complex; P, phosphate group; S5:7, the SMG-5:SMG-7 complex, S6, SMG-6.
In the present study, we have identified T28 as a novel SMG-1-mediated in vivo phosphorylation site of Upf1 (Figure 4). The sequence surrounding T28 is conserved among higher eukaryotes, including plants, supporting the importance of phosphorylation at this site (Supplementary Figure S3). An okadaic acid sensitive phosphatase, PP2A, dephosphorylates P-T28 of Upf1 according to SMG-5, SMG-6 and SMG-7 dependent mechanisms (Figure 4E), which is similar to other SMG-1-mediated phosphorylation sites, S1078 and S1096 (Figure 4D and E). However, ATPase-deficient Upf1 is highly phosphorylated (19) and binds efficiently with SMG-5, SMG-6 and SMG-7 (44). These observations suggest that the binding of SMG-5, SMG-6 and SMG-7 to phospho-Upf1 is not sufficient to promote dephosphorylation even though they bind to PP2A (30,33,35). Future studies will reveal the mechanism of Upf1 dephosphorylation.
What is the role of T28 phosphorylation? The following observations provide evidence that T28 phosphorylation induces the binding of SMG-6 via its 14-3-3-like domain during NMD. First, the binding of SMG-6 to Upf1 was strongly diminished in a Upf1-T28A mutant (Figure 3C). Second, the SMG-6 14-3-3-like domain mutation abolished the coprecipitation of phospho-Upf1 (Figure 1C). Third, overexpression of either Upf1-T28A or SMG-6-mt1433 mutant suppressed NMD in a dominant-negative manner (Figure 2 and 6). Another Upf1 phosphorylation site, at S1096, whose phosphorylation is mediated by SMG-1, formed the binding site for the SMG-5:SMG-7 complex (Figure 5B). SMG-7 is presumably responsible for the binding of the SMG5:SMG7 complex to phospho-S1096 (Figure 5D). However, it remains unclear why we failed to detect an increase of SMG-7 coprecipitation with phospho-Upf1 in SMG-5 knockdown cells, whereas SMG-5 knockdown accumulates the phosphorylation of S1096 in a similar manner to SMG-6 knockdown that induces accumulation of the SMG-5:SMG-7 complex with phospho-Upf1 (Figure 7) Hetero dimer formation might be required for the full binding activity of the SMG-5:SMG-7 complex. The dominant-negative effect of the Upf1-S1096A mutant suggested that this phospho-mediated binding of the SMG-5:SMG-7 complex is required for NMD (Figure 6). The involvement of two different phosphorylation sites for the SMG-5:SMG-7 complex and SMG-6 also suggests that they can simultaneously bind to phospho-Upf1, although they have been expected to compete (2,3). Consistent with this notion, the SMG-5:SMG-7 complex accumulated in the phospho-Upf1 complex together with the SMG-6dCT mutant (Figure 8C). These results indicate that simultaneous binding of the SMG-5:SMG-7 complex and SMG-6 to phospho-Upf1 are required for NMD. This is consistent with the observation that SMG-5, SMG-6 and SMG-7 are non-redundant (32,36,43,46).
Although SMG-5, SMG-6 and SMG-7 share 14-3-3-like domains (30,37), different phospho-S/T binding properties between the SMG-5:SMG-7 complex and SMG-6 exist; while the SMG5:SMG-7 complex binds to P-S1096 but not to P-T28 (Figure 5B and C), SMG-6 binds to P-T28 but not to P-S1096 (Figure 3C). Upf1-S1078 is dispensable in vivo, although SMG-7 can bind phospho-S1078 in vitro. Heterodimer formation of the SMG-5:SMG-7 complex might affect their specificity in vivo. Future studies, such as structural analysis, will clarify their specificity for phospho-S/TQ motifs. SMG-6 can associate with the mRNA surveillance complex not only through phospho-Upf1, but also through its ability to directly bind the EJC (39) (Supplementary Figure S2) and/or via direct RNA binding (47). These associations might be involved in EJC recycling, similar to PYM (48), or in some specific mRNA degradation (39).
While S1078 and S1116 of Upf1 are phosphorylated in vivo (22,30,31), they are likely to be dispensable for NMD (Figure 6). It is possible that they are involved with the other SQ-directed kinases, such as ATR, ATM and DNA-PKcs, which mediate phosphorylation events for various cellular functions of Upf1, such as histone mRNA decay or genome stability (49–51).
Our results presented in this study revealed that inactivation of either SMG-5 or SMG-6 induces accumulation of distinct phospho-Upf1 complexes on mRNP together with CBP80 (cap) and PABPC1/C4 [poly(A)] (Figures 7 and 8; Supplementary Figure S4). This suggests that phospho-specific binding of the SMG-5:SMG-7 complex and of SMG-6 to Upf1 are required for mRNP remodeling to dissociate Upf1 from mRNA and to promote mRNA decay.
On the other hand, the complexes accumulated by SMG-5 or SMG-6 inactivation are different. For instance, SMG-5 knockdown and overexpression of SMG-5-dCT resulted in the accumulation of phospho-Upf1 complex containing ribosome, release factor and EJC (Figures 7 and 8). Importantly, the SMG-5-dCT mutant can accumulate phospho-Upf1 complex containing ribosomal proteins and release factor. These results indicate that SMG-5 binding to phospho-Upf1 is required but not sufficient for the dissociation of ribosome and release factor from the DECID complex. The C-terminal PIN domain of SMG-5 is required for this action and SMG-5 might be involved in ribosome dissociation by recruitment and/or activation of ribosome dissociation factors, such as eEF2, eIF3 and/or ABCE1 (52,53), through its PIN domain (Figure 9).
In contrast to SMG-5, SMG-6 knockdown and overexpression of SMG-6-dCT resulted in the accumulation of phospho-Upf1 complex containing EJC and the SMG-5:SMG-7 complex, but not ribosomal proteins or release factors (Figures 7 and 8). These results suggested that, SMG-6 is likely to be dispensable for ribosome and release factor dissociation. However, SMG-6 and its PIN domain are still required for Upf1 dephosphorylation, NMD and Upf1 dissociation from mRNA (Figure 8 and Supplementary Figure S4). These results suggest that ribosome and release factor dissociation from the Upf1 complex precedes but is not sufficient to promote Upf1 dephosphorylation, NMD and probably Upf1 dissociation from mRNA. Because ATPase activity of Upf1 is required for the dissociation of Upf1 from mRNA (44) (Supplementary Figure S5), the SMG-6 PIN domain might be involved in Upf1 ATPase activation. It is also possible that the SMG-6 PIN domain-mediated endocleavage of PTC-mRNA is a prerequisite for Upf1 ATPase activation. In addition, even though ATP binding/ATPase-deficient Upf1 binds with SMG-5, SMG-6, SMG-7 and mRNA decay enzymes (19,44,54), they fail to efficiently promote mRNA decay (19,32,44). Consistent with this finidng, the RNase A sensitive accumulation of CBP80, an indicator of intact cap structure, is observed in Upf1-KQ mutant precipitate (Supplementary Figure S5). Taken together with the RNaseA sensitive coprecipitation of CBP80 with Upf1 in SMG-5 or SMG-6 inactivated conditions, these results suggested that both decapping and endocleavage do not occur before (i) SMG-5, SMG-6 and SMG-7 have all bound to Upf1 and (ii) Upf1 ATPase activation. Upf1 ATPase activity might regulate decapping enzyme and SMG-6 endonuclease activation (Figure 9). Further reconstitution analysis is required for resolving these issues.
The SMG-5:SMG-7 complex can bind to the N-terminal conserved region of Upf1 containing residue T28 in phospho-independent manner (30). Thus, it is possible that the SMG-5:SMG-7 complex might either inhibit the SMG-1-mediated phosphorylation of T28 of Upf1 or inhibit the binding of SMG6 to the phosphorylated T28 residue. One possible explanation that the phospho-spesific binding of SMG6 to Upf1 might occur after the SMG-5:SMG-7 complex are sequestered by the C-terminal phospho-S1096 of Upf1 (Figure 9). Simultaneous binding of the SMG-5:SMG-7 complex and SMG-6 to N-terminal conserved region of Upf1 is also possible since Upf1-4SA mutants, which lose the pohspho-S1096 binding of the SMG-5:SMG-7 complex can bind SMG-6 (Figure 3C).
EJC components that trigger Upf1 phosphorylation are also involved in splicing junction-independent NMD, although the mechanism is unknown, (27,29) (A.Yamashita, our unpublished data). Intriguingly, SMG-1 and its co-factors are also needed for the downstream splicing-junction independent NMD (A.Yamashita, our unpublished data). Thus, the simultaneous binding of the SMG-5:SMG-7 complex and SMG-6 to phospho-Upf1 described in this study seems to be generally involved in mammalian NMD.
Even though Upf1 sites phosphorylated by SMG-1 are conserved, SMG-5, SMG-6 and SMG-7 are highly diverse among higher eukaryotes. For instance, vertebrates and nematodes have all these molecules, while flies do not have SMG-7, and plants have two non-redundant SMG-7s, but not SMG-5 and SMG-6 (30,33,36,55). These differences might reflect the distinct mRNA decay mechanisms among these species. In particular, flies mainly use an SMG-6-mediated endocleavage pathway, and mammals use both endonucleolytic cleavage and deadenylation-initiated decapping pathways (40,41,56). No endonucleolytic cleavage of PTC-mRNA has been reported in plants, which is consistent with the absence of SMG-6 endonuclease (57). Intriguingly, at least two SMG-5/6/7 related molecules exist among higher eukaryotes and the two phosphorylation sites of Upf1, which are essential for binding with them are conserved among higher eukaryotes. These SMG-5/6/7 related molecules might have similar sequence and functional specificity to that of mammals.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1–S5.
FUNDING
The Japan Society for the Promotion of Science (to S.O. and N.I.); the Japan Science and Technology Corporation (to A.Y. and S.O.); the Ministry of Education, Culture, Sports, Science and Technology of Japan [a Grant-in-Aid for Scientific Research (A) (to S.O.), Young Scientists (A) (to A.Y.), Scientific Research on Innovative Areas ‘RNA regulation’ (to A.Y.) and Scientific Research on Innovative Areas ‘Functional machinery for non-coding RNAs’ (to A.Y.)]; Takeda Science Foundation (to S.O.); Mitsubishi Foundation (to S.O.), Uehara Memorial Foundation (to S.O.) and the Yokohama Foundation for Advancement of Medical Science (to A.Y.). Funding for open access charge: The Japan Science and Technology Corporation (to A.Y.)
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ms Yumi Bamba, Ms Reiko Muramatsu, Dr Tetsuo Ohnishi and Dr Isao Kashima for their technical assistance. Y.O.-K. and A.Y. designed and performed experiments, analyzed data and wrote the article; K.K. and N.I. performed the experiments; F.H. supervised the project; S.O. supervised the project and participated in the preparation of the article.
REFERENCES
- 1.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell. Mol. Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hwang J, Maquat LE. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr. Opin. Genet. Dev. 2011;21:422–430. doi: 10.1016/j.gde.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 5.Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Usuki F, Yamashita A, Fujimura M. Post-transcriptional defects of antioxidant selenoenzymes cause oxidative stress under methylmercury exposure. J. Biol. Chem. 2011;286:6641–6649. doi: 10.1074/jbc.M110.168872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 8.Usuki F, Yamashita A, Higuchi I, Ohnishi T, Shiraishi T, Osame M, Ohno S. Inhibition of nonsense-mediated mRNA decay rescues the phenotype in Ullrich's disease. Ann. Neurol. 2004;55:740–744. doi: 10.1002/ana.20107. [DOI] [PubMed] [Google Scholar]
- 9.Usuki F, Yamashita A, Kashima I, Higuchi I, Osame M, Ohno S. Specific inhibition of nonsense-mediated mRNA decay components, SMG-1 or Upf1, rescues the phenotype of ullrich disease fibroblasts. Mol. Ther. 2006;14:351–360. doi: 10.1016/j.ymthe.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Pastor F, Kolonias D, Giangrande PH, Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature. 2010;465:227–230. doi: 10.1038/nature08999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linde L, Kerem B. Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet. 2008;24:552–563. doi: 10.1016/j.tig.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. NMD: RNA biology meets human genetic medicine. Biochem. J. 2010;430:365–377. doi: 10.1042/BJ20100699. [DOI] [PubMed] [Google Scholar]
- 13.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson P. NMD in Caenorbabditis elegans. In: Maquat L, editor. Nonsense-Mediated mRNA Decay. Austin, TX: Landes bioscience; 2006. pp. 139–150. [Google Scholar]
- 15.Culbertson MR, Leeds PF. Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 2003;13:207–214. doi: 10.1016/s0959-437x(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 16.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 17.Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamashita A, Kashima I, Ohno S. The role of SMG-1 in nonsense-mediated mRNA decay. Biochim. Biophys. Acta. 2005;1754:305–315. doi: 10.1016/j.bbapap.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S. AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci. Signal. 2010;3:ra27. doi: 10.1126/scisignal.2000468. [DOI] [PubMed] [Google Scholar]
- 22.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez IS, Yamashita A, Arias-Palomo E, Bamba Y, Bartolome RA, Canales MA, Teixido J, Ohno S, Llorca O. Characterization of SMG-9, an essential component of the nonsense-mediated mRNA decay SMG1C complex. Nucleic Acids Res. 2011;39:347–358. doi: 10.1093/nar/gkq749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arias-Palomo E, Yamashita A, Fernandez IS, Nunez-Ramirez R, Bamba Y, Izumi N, Ohno S, Llorca O. The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes Dev. 2011;25:153–164. doi: 10.1101/gad.606911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimson A, O'Connor S, Newman CL, Anderson P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans. Mol. Cell. Biol. 2004;24:7483–7490. doi: 10.1128/MCB.24.17.7483-7490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogg JR, Goff SP. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell. 2010;143:379–389. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuda D, Hosoda N, Kim YK, Maquat LE. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat. Struct. Mol. Biol. 2007;14:974–979. doi: 10.1038/nsmb1297. [DOI] [PubMed] [Google Scholar]
- 30.Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 31.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 32.Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders KR, Grimson A, Anderson P. SMG-5, required for C.elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 2003;22:641–650. doi: 10.1093/emboj/cdg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wittmann J, Hol EM, Jack HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiu SY, Serin G, Ohara O, Maquat LE. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. Rna. 2003;9:77–87. doi: 10.1261/rna.2137903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 38.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 39.Kashima I, Jonas S, Jayachandran U, Buchwald G, Conti E, Lupas AN, Izaurralde E. SMG6 interacts with the exon junction complex via two conserved EJC-binding motifs (EBMs) required for nonsense-mediated mRNA decay. Genes Dev. 2010;24:2440–2450. doi: 10.1101/gad.604610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. Rna. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 42.Yamashita A, Ohno S. Analysis of nonsense-mediated mRNA decay by monitoring mRNA half-lives in mammalian cells. Cold Spring Harb. Protoc. 2010;2010 doi: 10.1101/pdb.prot5386. pdb. prot5386. [DOI] [PubMed] [Google Scholar]
- 43.Johns L, Grimson A, Kuchma SL, Newman CL, Anderson P. Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons. Mol. Cell. Biol. 2007;27:5630–5638. doi: 10.1128/MCB.00410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell. 2010;143:938–950. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baliga BS, Pronczuk AW, Munro HN. Mechanism of cycloheximide inhibition of protein synthesis in a cell-free system prepared from rat liver. J. Biol. Chem. 1969;244:4480–4489. [PubMed] [Google Scholar]
- 46.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 47.Redon S, Reichenbach P, Lingner J. Protein RNA and protein protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res. 2007;35:7011–7022. doi: 10.1093/nar/gkm724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009;137:536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 49.Muller B, Blackburn J, Feijoo C, Zhao X, Smythe C. DNA-activated protein kinase functions in a newly observed S phase checkpoint that links histone mRNA abundance with DNA replication. J. Cell. Biol. 2007;179:1385–1398. doi: 10.1083/jcb.200708106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 51.Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 2006;16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 52.Demeshkina N, Hirokawa G, Kaji A, Kaji H. Novel activity of eukaryotic translocase, eEF2: dissociation of the 80S ribosome into subunits with ATP but not with GTP. Nucleic Acids Res. 2007;35:4597–4607. doi: 10.1093/nar/gkm468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benkovics AH, Nyiko T, Merai Z, Silhavy D, Bisztray GD. Functional analysis of the grapevine paralogs of the SMG7 NMD factor using a heterolog VIGS-based gene depletion-complementation system. Plant Mol. Biol. 2011;75:277–290. doi: 10.1007/s11103-010-9726-0. [DOI] [PubMed] [Google Scholar]
- 56.Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 57.Kerenyi Z, Merai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 2008;27:1585–1595. doi: 10.1038/emboj.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.