Abstract
The yeast RNA helicase Dhh1p has been shown to associate with components of mRNA decay and is involved in mRNA decapping and degradation. An RNA-binding protein, Rbp1p, is known to bind to the 3′-UTR of porin (POR1) mRNA, and induces mRNA decay by an uncharacterized mechanism. Here, we show that Dhh1p can associate with POR1 mRNA and specifically promote POR1 mRNA decay via its interaction with Rbp1p. As compared to its mammalian homolog RCK/p54/DDX6, Dhh1p has a unique and long extension at its C-terminus. Interestingly, this non-conserved C-terminal region of Dhh1p is required for interaction with Rbp1p and modulating Rbp1p-mediated POR1 mRNA decay. Notably, expression of a C-terminal 81-residue deleted Dhh1p can fully complement the growth defect of a dhh1Δ strain and retains its function in regulating the mRNA level of an RNA-binding protein Edc1p. Moreover, mammalian DDX6 became capable of interacting with Rbp1p and could confer Rbp1p-mediated POR1 mRNA decay in the dhh1Δ strain upon fusion to the C-terminal unique region of Dhh1p. Thus, we propose that the non-conserved C-terminus of Dhh1p plays a role in defining specific interactions with mRNA regulatory factors that promote distinct mRNA decay.
INTRODUCTION
Regulation of mRNA stability is an important step that modulates the cellular abundance of a transcript (1). In the yeast Saccharomyces cerevisiae, degradation of mRNA occurs through two general pathways, both of which are initiated by shortening of the poly(A) tail by the major deadenylase complex. Following deadenylation, mRNA can undergo 3′–5′ exonucleolytic decay, which is catalyzed by the exosome and the SKI complex. Alternatively, the 5′-cap structure of an mRNA can be removed by the decapping enzyme Dcp1p/Dcp2p, followed by 5′–3′ cytoplasmic exonuclease Xrn1p digestion of the remainder of the mRNA (2,3). Decapping is thought to be an irreversible step and a site of regulatory signal inputs (4). Several proteins function in modulating decapping during mRNA decay, including Dhh1p, Pat1p, Lsm1-7p complex and Edc1/2p. These proteins are evolutionarily conserved and each plays a distinct role in modulating the decapping process (5–9).
One of these modulators of decapping, Dhh1p is a member of the DEAD-box protein family of RNA helicases. RNA helicases are ubiquitous, highly conserved enzymes that participate in multiple aspects of RNA metabolism, from transcription to degradation (10–13). These proteins bind to and remodel RNA or RNA–protein complexes in an ATP-dependent fashion (14). Many RNA helicases are required for specific post-transcriptional processes (11,13). However, in vitro assays for RNA helicase activities performed with purified RNA helicase proteins typically show little or no RNA sequence specificity. Dhh1p contains nine conserved motifs characteristic of DEAD-box proteins, which fold into two RecA-like catalytic domains (11,15). Within the Dhh1p helicase subfamily, there are significant differences related to the extensions of the N- and C-termini of amino acid sequences (16). For example, Dhh1p has a unique and long extension at its C-terminus and a short extension at its N-terminus. In contrast, the mammalian homolog RCK/p54/DDX6 has a long N-terminal extension and only a short C-terminal region. Therefore, it is thought that these diverse regions could interact with specific protein complexes to provide target specificity of RNA helicases in vivo (16,17). Dhh1p interacts with the decapping and deadenylase complexes (7) and along with other proteins involved in decapping and mRNA degradation localizes to discrete cytoplasmic foci known as processing bodies (P-bodies) (18). P-bodies are speculated to be cytoplasmic locations of mRNA repression and decay. Reijns et al. (19) have suggested that Dhh1p contains a glutamine–proline-rich C-terminus and this region contributes to efficient accumulation of Dhh1p in P-bodies under stress conditions. Interestingly, in addition to the implication of its function in mRNA decapping and decay, Dhh1p homologs in Drosophila (Me31B), Caenorhabditis elegans (cgh-1) and Xenopus (Xp54) have been implicated in translational repression and storage of maternal mRNA (20–23).
We have previously identified an RNA-binding protein encoded by RBP1 (24) that can bind to and regulate the stability of POR1 mRNA in S. cerevisiae (25). Rbp1p contains three RNA recognition motifs (RRMs), two glutamine-rich stretches and a C-terminal asparagine–methionine–proline-rich (NMP) region. It binds to the 3′-untranslated region (UTR) of POR1 mRNA through its RNA recognition motifs and accelerates porin mRNA turnover. Rbp1p-mediated POR1 mRNA decay is disrupted in an xrn1Δ strain, indicating that POR1 mRNA undergoes degradation through an Xrn1p-dependent pathway following decapping (26). It is unclear whether Rbp1p can recruit the general 5′–3′ decay machinery to promote POR1 mRNA decay. Using a yeast two-hybrid assay, we have found that Dhh1p interacts with Rbp1p, but the biological significance of such an interaction has not characterized.
In this article, we provide evidence that Dhh1p cooperates with Rbp1p to regulate a distinct mRNA decay. We show that the non-conserved C-terminal region of Dhh1p is required for interaction with Rbp1p and this interaction is required for Rbp1p-mediated POR1 mRNA decay. Moreover, the mammalian Dhh1p ortholog, DDX6, which could neither interact with Rbp1p nor promote POR1 mRNA decay, gains the ability to interact with Rbp1p and elicit Rbp1p-mediated POR1 mRNA decay in a dhh1Δ strain when fused with the C-terminal unique region of Dhh1p. Through our study, we propose that Dhh1p is recruited to specific mRNAs and promotes distinct mRNA decay by interacting with RNA-binding protein complexes through its non-conserved C-terminal region.
MATERIALS AND METHODS
Growth medium and yeast strains
Genotypes for the yeast strains used in this study are listed in Supplementary Table SI. Yeast cells were grown either in rich medium containing 1% yeast extract, 2% peptone and 2% glucose or in synthetic media containing 0.67% yeast nitrogen base (without amino acids) and 2% glucose supplemented with the appropriate nutrients. Yeasts were transformed by the lithium acetate method. The DHH1 gene was disrupted in YTC345 using a Kan disruption cassette amplified by PCR from pFA6-kanMX6 (27). Strains expressing Dcp2p-mCh, Dhh1p-GFP, Dhh1p-dC81-GFP, Dhh1p-dC106-GFP, Dhh1p–3HA, Dhh1p-dC81-3HA, or Rbp1p–3HA were obtained through insertion of an mCherry-kan cassette amplified from pBS34 (Roger Tsien lab), a GFP-HIS cassette amplified from pFA6a-GFP(S65T)-His3MX6, or a 3HA-HIS cassette amplified from pFA6a-3HA-His3MX6 (27). Disruption or insertion of each cassette was verified by western blotting.
Plasmid construction
Plasmids used in this study are listed in Supplementary Table SII. Escherichia coli DH5α was used for DNA manipulations. Plasmids were constructed by standard protocols. For expression in yeast, the plasmid pVT101U-HA-RBP1 was used, as previously described (25), and HA-RBP1 containing the ADH1 promoter was also cloned into YEplac181. The full-length DHH1, DHH1-dC81 and DHH1-dC106 were amplified by PCR from yeast genomic DNA using specific primers and cloned into the pVT101U vector. These plasmids were digested with restriction enzymes to isolate the DNA fragment containing the ADH1 promoter and the open reading frame. These fragments were subcloned into corresponding sites in YCplac111 to generate a series of Dhh1p expression plasmids. The C85 PCR fragment from the DHH1 gene, encoding the C-terminal 85 amino acids of Dhh1p, was amplified using specific chimera primers; with the 5′-end aligned with DDX6 and the 3′-end aligned with DHH1. Full-length DDX6 and DDX6-C85 were amplified by PCR from HeLa cDNA using specific primers and C85 PCR fragments, and then cloned into the expression vectors. For yeast two-hybrid assay, full-length RBP1 and DHH1 were separately amplified by PCR from yeast genomic DNA using specific primers and cloned into the pJG4-5 vector in-frame with the Gal4AD domain. Analogously, full-length and mutant RBP1 or DHH1 were generated and cloned into pEG202 in frame with the LexA DNA-binding domain. For in vitro GST pull-down assays, the coding sequence for full length and amino acid 1–425 of Dhh1p and for C-terminal fragment (CF) of Rbp1p were amplified by PCR from yeast genomic DNA with specific primers and subcloned into the pET15b or pGEX4T-1 expression vector, respectively.
Yeast two-hybrid assay
The yeast strain YEM1α was co-transformed with different combinations of bait (pEG202) and prey (pJG4-5) plasmids and β-galactosidase plate assays were performed by streaking transformants onto SC-Trp-His plates containing 2% galactose and 80 µg/ml X-Gal (5-bromo-4chloro-3-indolyl-β-d-galactoside). The plates were then incubated at 30°C for 2–3 days.
Yeast cell extracts preparation and western blotting
Extracts were obtained from ∼2 OD600 of yeast cells, suspended in 5% TCA and processed by vigorous vortexing with glass beads. Cell debris was collected by centrifugation at 13 000 rpm for 10 min, washed with water to remove residual TCA, followed by centrifugation at 13 000 rpm for 10 min, suspended in SDS-loading buffer and then heated at 95°C for 5–10 min. For western blotting, all cell extracts were run on 9% SDS–polyacrylamide gels. Proteins were then transferred to nitrocellulose membrane and probed with indicated antibodies. Act1p was used as a loading control.
In vitro GST pull-down experiments
Escherichia coli strain BL21 (DE3) (Novagen) was transformed with plasmids pET15b-Dhh1p, pET15b-Dhh1p-dC81, pGEX6T-1 or pGEX6T-1-Rbp1p-CF. Recombinant GST, GST-Rbp1p-CF, His-Dhh1p and His-Dhh1p-dC81 were expressed in BL21 (DE3) by induction with 0.4 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) at 25°C for 4 h. GST-fusion proteins or His-tagged proteins were purified from E. coli lysates using glutathione–Sepharose 4B (GE Healthcare) or nickel-affinity resins (Qiagen), respectively, according to the manufacturers’ instructions. For the pull-down assays, GST or GST-Rbp1p proteins were immobilized on glutathione agarose beads and incubated with His-Dhh1p or His-Dhh1p-dC81 in binding buffer (PBS, pH 7.4, 0.01% Triton X-100) for 2 h at 4°C. The beads were collected and washed four times with 1 ml of binding buffer. Bound protein was then analyzed by 9% SDS–PAGE and western blotting using anti-His monoclonal (BD Biosciences) and anti-GST polyclonal antibodies.
Fluorescence microscopy
Cells were grown in YP-rich medium containing 2% glucose to log phase. For glucose starvation, cells were centrifuged and quickly washed with YP medium lacking glucose and incubated for 20 min in a shaking incubator at 30°C. Images were acquired with a Zeiss Axioskop microscope (Germany) using a Plan-NeoFluar 100×/1.32 objective and Cool snap fx CCD camera (Photometrics) driven by Image-Pro Plus software. For quantification of co-localization of Dhh1p-GFP or its mutants to P-bodies, we randomly selected images, which contained at least 50 cells with Dcp2p-mCh foci in three independent experiments. P-bodies were identified by the presence of Dcp2p-mCh foci. For each GFP-fusion protein, 50 cells with Dcp2p-mCh foci were counted the number of cells with co-localization foci. The percentage of Dcp2p-mCh foci containing cells showing GFP-fusion proteins in foci after glucose starvation was determined. Data are represented as mean of three experiments ± SD.
Northern blotting and mRNA decay assay
For steady-state mRNA analysis, cells were grown in synthetic medium lacking the indicated nutrients and containing 2% glucose to log phase. For mRNA decay analysis, the yeast strain YTC345 carrying a temperature-sensitive RNA polymerase II allele (rpb1-1) (28) was grown at 25°C in synthetic medium lacking the indicated nutrients and containing 2% glucose until an OD600 of ∼1.25 was attained and then shifted to a 37°C water bath shaker to block transcription activity of RNA polymerase II. Aliquots were collected at the indicated time points after transcription shut-off for total RNA isolation and northern blot analysis. Total RNA was prepared by the hot acid phenol method (25) and 10 µg of each total RNA sample was separated by 1.2% agarose gel electrophoresis in the presence of 3.7% formaldehyde. Transfer to nylon membrane (Millipore) was achieved by capillary action with 20× SSC. Blots were probed with 32P-radiolabeled riboprobes directed against the genes as indicated. The level of mRNA in the northern blots was determined by quantifying the intensity of bands using Image J software, in three independent experiments, normalized against the intensity of SCR1 RNA, which is a stable polymerase III transcript (29), and graphed with Microsoft Excel. For quantification of relative POR1 mRNA levels, the values were set to 1 in wild-type yeast. For relative EDC1 mRNA levels, the values were set to 1 in dhh1Δ mutant yeast. Mean values ± SD are shown.
Immunoprecipitation and POR1 mRNA detection by RT–PCR
Exponentially growing cells (OD600 ∼ 10) were disrupted with glass beads in 0.4 ml of extraction buffer [25 mM HEPES–KOH, pH 7.5, 75 mM KCl, 2 mM MgCl2, 0.1% NP-40, 1 mM DTT, 0.2 mg/ml heparin, 20 U/ml DNase (TaKaRa) and 10 mg/ml aprotinin, leupeptin, and pepstatin]. Extracts were cleared by centrifugation at 4000 g for 10 min. Monoclonal anti-HA antibody-conjugated agarose beads (mouse monoclonal anti-HA-Agarose antibody) (Sigma #A2095) was added to the cleared extracts and incubated at 4°C for 4 h. Beads were washed four times with wash buffer (25 mM HEPES–KOH, pH 7.5, 75 mM KCl, 2 mM MgCl2, 0.1% NP-40) and the bound complexes were eluted with 50 mM Tris–HCl, pH 8.0, 100 mM NaCl, 10 mM EDTA, 1% SDS for 10 min at 65°C. The RNA from cell extract (Input) and immunoprecipitates (IP) were extracted by the hot acid phenol method and used as template for RT–PCR. POR1, COR1, or SED1 mRNA was detected by RT–PCR with equal amounts of RNA from each sample using a RvertAid H minus First Strand cDNA Synthesis kit (Fermentas) following conditions suggested by the manufacturer. The number of amplification cycles was adjusted to avoid reaching a plateau phase during PCR. One-third of each reaction was separated on a 1.5% agarose gel and stained with ethidium bromide. The RT–PCR products from precipitated POR1 mRNA were quantified and normalized to RT–PCR products from input mRNA by Image J software. Values represent the mean of three independent experiments ± SD. Statistical significance was assessed by the t-test (**P < 0.001; ***P < 0.0001). Endogenous Rbp1p or HA-tagged proteins from cell extract and immunoprecipitate was separated on a 9% SDS–PAGE gel, blotted and hybridized with anti-Dhh1p or anti-Rbp1p antibody for the presence of proteins.
RESULTS
Dhh1p is involved in Rbp1p-mediated porin mRNA decay
We have previously described that Rbp1p specifically binds to the 3′-UTR of POR1 mRNA and promotes its specific degradation in an Xrn1p-dependent pathway (25, 26). To explore the potential participation of decapping activators Dhh1p, Pat1p, or Lsm1p in Rbp1p-mediated POR1 mRNA decay, we overexpressed Rbp1p in wild-type, dhh1Δ, pat1Δ, lsm1Δ, and xrn1Δ mutant cells, and examined their steady-state POR1 mRNA levels. Northern blot analysis (Figure 1A) shows that overexpression of Rbp1p in dhh1Δ or xrn1Δ mutant cells had no effect on the level of POR1 mRNA, whereas the POR1 mRNA level was reproducibly decreased in Rbp1p-overexpressing pat1Δ, lsm1Δ, or wild-type cells. This result suggests that Dhh1p, but not other decapping activators, participates in Rbp1p-meidated POR1 mRNA decay. To test if Rbp1p and Dhh1p could act on POR1 mRNA decay in the same pathway, we examined the POR1 mRNA level in dhh1Δ and rbp1Δ dhh1Δ strains. Figure 1B shows a significant increase in the POR1 mRNA level in the dhh1Δ strain, but no additional increase in rbp1Δ dhh1Δ double mutant cells, suggesting that Rbp1p and Dhh1p may regulate POR1 mRNA decay cooperatively.
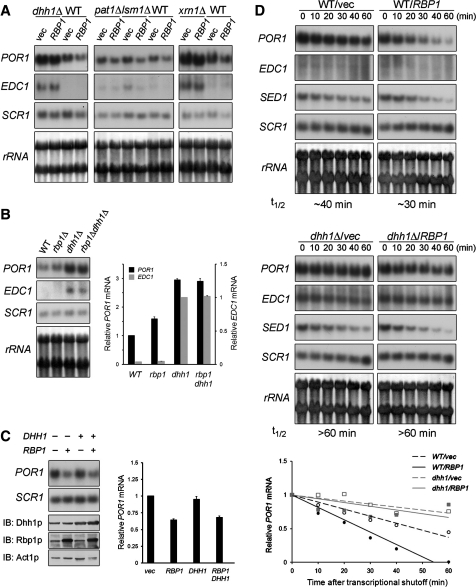
Figure 1.
Rbp1p requires Dhh1p to elicit porin mRNA decay. (A) The steady-state POR1 mRNA levels in dhh1Δ, pat1Δ, lsm1Δ, and xrn1Δ mutant cells overexpressing Rbp1p. BY4741 wild-type or indicated mutant strains transformed with pVT101U or pVT101U-HA-RBP1 plasmid were grown to log phase. Total RNA of these cells was extracted and analyzed by northern blotting. (B) Steady-state levels of POR1 mRNA in wild-type, rbp1Δ, dhh1Δ, and rbp1Δ dhh1Δ mutant cells. BY4741 strains were grown to log phase. Total RNA samples were isolated and analyzed by northern blot. (C) Effect of Dhh1p overexpression on POR1 mRNA steady-state level. BY4741 wild-type strain were transformed with pVT101U plus YEplac181, pVT101U plus YEplac181-HA-RBP1, pVT101U-DHH1 plus YEplac181, or pVT101U-DHH1 plus YEplac181-HA-RBP1 and grown to log phase. Total RNA samples were isolated and analyzed by northern blot. Total proteins were precipitated by TCA and analyzed by western blotting. (D) POR1 mRNA turnover in dhh1Δ strain overexpressing Rbp1p. YTC345 wild-type or dhh1Δ strain transformed with either pVT101U, or pVT101U-HA-RBP1 plasmid was grown to log phase at 25°C and then shifted to 37°C. Total RNA was extracted at each indicated time point after temperature shift and analyzed. t1/2 indicated the half-life of POR1 mRNA. Graphical representation of the POR1 mRNA decay kinetics is shown. The levels of the mRNAs in (B–D) were quantitated as described in ‘Materials and Methods’ section. Mean values ± SD are shown.
We next examined whether overexpression of Dhh1p can decrease the POR1 mRNA level. Figure 1C shows that the POR1 mRNA level was affected by overexpression of Rbp1p, but not Dhh1p, in wild-type cells. In addition, the extent of POR1 mRNA decrease in yeast co-expressing Rbp1p and Dhh1p is similar to that in yeast overexpressing Rbp1p (Figure 1C). These results indicate that Dhh1p alone is not sufficient to decrease POR1 mRNA levels and suggest that Rbp1p plays as a primary regulator for the degradation of POR1 mRNA.
We next examined whether the failure of overexpressing Rbp1p to decrease the steady-state level of POR1 mRNA in the dhh1Δ strain (Figure 1A) is due to its inability to promote POR1 mRNA decay. We took advantage of an RNA polymerase II temperature-sensitive mutant (rpb1-1) strain (28), which allows transcriptional shutoff by temperature shift, and evaluated the mRNA turnover rate in the presence of overexpressed Rbp1p. The half-life of POR1 mRNA in these cells was measured by northern blotting. As shown in Figure 1D, the half-life of POR1 mRNA is prolonged in dhh1Δ mutant cells (∼60 min) and overexpression of Rbp1p in dhh1Δ mutant cells had no effect on the half-life of POR1 mRNA. The turnover of SED1 mRNA, which encodes an abundant cell-wall protein (30), served as a control, indicating that the decay machinery was not impaired by DHH1 deletion. The pronounced stabilization of POR1 mRNA in the dhh1Δ strain overexpressing Rbp1p provides evidence that Dhh1p participates in Rbp1p-mediated POR1 mRNA decay. Together, our data demonstrate that Dhh1p is required for the efficiency of Rbp1p-mediated POR1 mRNA decay.
Dhh1p-mediated EDC1 mRNA level is independent of Rbp1p
Previous studies showed that DHH1, but not PAT1 and LSM1, is involved in regulating the level of EDC1 mRNA (15,31). Therefore, we examined whether Rbp1p is also involved in regulation of EDC1 mRNA decay. Consistent with previous reports (15,31), Figure 1A and B show that the steady-state level of EDC1 mRNA is significantly increased in the dhh1Δ and xrn1Δ strains and the half-life of EDC1 mRNA in the dhh1Δ strain is prolonged for >60 min (Figure 1D). However, unlike POR1 mRNA, the steady-state level and turnover rate of EDC1 mRNA were not altered in cells overexpressing Rbp1p (Figure 1A and D). In addition, the steady-state level of EDC1 mRNA is not affected in rbp1Δ mutant cells (Figure 1B). These results indicate that Rbp1p is not involved in Dhh1p-mediated down-regulation of EDC1 mRNA.
Growth impairment caused by RBP1 overexpression is partially rescued in dhh1Δ mutant cells
Previous results have shown that overexpression of Rbp1p impairs cell growth (24), however, overexpression of the RNA binding-defective RBP1-RRM mutants had no such effect. Therefore, cell growth defects caused by RBP1 overexpression might be a consequence of aberrant biogenesis of specific mRNAs regulated by Rbp1p. To reproduce these observations, we genetically manipulated the expression of RBP1 under the control of a galactose-inducible GAL1 promoter. As expected, no obvious difference in growth rate was observed between wild-type cells expressing RBP1 by its own promoter or by GAL1 promoter when grown on glucose-containing medium; whereas a significant growth defect was observed for cells carrying GAL1 promoter-controlled Rbp1p when grown in galactose-containing medium plates or liquid cultures, as compared to the wild-type cells (Supplementary Figure S1A and S1B). If Dhh1p is required for facilitating specific Rbp1p-mediated mRNA decay, the impaired growth caused by Rbp1p overexpression may be rescued in dhh1Δ mutant cells. We next assayed the effect of Rbp1p overexpression on the growth rate of yeast strains lacking Dhh1p. Although Dhh1p is involved in Rbp1p-mediated POR1 mRNA decay, we found that deletion of DHH1 can only partially rescue cell growth impairment caused by Rbp1p overexpression (Supplementary Figure S1A and S1B). Quantification of differential growth after 25-h incubation in liquid cultures is shown in Supplementary Figure S1C. Furthermore, using the same GAL1 promoter-controlled Rbp1p expression assay, we found that cell growth impairment caused by overexpressing Rbp1p was not affected when PAT1 or LSM1 gene was deleted (Supplementary Figure S1A). These results suggest that Dhh1p, but not Pat1p and Lsm1p, may play a partial role in the mechanism of Rbp1p-mediated mRNA metabolism.
The non-conserved C-terminus of Dhh1p is required for its interaction with Rbp1p
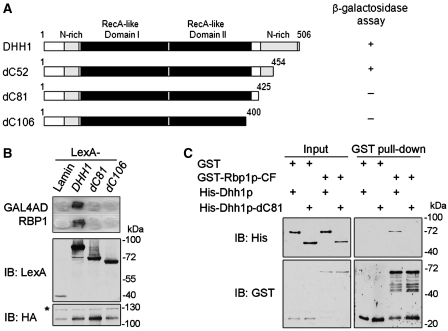
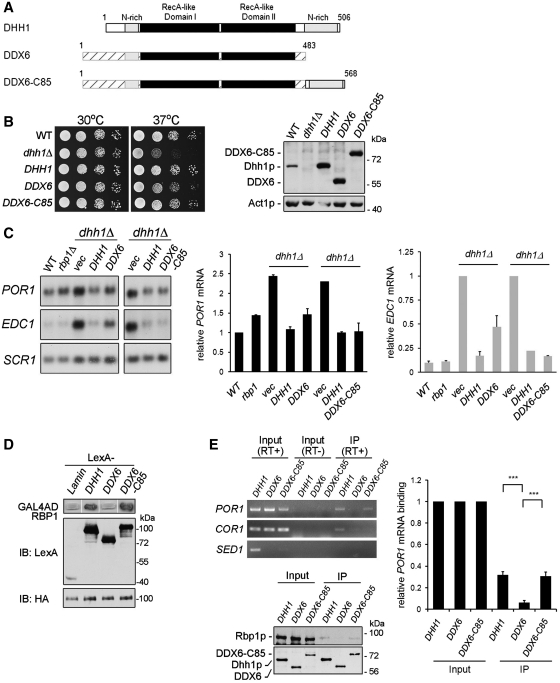
Our previous study has shown that Dhh1p interacted with Rbp1p [(26) and data not shown]. We speculated that Rbp1p could recruit Dhh1p to Rbp1p-specific target mRNAs and then promote their degradation via the general mRNA turnover pathway. Dhh1p consists of two RecA-like domains, like all DEAD-box family members and also contains amino- and carboxy-terminal glutamine-rich extensions [(15) and Figure 2A]. Here, we further narrowed down the interaction region of the Dhh1p C-terminus using yeast two-hybrid assays. By a series of C-terminal deletion mutants of Dhh1p (Figure 2A), and showed that deletion of the C-terminal 81 amino acids of Dhh1p, in the glutamine-rich region, completely abolished the interaction of Dhh1p with Rbp1p (Figure 2B). However, the C-terminal 81 residues of Dhh1p were not sufficient for interaction with Rbp1p (data not shown). To confirm these observations, we also performed in vitro GST pull-down assays using purified recombinant GST-Rbp1p-CF protein, which is a C-terminal fragment of Rbp1p and interacts with Dhh1p (data not shown). Figure 2C shows that purified His-tagged Dhh1p, but not His-tagged Dhh1p-dC81, was efficiently pulled down by GST-Rbp1p-CF. Together, these results indicate that the C-terminal 81 residues mediate the Dhh1p–Rbp1p interaction, which may allow the recruitment of Dhh1p by Rbp1p to specific mRNAs.
Figure 2.
The non-conserved C-terminal 81 residues of Dhh1p are required for its interaction with Rbp1p. (A) Schematic representation of the Dhh1p protein domain structure and C-terminal truncated variants used in yeast two hybrid and in vitro pull-down assays. (B) Dhh1p interacts with Rbp1p through C-terminal 81 residues in yeast two-hybrid assay. YEM1α cells expressing LexA- and Gal4AD-fusion proteins as indicated were used to perform β-galactosidase reporter assay. Immunoblotting shows the expression level of indicated proteins. (C) Dhh1p directly binds Rbp1p. In vitro pull-down assay between purified GST-tagged Rbp1p-CF and His-tagged Dhh1p or Dhh1p-dC81. Anti-His and anti-GST antibodies were used to detect indicated fusion proteins in western blotting. The same amounts of fusion proteins in binding reaction were loaded as input controls.
The non-conserved C-terminus of Dhh1p is involved in regulating Rbp1p-mediated POR1 mRNA decay, but not the EDC1 mRNA level
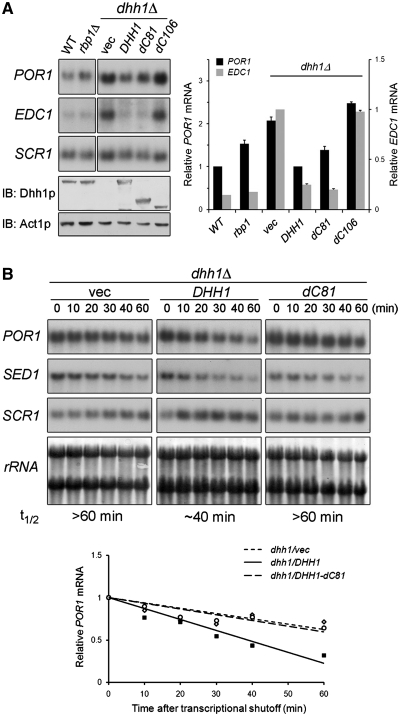
To determine the significance of the in vitro direct interaction between Dhh1p and Rbp1p on POR1 and EDC1 mRNA decay, we examined the levels of these mRNAs in dhh1Δ mutant cells expressing full-length Dhh1p, Dhh1p-dC81, or Dhh1p-dC106. We hypothesized that Dhh1p lacking the C-terminal 81 residues would not restore POR1 mRNA levels as sufficient as wild-type protein caused by loss of its interaction with endogenous Rbp1p. As shown in Figure 3A, the POR1 mRNA level in the dhh1Δ strain expressing Dhh1p-dC81 was not fully restored as compared with the dhh1Δ strain expressing wild-type protein. Compared with the wild-type strain, there was a slight increase in the level of POR1 mRNA in the dhh1Δ strain carrying Dhh1p-dC81, which was comparable to the level observed in the rbp1Δ strain. In contrast to POR1, the level of EDC1 mRNA in the dhh1Δ strain was restored by expression of Dhh1p or Dhh1p-dC81 (Figure 3A). These results indicate that Dhh1p-dC81 failed to induce Rbp1p-mediated regulation of POR1, but was capable of regulating the mRNA level of EDC1. Dhh1p-dC106, lacking the entire non-conserved C-terminal region and part of RecA-like domain II that resulted in a loss of interaction with Rbp1p (Figure 2), could restore the mRNA levels of neither POR1 nor EDC1 (Figure 3A). Western blots indicate that this effect is not due to differences in protein expression level (Figure 3A, lower panel). We further determined whether the incomplete restoration of the POR1 mRNA level in the dhh1Δ strain expressing Dhh1p-dC81 was related to a failure to mediate POR1 mRNA decay. We determined the POR1 mRNA turnover in an rpb1-1 temperature-sensitive mutant strain in which the DHH1 gene was deleted. We expressed single-copy DHH1 or DHH1-dC81 in rpb1-1 dhh1Δ strain and analyzed POR1 mRNA after shutoff transcription. Northern blots (Figure 3B) show that the half-life of POR1 mRNA is prolonged in dhh1Δ mutant cells. The turnover rate of POR1 mRNA was restored to normal (∼40 min) in dhh1Δ mutant cells expressing wild-type Dhh1p, but not in the dhh1Δ strain carrying an empty vector or expressing Dhh1p-dC81 (Figure 3B). Consistent with the steady-state level (Figure 3A), the turnover rate of EDC1 mRNA in the dhh1Δ strain expressing Dhh1p-dC81 was similar to that in cells expressing Dhh1p (data not shown), indicating that Dhh1p-dC81 does not affect the stability of EDC1 mRNA. These results suggest that the insufficient restoration of POR1 mRNA degradation in dhh1Δ mutant cells by Dhh1p-dC81 may be due the loss of interaction with Rbp1p. Taken together, these data indicate that non-conserved C-terminal region of Dhh1p is not critical for regulating EDC1 mRNA level, but is required for Rbp1p-mediated POR1 mRNA decay.
Figure 3.
The C-terminal 81 residues of Dhh1p are required for Rbp1p-mediated POR1 mRNA decay. (A) Steady-state POR1 mRNA levels in dhh1Δ strain expressing Dhh1p, Dhh1p-dC81, or Dhh1p-dC106. BY4741dhh1Δ strain carrying YCplac111, YCplac111-DHH1, YCplac111-DHH1-dC81, or YCplac111-DHH1-dC106 was grown to log phase and total RNA were extracted and analyzed by northern blotting. Total proteins were precipitated by TCA and analyzed by western blotting. (B) Turnover of POR1 mRNA in dhh1Δ strain expressing Dhh1p or Dhh1p-dC81. YTC345dhh1Δ strain carrying YCplac111, YCplac111-DHH1, or YCplac111-DHH1-dC81 was grown to log phase at 25°C and then shifted to 37°C. Total RNA was extracted at each indicated time point after temperature shift and analyzed. t1/2 indicated the half-life of POR1 mRNA. Graphical representation of the POR1 mRNA decay kinetics is shown. The levels of the mRNAs were quantitated as described in ‘Materials and Methods’ section. Mean values ± SD are shown.
The non-conserved C-terminus of Dhh1p is dispensable for the conserved function and localization of Dhh1p
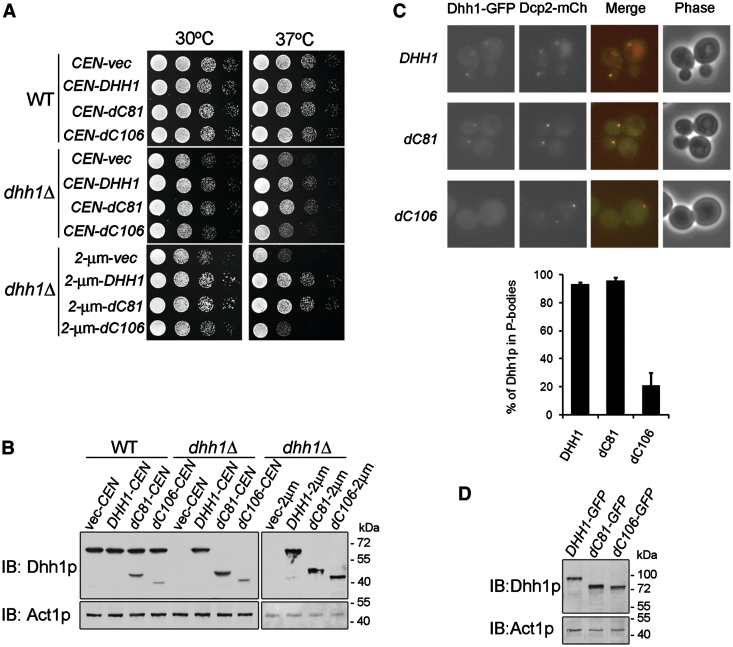
Although the DHH1 gene is not essential for yeast cell viability, the dhh1Δ strain has been shown to display a temperature-sensitive growth phenotype (32). To examine whether the non-conserved C-terminus of Dhh1p possesses other in vivo functions, we expressed C-terminal deletions of Dhh1p in the dhh1Δ strain and determined the functional complement of the temperature-sensitive phenotype. We observed that Dhh1p-dC81, like Dhh1p, fully complemented the capacity of dhh1Δ mutant cells to grow at a non-permissive temperature (Figure 4A). In contrast, expression of Dhh1p-dC106 failed to complement the growth defect in the dhh1Δ strain (Figure 4A). Even though Dhh1p-dC106 was expressed from a high-copy 2 µ plasmid to increase protein levels similar to wild-type Dhh1p (Figure 4B), the complementation of the dhh1Δ strain to grow at the non-permissive temperature still failed (Figure 4A). These data indicate that the non-conserved C-terminal 81 residues are not needed for function of Dhh1p in cell proliferation.
Figure 4.
Dhh1p-dC81 complements the temperature-sensitive phonotype of dhh1Δ strain and efficiently accumulates to P-bodies. (A) Dhh1p-dC81 complements the growth defect of dhh1Δ mutant strain. BY4741 wild-type or dhh1Δ strain expressing Dhh1p, Dhh1p-dC81, or Dhh1p-dC106 from CEN or 2 µ plasmid was grown to log phase, serially diluted and spotted on two plates, which were then separately incubated at 30°C or 37°C for 2 days. (B) Isometric expression level of various Dhh1p mutants. Immunoblotting confirmed the protein expression of various Dhh1p mutants in panel A. (C) Dhh1p-dC81-GFP localizes to P-bodies under glucose deprivation. BY4741 wild-type cells chromosomally expressing C-terminal tagged GFP-fusion Dhh1p, Dhh1p-dC81 or Dhh1p-dC106 with the P-bodies marker Dcp2p-mCherry were grown in YPD medium to log phase and then shifted to medium lacking glucose for 20 min. Co-localization of fluorescence fusion proteins was quantitated as described in ‘Materials and Methods’ section (D) Isometric expression level of various GFP-fusion Dhh1p mutants. Immunoblotting confirms the protein expression of GFP-fusion Dhh1p, Dhh1p-dC81 and Dhh1p-dC106.
Since Dhh1p is a component of cytoplasmic P-bodies and accumulates with Dcp2p in P-bodies under different cell stress conditions (33), we therefore asked whether the non-conserved C-terminal 81 residues of Dhh1p are required for its localization to P-bodies. We expressed C-terminal GFP-fusion Dhh1p, Dhh1p-dC81, or Dhh1p-dC106 in yeast with a P-bodies marker, Dcp2p-mCherry. We examined P-body formation under glucose deprivation, which leads to rapid localization of Dhh1p to Dcp2p-marked P-bodies (33). As shown in Figure 4C, GFP-fusion Dhh1p and Dhh1p-dC81 showed accumulation to cytoplasmic foci and co-localization with the P-bodies marker Dcp2p-mCh under stress. However, the GFP-fusion Dhh1p-dC106 showed reduced accumulation with the P-bodies marker Dcp2p-mCh under the same treatment (Figure 4C). As the protein level of the GFP-fusion Dhh1p-dC106 was equal to that of Dhh1p, we ruled out that its reduced P-bodies localization is due to lower protein levels (Figure 4D). These results indicate the unique C-terminal domain of yeast Dhh1p was not critical for its P-body localization.
Dhh1p associates with POR1 mRNA in vivo via its interaction with Rbp1p
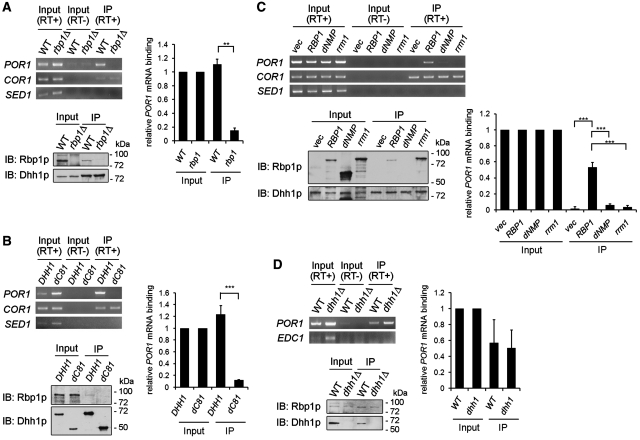
To determine whether Dhh1p can associate with POR1 mRNA in vivo, we precipitated Dhh1p-containing mRNP complexes and detected the presence of POR1 mRNA by RT–PCR. Cell extracts prepared from a strain carrying the chromosomal DHH1 tagged with 3HA was used for immunoprecipitation using anti-HA antibody-conjugated beads. As shown in Figure 5A, anti-HA beads efficiently precipitated the Dhh1p–3HA protein from yeast extracts and endogenous POR1 mRNA was detected from immunoprecipitates of Dhh1p–3HA (Figure 5A).
Figure 5.
Dhh1p associated with POR1 mRNA in vivo requires its interaction with Rbp1p. (A) Dhh1p association with POR1 mRNA depends on the presence of Rbp1p. Dhh1p chromosomally tagged with three HA epitopes in wild-type or rbp1Δ strain was immunoprecipitated and used to perform RT–PCR as described in ‘Materials and Methods’ section. (B) Dhh1p associates with POR1 mRNA through C-terminal 81 amino acids. Dhh1p or Dhh1-dC81 chromosomally tagged with three HA epitopes was immunoprecipitated and used to perform RT–PCR as described in ‘Materials and Methods’ section. (C) Dhh1p association with POR1 mRNA in rbp1Δ mutant was rescued by Rbp1p, but not Rbp1p-dNMP or Rbp1p-rrm1. DHH1-3HA rbp1Δ strains carrying pVT101U, pVT101U-RBP1, pVT101U-RBP1-dNMP, or pVT101U-RBP1-rrm1 were grown and then used to perform immunoprecipitation and RT–PCR as described in ‘Materials and Methods’ section. (D) Rbp1p specifically associates with POR1 mRNA independent of Dhh1p. Rbp1p chromosomally tagged with three HA epitopes in wild-type or dhh1Δ strain was immunoprecipitated and used to perform RT–PCR as described in ‘Materials and Methods’ section. The presence of Dhh1p or Rbp1p was shown in western blots. The level of the RT–PCR products of POR1 mRNA was quantitated as described in ‘Materials and Methods’ section. Mean values ± SD are shown.
We further examined whether POR1 mRNA association with Dhh1p is dependent on the presence of Rbp1p. To address this, we immunoprecipitated Dhh1p–3HA protein complexes from an rbp1Δ strain and analyzed the presence of POR1 mRNA. In contrast to the DHH1-3HA wild-type strain, we did not detect POR1 mRNA from Dhh1p–3HA precipitates in the rbp1Δ strain (Figure 5A). The PCR product was not detected when reverse transcriptase was omitted from the input fraction, indicating that formation of bands in the immunoprecipitate fraction is from cDNA. The interaction of Dhh1p with endogenous Rbp1p was confirmed in Dhh1p–3HA precipitates by immunoblotting using anti-Rbp1p antibody (Figure 5A). Our observations indicate that Dhh1p can in vivo associate with POR1 mRNA and Rbp1p, and suggest that the association of POR1 mRNA to Dhh1p might be through Rbp1p.
It has been suggested that the target RNA specificity of DEAD-box proteins can be determined by their interacting partners (14,16,17). The above observations indicate that the association of the POR1 mRNA with Dhh1p was dependent on its interacting partner Rbp1p. Therefore, it is possible that the C-terminal Rbp1p interacting region of Dhh1p is required for its association with POR1 mRNA. To verify this possibility, we precipitated Dhh1p–3HA or Dhh1p-dC81-3HA protein from DHH1-3HA and DHH1-dC81-3HA strains. Using RT–PCR, we detected POR1 mRNA from the precipitates of DHH1-3HA but not from that of DHH1-dC81-3HA (Figure 6B). Endogenous Rbp1p was only detected in the Dhh1p–3HA protein complex but not the Dhh1p-dC81-3HA complex (Figure 5B). From these data, we propose that Dhh1p associates with POR1 mRNA via its interaction with Rbp1p.
Figure 6.
Mammalian DDX6 fused with the non-conserved C-terminal 85 residues of Dhh1p gains function to confer specific regulation for Rbp1p-mediated porin mRNA. (A) Schematic representation of the Dhh1p, DDX6 and DDX6 chimera protein domain structure. Portion of DDX6 chimera protein, DDX6-C85, derived from Dhh1p are shown. (B) DDX6 and its chimera protein can complement the growth defect of dhh1Δ strain. BY4741dhh1Δ strain expressing Dhh1p, DDX6 or DDX6-C85 was used to perform growth assays as described in Figure 4A. Protein expression level was shown by western blotting. (C) Steady-state POR1 mRNA levels in dhh1Δ strain expressing DDX6 or DDX6-C85. BY4741dhh1Δ strain expressing DDX6 or DDX6-C85 from 2-µ plasmid was grown to log phase and total RNA were extracted and analyzed. The levels of the mRNAs were quantitated as described in ‘Materials and Methods’ section. (D) DDX6 chimera protein DDX6-C85, but not DDX6, interacts with Rbp1p in yeast two-hybrid assay. YEM1α cells expressing LexA- and Gal4AD-fusion proteins as indicated were used to perform β-galactosidase reporter assays. Immunoblotting shows the expression level of indicated proteins. (E) DDX6-C85 associates with POR1 mRNA in vivo through interaction with Rbp1p. BY4741dhh1Δ strain expressing Dhh1p-2HA, DDX6-2HA or DDX6-C85-2HA were used to perform immunoprecipitation and RT–PCR as described in ‘Materials and Methods’ section. The level of RT–PCR products of POR1 mRNA was quantitated as described in ‘Materials and Methods’ section.
To further support this hypothesis, we proposed that the inability of the POR1 mRNA to associate with Dhh1p in the rbp1Δ strain could be rescued by wild-type Rbp1p, but not the mutants of Rbp1p lacking the ability of interacting with Dhh1p or binding to POR1 mRNA. Two Rbp1p mutants were used in this test; Rbp1p-dNMP, a C-terminally deleted Rbp1p, which lost interaction with Dhh1p (Supplementary Figure S2A), and Rbp1p-rrm1, which is defective for POR1 mRNA-binding activity but still possesses ability to interact with Dhh1p [(25) and Supplementary Figure S2B]. We analyzed POR1 mRNA from Dhh1p protein complexes in rbp1Δ strains expressing exogenous wild-type Rbp1p, Rbp1p-dNMP or Rbp1p-rrm1. As shown in Figure 5C, Rbp1p, but not Rbp1p-dNMP, was co-precipitated with Dhh1p and could associate with POR1 mRNA in the rbp1Δ strain. This result supports the notion that interaction with Rbp1p is required for Dhh1p association with POR1 mRNA. Another mutant, Rbp1p-rrm1, also failed to restore the association of POR1 mRNA with Dhh1p in the rbp1pΔ strain, although it could interact with Dhh1p in vivo (Figure 5C). Furthermore, in the absence of Rbp1p, Dhh1p–3HA could associate in vivo with one of the known Dhh1p association mRNAs, COR1 mRNA (34), but not the control SED1 mRNA, whose expression is not sensitive to the dhh1Δ mutant (Figure 1D) and is not immunoprecipitated by Dhh1p–3HA (Figure 5A–C). These results indicate that our immunoprecipitation assay could detect specific in vivo Dhh1p–mRNP complexes. Together, these results indicate that proper POR1 mRNA association with Dhh1p not only required Dhh1p interaction with Rbp1p but also depended on the RNA-binding ability of Rbp1p.
To test if Rbp1p binding to POR1 mRNA requires Dhh1p, we fused three HA tags at the C-terminus of endogenous Rbp1p in dhh1Δ or wild-type strains. Yeast extracts were prepared and precipitated as described above. POR1 mRNA could be detected in Rbp1p–3HA precipitates from both wild-type and dhh1Δ strains (Figure 5D), indicating a Dhh1p-independent association of Rbp1p and POR1 mRNA in vivo. In contrast to POR1 mRNA, EDC1 mRNA was not precipitated from the Rbp1p–3HA protein complex even though its level highly increased in the absence of Dhh1p. We also identified that Dhh1p was co-precipitated from the Rbp1p protein complex (Figure 5D), indicating that POR1 mRNA, but not EDC1 mRNA, specifically associates with the Rbp1p–Dhh1p protein complex.
Mammalian DDX6 fused with the C-terminal non-conserved region of Dhh1p confers Rbp1p-mediated porin mRNA decay in dhh1Δ mutant cells
The Dhh1p protein and its homologs of different species are highly conserved in function, because Xenopus Xp54 (32) and mammalian RCK/p54/DDX6 (35–37) could complement the growth defects of a yeast dhh1Δ mutant. We therefore asked if human DDX6 is capable of substituting for Dhh1p in mediating POR1 mRNA decay in yeast cells. As shown in Figure 6C, the POR1 mRNA level was not fully restored in the dhh1Δ strain expressing human DDX6. This result was similar to the effect of POR1 mRNA level in the dhh1Δ strain expressing Dhh1p-dC81 (Figure 3A). Although the non-conserved C-terminal region of Dhh1p was not sufficient to interact with Rbp1p, we proposed that it contains a critical element to determine the specific interacting structure for Dhh1p. We further tested whether the human DDX6 fused with the non-conserved C-terminus of Dhh1p can confer Rbp1p-mediated POR1 mRNA decay in dhh1Δ strain. We generated a DDX6-Dhh1p(C-terminus) fused protein, DDX6-C85(Dhh1p), containing the C-terminal 85 residues of Dhh1p appended to the C-terminus of DDX6, as illustrated in Figure 6A. Northern blotting shows that, in contrast to DDX6, DDX6-C85(Dhh1p) can restore the level of POR1 mRNA in dhh1Δ strain (Figure 6C). Interestingly, the level of EDC1 mRNA in the dhh1Δ strain was restored by DDX6 and DDX6-C85(Dhh1p) although at varied degrees (Figure 6C), suggesting that a certain specific motif in Dhh1p might be important to regulate the EDC1 mRNA level. These data indicate that DDX6 fused with the non-conserved C-terminus of Dhh1p could complement the function of Dhh1p in dhh1Δ mutant cells. We also confirmed prior reports for phenotype complementation (35–37) that DDX6 and DDX6-C85(Dhh1p) restored the capacity of dhh1Δ strain to grow at a non-permissive temperature (Figure 6B).
The ability of DDX6-C85(Dhh1p) to mediate porin mRNA decay suggested that it might possess the ability to interact with Rbp1p and subsequently to associate with POR1 mRNA in vivo. To test this hypothesis, we assayed the interaction between Rbp1p and DDX6-C85(Dhh1p) using the yeast two-hybrid assay. Figure 6D shows that DDX6-C85(Dhh1p) could interact with Rbp1p. We further examined what besides the C-terminal region is sufficient for the Rbp1p interaction. Based on the structural regions of Dhh1p (15,16), we constructed two deletion mutants of Dhh1p, Dhh1p-VI and Dhh1p-Ct, which contain motif VI or RecA-like domain II, adjacent to non-conserved C-terminal region, respectively (Supplementary Figure S3A). We found that Dhh1p-Ct, but not Dhh1p-VI, had similar interaction intensity as full-length Dhh1p (Supplementary Figure S3B). We also demonstrated that DDX6-Ct-C85, which contains RecA-like domain II fused with non-conserved C-terminal region of Dhh1p, but not DDX6-Ct, is sufficient for the Rbp1p interaction. This result indicates that the RecA-like domain II contributes to the interaction between Dhh1p and Rbp1p. Next, we performed RNA immunoprecipitation in the dhh1Δ strain expressing Dhh1p-2HA, human DDX6-2HA or DDX6-C85(Dhh1p)-2HA. As shown in Figure 6E, we detected POR1 mRNA from Dhh1p–2HA and DDX6-C85(Dhh1p)-2HA, but not from DDX6–2HA precipitates. Immunoblotting shows that endogenous Rbp1p can be detected in Dhh1p–2HA and DDX6-C85(Dhh1p)–2HA, but not DDX6–2HA precipitates (Figure 6E). Furthermore, the interaction between Dhh1p or DDX6-C85 and endogenous Rbp1p was insensitive to RNase treatment (Supplementary Figure S4), indicating that the interaction is not due to RNA bridging. These results suggest that DDX6-C85(Dhh1p), but not DDX6, can mediate the POR1 mRNA decay through interaction with endogenous Rbp1p via its fusion with the non-conserved C-terminus of Dhh1p.
DISCUSSION
In this report, we show that the non-conserved C-terminus of Dhh1p has a role in defining specific interactions with Rbp1p and promotes POR1, but not EDC1 mRNA decay. Moreover, mammalian DDX6, after being fused with the non-conserved C-terminus of Dhh1p, possesses the ability to interact with Rbp1p and to confer Rbp1p-mediated POR1 mRNA decay in a dhh1Δ strain. We propose that promoting a distinct mRNA decay by Dhh1p may be defined by specific RNA-binding proteins and/or regulating factors that interact with its non-conserved extended C-terminus.
Dhh1p participates in Rbp1p-mediated POR1 mRNA decay
By analyzing the POR1 mRNA level in deletion mutants, we found that the magnitude of the effect of DHH1-deletion, but not PAT1- or LSM1-deletion, on the POR1 mRNA level was comparable to that of XRN1-deletion. In addition, only in dhh1Δ, but not pat1Δ or lsm1Δ mutant cells, overexpression of Rbp1p failed to destabilize POR1 mRNA. Dhh1p physically interacts with Pat1p (7). Although both proteins carry out similar functions, they now appear to operate through separate mechanisms, which are not known in detail (16). Consistent with this, our data show that POR1 mRNA levels are not affected in PAT1-deleted yeast. Interestingly, although both dhh1Δ and rbp1Δ strains showed delays in the POR1 mRNA decay rate, only the dhh1Δ strain exhibited a corresponding large increase in steady-state POR1 mRNA levels. This discrepancy between decay rates and steady-state levels is similar to what has been reported for EDC1 mRNA in the dhh1Δ strain and some mRNAs in dcp1Δ strains (31,38). This result suggests that there might be Dhh1p-dependent and Rbp1p-independent pathways for POR1 mRNA degradation. It also implies that other mRNA-binding regulator(s) can cooperate with Dhh1p to regulate POR1 mRNA degradation. Furthermore, overexpression of Dhh1p does not lead to destabilization of POR1 mRNA. This indicates that Dhh1p is not the limiting factor in POR1 mRNA decay, and that Dhh1p requires Rbp1p or other RNA-binding molecules to promote POR1 mRNA degradation.
In agreement with a previous report (31), we observed a large increase in EDC1 mRNA levels in the dhh1Δ strain. Our preliminary data showed that the decrease in RNA decay efficiency of both POR1 and EDC1 mRNA in the dhh1Δ strain was rescued by wild-type, but not helicase mutants of Dhh1p, signifying the importance of Dhh1p helicase activity in the degradation of both mRNAs. However, unlike POR1 mRNA, the level of EDC1 mRNA was not affected by Rbp1p. Moreover, deletion of DHH1 can only partially rescue cell growth impairment caused by Rbp1p overexpression, suggesting that Dhh1p only plays a partial role in Rbp1p-mediated mRNA metabolism. Together, we believe that Rbp1p is not a universal partner for the Dhh1p-dependent mRNA degradation pathway and that Dhh1p might modulate the decay of different mRNAs through cooperating with different molecules.
The non-conserved C-terminus of Dhh1p is involved in Rbp1p-mediated POR1 mRNA
The extended C-terminal non-conserved region of Dhh1p has been proposed to contribute to protein–protein interactions and confer substrate specificity, thereby facilitating unique functions (13,14,16). In this report, we found that the C-terminal non-conserved region is involved in Rbp1p-mediated POR1 mRNA and is necessary for its interaction with Rbp1p. Previous study has shown that a Dhh1p deleted C-terminal 81 residues still conforms to a typical helicase structure and retains its helicase-dependent function (15), suggesting that the interaction loss of Dhh1p-dC81 with Rbp1p was not due to disruption of overall protein conformation.
An interesting issue is the diversity of interactions that may allow mRNA specific-binding proteins to recruit the mRNA degradation machinery. Several studies have shown that some mRNA-specific regulatory proteins recruit the general repression and decay machinery to specific transcripts, leading to transcript destabilization. Yeast Cth2p was shown to interact with the C-terminal catalytic core domain and non-conserved region of Dhh1p and recruit it to ARE-containing mRNAs to promote mRNA decay (39). The similar region of Dhh1p also physically interacts with the Dcp1p–Dcp2p decapping complex and the Edc3p scaffold protein, thereby promoting the transition of mRNAs from translation to decapping and 5′–3′ degradation at specific intracellular sites known as processing bodies (P-bodies) (40). Mpt5p was reported to interact with Pop2p, thereby inducing the recruitment of the Ccr4p, Dhh1p and Dcp2p to specific mRNAs to promote mRNA deadenylation and decay (41). Our data, consistent with previous reports, show that the recruitment of Dhh1p to Rbp1p is mediated by its non-conserved C-terminus, thereby promoting the Rbp1p-mediated POR1 mRNA decay.
The non-conserved C-terminus of Dhh1p is dispensable for conserved function of Dhh1p
Cell growth analyses and RNA decay assays show that Dhh1p-dC81 is sufficient to complement the growth defect and restore EDC1 mRNA degradation in a dhh1Δ mutant strain. This observation is parallel to previous reports that the vertebrate orthologs Xenopus Xp54 (32) and mammalian RCK/p54/DDX6 (36), which do not possess a C-terminal extension like Dhh1p, can complement the growth defect in a dhh1Δ mutant strain. Furthermore, expression of DDX6 in a dhh1Δ mutant strain can partially restore the efficiency of degradation of EDC1 mRNA. Therefore, we suggest that the non-conserved C-terminal region of Dhh1p is not required for known conserved DEAD-box helicase Dhh1p function.
The non-conserved C-terminal region of Dhh1p is a Q/N-rich stretch. Some mRNA-specific regulatory proteins that recruit the translation repression and 5′ to 3′ decay machinery and localize to cytoplasmic P-bodies also contain similar Q/N-rich sequences. These Q/N-rich regions within those proteins have been reported to be both required and sufficient for protein aggregation into foci under certain stress conditions (19). However, Reijns et al. (19) found that GFP-tagged C-terminal non-conserved region of Dhh1p do not aggregate under stresses and GFP-tagged Dhh1p-dC (1–427) could still accumulate to the P-bodies. Among orthologs of Dhh1p, neither Drosophila Me31B nor Caenorhabditis CGH1 contain such Q/N-rich sequences at their C-termini. Nevertheless, these proteins could localize to granule-like subcellular structures similar to P-bodies (21,22). Compatibly, our data demonstrate that the C-terminal non-conserved Q/N-rich region of Dhh1p is not responsible for accumulation into P-bodies under stress conditions.
Dhh1p–Rbp1p interaction is required for the association of POR1 mRNA with Dhh1p
DEAD-box RNA helicase proteins are believed to modulate the structure of RNAs and ribonucleoprotein complexes by disrupting RNA helices and RNA–protein interactions. In addition to duplex unwinding, RNA helicases display an array of additional activities. Most prominently, several RNA helicases have been directly shown to displace other proteins from RNA in an active, ATP-dependent fashion (42). Protein displacement or RNP remodeling is thought to be central to the physiological function of RNA helicases, because RNAs are generally bound to other proteins in vivo (10). DEAD-box helicase proteins generally possess in vitro RNA-binding ability (15), however, in vivo specific RNA substrates are not known for most of the DEAD-box helicase proteins (11). Hogan et al. (34) have systematically searched for RNAs directly associated with purified putative RNA-binding proteins. Less than 10 RNAs were identified to be associated directly with Dhh1p. This lack of RNA specificity in vitro may reflect the fact that the biologically relevant substrate is an RNP, a rare RNA conformation, or an RNA that is recognized only in the context of a larger complex (43).
We previously have shown that Rbp1p exhibits obvious characteristics of known RNA-binding motifs, RRMs and mutation of conserved residues in these motifs result in defective POR1 mRNA binding (25). Analysis from immunoprecipitation of RNP complexes showed that Dhh1p association with POR1 mRNA in vivo requires its interaction with Rbp1p. In an rbp1Δ strain or cells expressing only RNA-binding deficient Rbp1p, Dhh1p failed to associate with POR1 mRNA. This result indicates that Dhh1p may not be able to form a stable complex with mRNAs in vivo by itself. Our data support the notion that proper POR1 mRNA association with Dhh1p not only requires Dhh1p interaction with Rbp1p but also depends on the RNA-binding ability of Rbp1p.
Highly conserved DEAD-box domain in Dhh1p/RCK/Xp54/DDX6 mediates non-conserved C-terminus of Dhh1p to modulate transcript-specific decay
Dhh1p belongs to a highly conserved DEAD-box helicase subfamily that includes human RCK/p54/DDX6, Xenopus Xp54, Drosophila Me31B and Caenorhabditis CGH-1 (16). These proteins share high sequence homology except at the extended N- and C-termini flanking the conserved catalytic domains (16). The non-conserved C-terminal region of Dhh1p is a unique extension sequence in yeast. In contrast, the mammalian RCK/p54/DDX6 has a long N-terminal extension and only a short C-terminal region.
Through expressing DDX6 in a dhh1Δ strain, we found that in addition to complementing the growth defect of the dhh1Δ strain, DDX6 also partially promoted the efficiency of degradation of EDC1 mRNA; whereas, it lacks the ability to interact with Rbp1p and elicit Rbp1p-mediated POR1 mRNA degradation. Interestingly, we found that the addition of the C-terminal non-conserved region of Dhh1p to DDX6 is sufficient to interact with Rbp1p and introduce POR1 mRNA as a target in yeast. Because the RecA-like domain II of DDX6 fused with the non-conserved C-terminal region of Dhh1p can interact with Rbp1p, we reason that the evolutionarily conserved helicase domain may assist the C-terminal region of Dhh1p to interact with other factors, thereby modulating transcript-specific decay.
In S. cerevisiae, a number of mechanisms exist by which the decapping machinery can be selectively recruited to specific mRNA substrates (31,39,44,45). It is interesting to note that the non-conserved C-terminal domain of Dhh1p may be responsible for the interaction with Dcp2p, Edc3p, Cth2p and Rbp1p, which raises the question of whether these interactions can be regulated by cellular signaling. Consistent with this notion, our preliminary data show that Rbp1p is phosphorylated at more than 10 residues. It would also be interesting to elucidate which phosphorylation sites on Rbp1p function as mRNA decay activation domains.
Orthologs of Dhh1p in various organisms have been shown to carry out a diverse array of cellular processes while functional conservation has also been observed. Our findings suggest that the unique C-terminal extension of Dhh1p facilitates a species-specific function in yeast. Whether other non-conserved N- and C-terminal extensions of each ortholog could interact with certain RNA-binding proteins to carry out more species-specific functions and when these specific functions emerge or diminish during evolution would be an interesting future subject. On the other hand, some mRNA specific-binding proteins may be responsible for conserved functions of Dhh1p and DDX6, recruiting them to mRNAs such as EDC1. In conclusion, the data presented in this report elucidate the role of the species-specific non-conserved C-terminus of Dhh1p in defining specific interaction with an RNA-binding protein, Rbp1p, to promote POR1 mRNA decay. The challenge for future studies will be to understand how mRNA-specific regulators recruit Dhh1p and coordinate multiple mechanisms of post-transcriptional regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables I–II, Supplementary Figures 1–4.
FUNDING
Funding for open access charge: National Health Research Institutes, Taiwan, R.O.C. (NHRI-EX94-9222BI, NHRI-EX96-9513SI) and Yung-Shin Biomedical Research Funds (to F.-J.S.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Woan-Yuh Tarn, Tien-Hsien Chang, Randy Haun and Chun-Fang Huang for critical reading of the manuscript.
REFERENCES
- 1.Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 2.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 3.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell. Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 4.Coller J, Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 5.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol. Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 7.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer N, Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linder P. Dead-box proteins: a family affair–active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell. Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 12.Silverman E, Edwalds-Gilbert G, Lin RJ. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 13.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng Z, Coller J, Parker R, Song H. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA. 2005;11:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weston A, Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 2006;34:3082–3094. doi: 10.1093/nar/gkl409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minshall N, Thom G, Standart N. A conserved role of a DEAD box helicase in mRNA masking. RNA. 2001;7:1728–1742. doi: 10.1017/s135583820101158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- 22.Navarro RE, Shim EY, Kohara Y, Singson A, Blackwell TK. cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development. 2001;128:3221–3232. doi: 10.1242/dev.128.17.3221. [DOI] [PubMed] [Google Scholar]
- 23.Smillie DA, Sommerville J. RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles. J. Cell Sci. 2002;115:395–407. doi: 10.1242/jcs.115.2.395. [DOI] [PubMed] [Google Scholar]
- 24.Lee FJ, Moss J. An RNA-binding protein gene (RBP1) of Saccharomyces cerevisiae encodes a putative glucose-repressible protein containing two RNA recognition motifs. J. Biol. Chem. 1993;268:15080–15087. [PubMed] [Google Scholar]
- 25.Buu LM, Jang LT, Lee FJ. The yeast RNA-binding protein Rbp1p modifies the stability of mitochondrial porin mRNA. J. Biol. Chem. 2004;279:453–462. doi: 10.1074/jbc.M309278200. [DOI] [PubMed] [Google Scholar]
- 26.Jang LT, Buu LM, Lee FJ. Determinants of Rbp1p localization in specific cytoplasmic mRNA-processing foci, P-bodies. J. Biol. Chem. 2006;281:29379–29390. doi: 10.1074/jbc.M601573200. [DOI] [PubMed] [Google Scholar]
- 27.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 28.Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felici F, Cesareni G, Hughes JM. The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol. Cell. Biol. 1989;9:3260–3268. doi: 10.1128/mcb.9.8.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimoi H, Kitagaki H, Ohmori H, Iimura Y, Ito K. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J. Bacteriol. 1998;180:3381–3387. doi: 10.1128/jb.180.13.3381-3387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muhlrad D, Parker R. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005;24:1033–1045. doi: 10.1038/sj.emboj.7600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tseng-Rogenski SS, Chong JL, Thomas CB, Enomoto S, Berman J, Chang TH. Functional conservation of Dhh1p, a cytoplasmic DExD/H-box protein present in large complexes. Nucleic Acids Res. 2003;31:4995–5002. doi: 10.1093/nar/gkg712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alves-Rodrigues I, Mas A, Diez J. Xenopus Xp54 and human RCK/p54 helicases functionally replace yeast Dhh1p in brome mosaic virus RNA replication. J. Virol. 2007;81:4378–4380. doi: 10.1128/JVI.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westmoreland TJ, Olson JA, Saito WY, Huper G, Marks JR, Bennett CB. Dhh1 regulates the G1/S-checkpoint following DNA damage or BRCA1 expression in yeast. J. Surg. Res. 2003;113:62–73. doi: 10.1016/s0022-4804(03)00155-0. [DOI] [PubMed] [Google Scholar]
- 37.Bergkessel M, Reese JC. An essential role for the Saccharomyces cerevisiae DEAD-box helicase DHH1 in G1/S DNA-damage checkpoint recovery. Genetics. 2004;167:21–33. doi: 10.1534/genetics.167.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muhlrad D, Parker R. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA. 1999;5:1299–1307. doi: 10.1017/s1355838299990829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedro-Segura E, Vergara SV, Rodriguez-Navarro S, Parker R, Thiele DJ, Puig S. The Cth2 ARE-binding protein recruits the Dhh1 helicase to promote the decay of succinate dehydrogenase SDH4 mRNA in response to iron deficiency. J. Biol. Chem. 2008;283:28527–28535. doi: 10.1074/jbc.M804910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell. Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 42.Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linder P, Tanner NK, Banroques J. From RNA helicases to RNPases. Trends Biochem. Sci. 2001;26:339–341. doi: 10.1016/s0968-0004(01)01870-9. [DOI] [PubMed] [Google Scholar]
- 44.Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 45.Dong S, Li C, Zenklusen D, Singer RH, Jacobson A, He F. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol. Cell. 2007;25:559–573. doi: 10.1016/j.molcel.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.