Abstract
RAD18, a RING-type ubiquitin ligase (E3) that plays an essential role in post-replication repair, possesses distinct domains named RING, UBZ, SAP and the RAD6-binding domain (R6BD) and forms a dimer. RAD6, an ubiquitin-conjugating enzyme (E2), stably associates with R6BD in the C-terminal portion. In this study, we established a method to distinguish between the two subunits of RAD18 by introduction of different tags, and analyzed mutant complexes. Our results, surprisingly, demonstrate that RAD6A and RAD18 form a ternary complex, RAD6A–(RAD18)2 and the presence of only one R6BD in the two RAD18 subunits is sufficient for ternary complex formation and the ligase activity. Interestingly, ligase activity of a mutant dimer lacking both R6BDs is not restored even with large amounts of RAD6A added in solution, suggesting a requirement for precise juxtaposition via interaction with R6BD. We further show that mutations in both subunits of either RING or SAP, but not UBZ, strongly reduce ligase activity, although inactivation in only one of two subunits is without effect. These results suggest an asymmetric nature of the two RAD18 subunits in the complex.
INTRODUCTION
Ubiquitin ligases (E3s) catalyze the transfer of ubiquitin from E2 (ubiquitin-conjugating enzyme)–ubiquitin conjugates to lysine residues in target proteins. A subset of E3s contains a RING (really interesting new gene) domain, which binds to E2–ubiquitin conjugates and seems to activate thioester bonds (1,2). Some RING-type E3s are known to form heterodimers such as BRCA1–BARD1 (3–5), Ring1b-Bmi1 (6,7) and MDM2–MDMX (8–10), while others like cIAP2 (11) and RNF4 (12) act as homodimers. The heterodimeric RING-type E3s are composed of active and inactive RING domains, and dimerization enhances the ligase activity, suggesting that the pairing itself is very important for enzyme function (5,6,9).
Through a RAD6-binding domain (R6BD) located in its C-terminal region, the RAD18 RING-type ubiquitin ligase forms a stable complex with a specific E2, RAD6 (13–17). Since RAD6 also contacts the RING domain near the N-terminal of RAD18 for catalytic function (2,15,18), it could interact with two distinct domains of RAD18 simultaneously. Such interactions between an E2 and an E3 are quite unique to RAD6–RAD18. Recently, it has been reported that interaction between R6BD and RAD6 blocks the intrinsic activity of RAD6 in forming ubiquitin chains, ensuring mono-ubiquitination of PCNA (19). Previous studies have also reported that RAD18 forms a dimer and suggested the RAD6–RAD18 complex to be a dimer of RAD6–RAD18 heterodimer (15,20,21), hereafter designated as (RAD6–RAD18)2.
RAD6 and RAD18 play an essential role in post-replication repair of damaged DNA via ubiquitination of proliferating cell nuclear antigen (PCNA) at Lys164 (22,23). The RAD6–RAD18 complex itself catalyzes mono-ubiquitination of PCNA in vitro (24–28). The mono-ubiquitinated PCNA appears to enhance lesion bypass replication by stimulation of entry of translesion DNA polymerases at stalled 3′-ends, through interactions between ubiquitin-binding domains of the polymerases and ubiquitin moieties of mono-ubiquitinated PCNA (24,26,29–31). Furthermore, RAD18 features a pol η-binding domain that is important for recruiting pol η to stalled 3′-ends (17,32).
RAD18 has two other domains UBZ (ubiquitin-binding zinc finger) (33–35) and SAP (SAF-A/B, Acinus and PIAS) (15,36–38). The former is required for accumulation of RAD18 at damage sites (34,37,39) and has affinity for Ub chains (33,34) suggesting specific binding to damage-associated poly-ubiquitinated proteins (39). The SAP domain possesses DNA-binding activity (15,38), although it appears to be unnecessary for accumulation of RAD18 at damage sites (37,39). Rather, it is crucial for pol η focus formation (37) possibly depending on PCNA ubiquitination, which is attributed to an essential role of the SAP domain in ligase activity (38,39).
Although multiple domains of RAD18 are clearly concerned with ligase activity, it is unknown how the distinct entities of the two RAD18 subunits interact with each other for enzyme function. In the present study, we established a method to analyze the structure and functions of the human RAD6A–RAD18 complex and demonstrated an asymmetric nature of the two RAD18 molecules in the complex.
MATERIALS AND METHODS
Plasmids
Expression plasmids for RAD6A–RAD18, E1, ubiquitin, PCNA and RFC were as described previously (26,40–42). For overproduction of human RAD6A, RAD6A–HisRAD18, FLAGRAD6A–HisRAD18, the genes were cloned into pET20b(+) (Novagen) to yield pET-RAD6A and pET-RAD6A/hisRAD18, pET-flagRAD6A/hisRAD18, respectively. To make an expression plasmid compatible with pET plasmids in Escherichia coli cells, the entire coding unit of pET-RAD6A/RAD18 (42) was cloned into pACYC Duet1 (Novagen) to yield pAC-RAD6A/RAD18. Expression plasmids for overproduction of RAD6A–FLAGRAD18 and FLAGRAD6A–FLAGRAD18, pAC-RAD6A/flagRAD18 and pAC-flagRAD6A/flagRAD18 were generated, respectively. In those plasmids, His-tagged sequences were taken from pET15 and all tagged sequences were attached to immediately before the start codons of the respective genes. Expression plasmids in human cells for RAD6 were cloned in pCMV, and for RAD18 in pCDNA3 flag and pCAGGS (43).
Proteins
Proteins used in this study were overproduced in E. coli cells. During all purification steps, monitoring was done by SDS–PAGE followed by staining with Coomassie Brilliant Blue R-250 (CBB), or western blotting. Protein concentrations were determined by Bio-Rad protein assay using BSA (Bio-Rad) as the standard. PCNA, RFC, E1, ubiquitin and RAD6A–RAD18 complex were purified as described previously (26,40,41). Detailed procedures for purification of recombinant proteins established in this study are described in Supplementary Data.
Sucrose density gradient sedimentation
Sucrose density gradient sedimentation was performed as described earlier (44). Purified RAD6A–RAD18 complexes (1.7 µg), were sedimented through 2 ml of 10–40% sucrose gradient in buffer A containing 300 mM NaCl by centrifugation at 55 000 rpm for 20 h in a TLS 55 rotor (Beckman) at 4°C and fractions (100 µl) were collected from the bottom of the tube and analyzed by SDS–PAGE. Gel bands were stained with CBB and quantified using Multi Gauge software Version 3.0 (FUJIFILM). Sedimentation coefficients were determined relative to those of standard proteins sedimented in parallel gradients.
Antibodies
To obtain polyclonal antibodies against RAD18, truncated His-tagged RAD18 proteins (127–255 amino acids) were expressed in BL21 (DE3) (45), purified and used to immunize rabbits. Anti-Penta-His monoclonal (Qiagen, 34660), anti-FLAG M2 monoclonal (Sigma, F3165), anti-RAD6 polyclonal (Abcam, ab31917) and anti-PCNA polyclonal (Santa Cruz, sc-7907) antibodies were purchased.
PCNA-mono-ubiquitination assays
The standard reaction mixture (25 µl) contained 20 mM HEPES–NaOH (pH 7.5), 50 mM NaCl, 0.2 mg/ml BSA, 1 mM DTT, 10 mM MgCl2, 1 mM ATP, 100 ng poly(dA)-oligo(dT), PCNA (1 pmol), RFC (350 fmol), E1 (850 fmol), ubiquitin (170 pmol) and the indicated amounts of RAD6A–RAD18 complex. Reaction mixtures were prepared on ice then incubated at 30°C for 10 min. The reactions were terminated by addition of 2× SDS sample buffer containing 25 mM EDTA. Ubiquitination of PCNA was assessed by western analysis, detected by an ECL chemiluminescence kit (GE Healthcare Life Science).
RESULTS
Physicochemical properties of the human RAD6A–RAD18 complex
To study the subunit composition of RAD6A–RAD18 complex, we first determined the Stokes' radius and sedimentation coefficient of the purified RAD6A–RAD18 complex (26) by gel filtration and sucrose density gradient centrifugation, respectively (Table 1). The obtained value of Stokes' radius (62 Å) corresponds to an apparent molecular mass of 490 kDa, but that of the sedimentation coefficient (5.0 S) corresponds to an apparent molecular mass of 70 kDa. The large difference between these two values suggests that the complex does not have a compact globular shape. Employing the method described by Siegel and Monty (46), we estimated the molecular mass of the RAD6A–RAD18 complex to be 131 kDa (Table 1). Since calculated molecular masses of RAD6A and RAD18 are 17.3 and 56.2 kDa, respectively, the total of molecular mass should be 73.5 kDa for RAD6A–RAD18 and 147 kDa for (RAD6A–RAD18)2. Although the value estimated from the experiments was close to that of (RAD6A–RAD18)2, we further investigated the stoichiometry of RAD6A and RAD18 in the complex.
Table 1.
Physicochemical properties of the RAD6A–RAD18 complex
aDetermined by Superdex 200 gel filtration using the size markers ferritin (61.0 Å), aldolase (48.1 Å), albumin (35.5 Å) ovalbumin (30.5 Å) and ribonuclease A (16.4 Å), and the data were based on A280 values monitored during the chromatography.
bDetermined with ferritin (17.6 S), catalase (11.3 S), aldolase (7.4 S), albumin (4.2 S) and ribonuclease A (1.8 S) as standards, and the data were based on the SDS–PAGE gel profile.
cEstimated from the Stokes' radius and the sedimentation coefficient assuming a partial specific volume of 0.73 (46).
Direct evidence that the RAD6A–RAD18 complex contains two RAD18 molecules
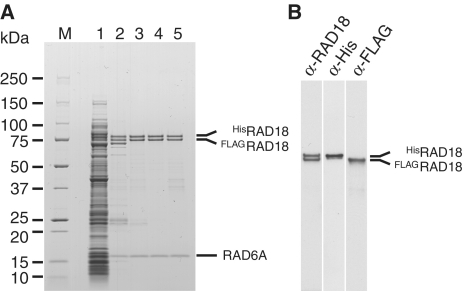
To obtain direct evidence that the RAD6A–RAD18 complex contains two molecules of RAD18, we co-expressed both His-tagged RAD18 and FLAG-tagged RAD18 genes together with RAD6A gene in the same E. coli cells. Three different complexes containing HisRAD18–HisRAD18, FLAGRAD18–FLAGRAD18 or HisRAD18–FLAGRAD18 would be expected if we assume that the complex should contain two molecules of RAD18. When the cell lysate was loaded to a Ni-chelating column, we found some FLAGRAD18 to be absorbed to the column and eluted together with approximately equal amounts of HisRAD18 at a lower imidazole concentration (Figure 1A, lane 2), whereas the remainder of the HisRAD18 was eluted at a higher imidazole concentration (see Supplementary Materials and Methods section). Since it was expected that the former was HisRAD18–FLAGRAD18 hetero complex and the latter was HisRAD18 homo complex, the former was further purified through a heparin column (Figure 1A, lane 3) and analyzed by gel filtration (Figure 1A, lane 4). The elution profile was the same as that of untagged RAD6A–RAD18 complex (Table 1 and see also Figure 6C), suggesting the tag sequences do not affect the overall structure of the complex. While western blotting with anti-Penta-His and anti-FLAG antibodies specifically detected HisRAD18 and FLAGRAD18 proteins, respectively (Figure 1B), blotting with anti-RAD18 antibodies detected the two proteins equally, thereby indicating that the purified complex contains HisRAD18 and FLAGRAD18 at a 1:1 ratio. To further verify that RAD6A, HisRAD18 and FLAGRAD18 form a complex, a fraction eluted from gel filtration was applied to FLAG-affinity chromatography. The result demonstrated that the three proteins were adsorbed to the anti-FLAG M2 affinity gel and eluted with the FLAG peptide (Figure 1A, lane 5). We thus conclude that the RAD6A–RAD18 complex contains two RAD18 molecules.
Figure 1.
Purification of the RAD6A–HisRAD18–FLAGRAD18 complex. (A) Pooled fractions eluted from respective columns were analyzed by SDS–PAGE followed by staining with CBB. Lane 1, cell lysate; lane 2, Ni-chelating column; lane 3, heparin column; lane 4, gel-filtration column; lane 5, anti-FLAG affinity column. Molecular masses of each marker (lane M) are shown to the left of the gel. (B) Western analysis of the pooled fraction eluted from the gel filtration column. Membranes were probed with the indicated antibodies.
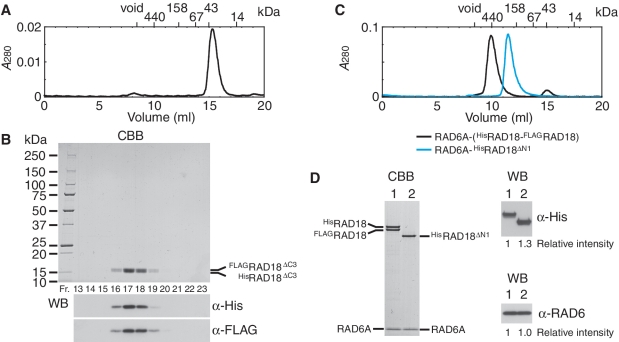
Figure 6.
Analysis of RING domain for dimerization of RAD18. (A) Elution profile of HisRAD18ΔC2–FLAGRAD18ΔC2 complexes from a Superdex 200 gel filtration column. The size markers, ferritin (440 kDa), aldolase (158 kDa), albumin (67 kDa), ovalbumin (43 kDa) and ribonuclease A (14 kDa) were eluted in 10.09, 12.26, 13.74, 14.98 and 17.56 ml, respectively. The complex was eluted in 15.35 ml, estimated the apparent molecular mass of the complex to be 37 kDa from a standard curve of the marker proteins. (B) Analysis of fractions eluted from a Superdex 200 gel filtration column (A). Fractions between 13 and 18.5 ml were analyzed by SDS–PAGE followed by staining with CBB and western blotting probed with the indicated antibodies. HisRAD18ΔC2 (15.1 kDa) migrated slightly faster than FLAGRAD18ΔC2 (14.2 kDa). (C) Elution profile of RAD6A–(HisRAD18–FLAGRAD18) and RAD6A–HisRAD18ΔN1 complexes from a Superdex 200 gel filtration column. Respective complexes were eluted in 9.77 and 11.45 ml, corresponding to 490 and 220 kDa of the apparent molecular masses, and 62 and 52 Å of Stokes' radius, estimated from a standard curve of the marker proteins. (D) Peak fractions of gel filtration chromatography (C) were analyzed by SDS–PAGE followed by CBB staining and western blotting probed with the indicated antibodies. Relative chemiluminescence signals detected with a CCD camera are shown. Lane 1, RAD6A–(HisRAD18–FLAGRAD18) (3.7 pmol as a trimer); lane 2, RAD6A–HisRAD18ΔN1 (3.7 pmol as a dimer).
Subunit composition of the RAD6A–RAD18 complex is RAD6A–(RAD18)2
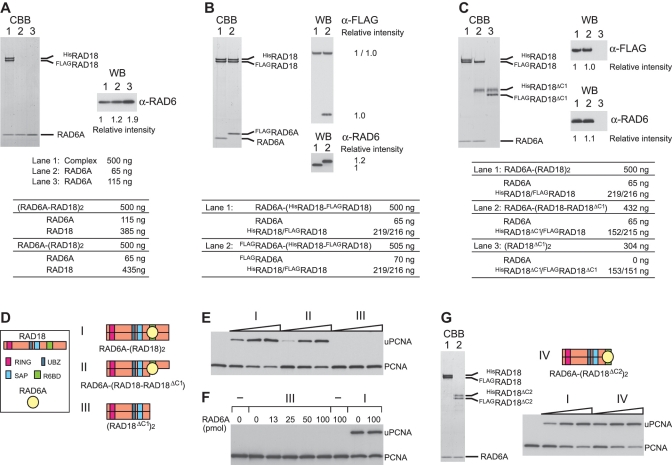
To examine the stoichiometry of RAD6A in the complex, we directly compared amounts of RAD6A in the complex with purified RAD6A protein as a reference. We applied 500 ng of the purified complex, RAD6A–HisRAD18–FLAGRAD18, in parallel with different amounts (65 or 115 ng) of RAD6A monomers to SDS–PAGE. The amounts of RAD6A in the complex should be 65 or 115 ng, if the complex is RAD6A–(RAD18)2 or (RAD6A–RAD18)2, respectively (Figure 2A). The results of CBB staining and western blotting showed the amount of RAD6A in the complex to be closer to 65 ng (Figure 2A), thus suggesting that the complex contains RAD6A and RAD18 at the ratio of 1:2. To confirm this, the FLAG-tag was introduced to RAD6A and the FLAGRAD6A–HisRAD18–FLAGRAD18 complex was purified as described above. We applied 505 ng of the purified complex to SDS–PAGE, in parallel with 500 ng of RAD6A–HisRAD18–FLAGRAD18 as a reference (Figure 2B). Western blotting with anti-RAD6 antibodies confirmed that both complexes contained equivalent amounts of RAD6A (Figure 2B), and blotting with an anti-FLAG antibody clearly demonstrated that the molecular ratio of FLAGRAD6A and FLAGRAD18 was 1:1 (Figure 2B), suggesting strongly the ratio of RAD6A to RAD18 to be 1:2.
Figure 2.
Subunit composition of the RAD6A–RAD18 complex and deletion analysis of RAD18. (A) The indicated amounts of purified RAD6A–RAD18 complex and RAD6A were analyzed by SDS–PAGE followed by staining with CBB and western blotting probed with anti-RAD6 antibodies. Relative chemiluminescence signals detected with a CCD camera are shown. Amounts of each subunit calculated for two different postulated subunit compositions are shown in the table. (B and C) Indicated amounts of purified RAD6A–RAD18 complexes were analyzed by SDS–PAGE followed by staining with CBB and western blotting probed with anti-FLAG and anti-RAD6 antibodies. Relative chemiluminescence signals detected with a CCD camera are shown. Amounts of each subunit calculated for the postulated subunit compositions are shown in the tables. (D) Schematic representations of the structures for respective RAD6A–RAD18 complexes. (E) Ligase activities of the respective RAD6A–RAD18 complexes. Increasing amounts of the complexes (0.5, 1 and 2 pmol) were subjected to standard assays. The reaction products were analyzed by western blotting with anti PCNA antibodies. I, RAD6A–(HisRAD18–FLAGRAD18); II, RAD6A–(HisRAD18ΔC1–FLAGRAD18); III, HisRAD18ΔC1–FLAGRAD18ΔC1. (F) Titration of RAD6A in the reaction with HisRAD18ΔC1–FLAGRAD18ΔC1. Indicated amounts of RAD6A were incubated with 2 pmol of HisRAD18ΔC1–FLAGRAD18ΔC1 (III) under standard assay conditions. As control reactions, 1 pmol of RAD6A–(HisRAD18–FLAGRAD18) (I) was incubated in the presence or absence of additional RAD6A. (G) Analysis of complex formation and ligase activity of another C-terminal deletion mutant of RAD18. Purified complexes (3.7 pmol as a trimer) were analyzed by SDS–PAGE followed by staining with CBB. Lane 1, RAD6A–(HisRAD18–FLAGRAD18) (I); lane 2, RAD6A–(HisRAD18ΔC2–FLAGRAD18ΔC2) (IV). Structure of the mutant complex (IV) was represented schematically. Assays were performed as described in (E).
R6BD is located within amino acid residues 340–395 of RAD18 (Figure 3D) (17,19). Using a deletion mutant consisting of amino acid residues 1–341 of RAD18 (hereafter designated as RAD18ΔC1, Figure 3D), we further examined the stoichiometry of RAD6 and RAD18. When HisRAD18ΔC1 and FLAGRAD18ΔC1 were co-produced with RAD6A in the same E. coli cells, a complex containing HisRAD18ΔC1 and FLAGRAD18ΔC1 was purified similarly as described above, but it did not contain RAD6A (Figure 2C, lane 3). This result indicates that the R6BD is required for complex formation with RAD6 but is dispensable for dimerization. In contrast, when HisRAD18ΔC1 was co-produced with FLAGRAD18 and RAD6A in the same E. coli cells, we successfully obtained a RAD6A–HisRAD18ΔC1–FLAGRAD18 ternary complex (Figure 2C, lane 2), indicating that only one R6BD in the two RAD18 subunits could form the complex with RAD6A. Again, amounts of each protein in these complexes were compared by CBB staining and western blotting. For Figure 2C, we applied 500 ng of RAD6A–HisRAD18–FLAGRAD18 (lane 1) and 432 ng of the RAD6A–HisRAD18ΔC1–FLAGRAD18 complex (lane 2) to SDS–PAGE. If stoichiometry of each component were 1:1:1 in both complexes, amounts of each FLAGRAD18 and each RAD6A in the two complexes should be identical (Figure 2C). The results of CBB staining and western blotting proved to be in good agreement with our estimations (Figure 2C, lanes 1 and 2), suggesting that the RAD18 dimer, as well as the RAD18 monomer, is capable of accommodating only one RAD6 molecule (Figure 2D). Very interestingly, the hetero complex RAD6A–(HisRAD18ΔC1–FLAGRAD18) was catalytically active in terms of PCNA ubiquitination (Figure 2E). As expected, HisRAD18ΔC1–FLAGRAD18ΔC1 was inactive (Figure 2E), and its ligase activity was hardly restored by addition of excess amounts of RAD6A in solution (Figure 2F). To see whether the defect is due to the loss of function of the C-terminal portion, another truncated RAD18 mutant consisting of amino acid residues 1–388 (hereafter designated as RAD18ΔC2, Figure 3D), was examined. It was expected that RAD18ΔC2 could form a complex with RAD6, since it retained R6BD (Figure 3D) (15). Indeed, it was successfully reconstituted into a ternary complex, RAD6A–HisRAD18ΔC2–FLAGRAD18ΔC2, and the complex exhibited ligase activity similar to the wild-type level (Figure 2G), indicating that the C-terminal region of 389–495 amino acid residues is dispensable for ligase activity. From all these results (Figures 1 and 2), together with the fact that RAD6 is essentially a monomer (see Supplementary Materials and Methods section) (19,47), we conclude the subunit composition of the RAD6A–RAD18 complex to be RAD6A–(RAD18)2 with a molecular mass of 130 kDa. Thus, the overall structure of the complex is asymmetric. Notably, this well matched the 131 kDa estimated molecular mass from the Stokes' radius and the sedimentation coefficient (Table 1).
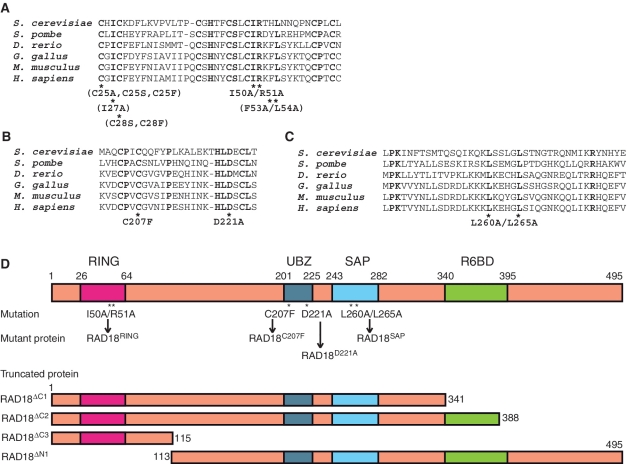
Figure 3.
RAD18 mutants analyzed in this study. (A) Multiple alignments of the RING domains of human RAD18 and its orthologs. Mutants analyzed in this study are shown below the alignments. Mutants shown in parentheses were eluted in void volumes of gel filtration chromatography. (B) Multiple alignments of UBZ of human RAD18 and its orthologs. (C) Multiple alignments of the SAP domains of human RAD18 and its orthologs. (D) Schematic representations of the structures of RAD18. Positions of the mutations are shown with designated names of the mutant proteins.
Evidence of the ternary complex, RAD6A–(RAD18)2, in vivo
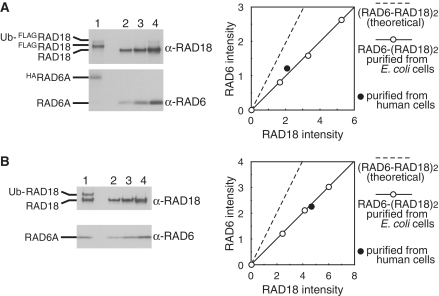
To obtain evidence of ternary complex formation in vivo, HA-tagged RAD6A and FLAG-tagged RAD18 were expressed together in human cells, and HARAD6A–FLAGRAD18 complexes were isolated using FLAG affinity and then HA affinity gels. The complexes were a mixture of unmodified RAD18 and mono-ubiquitinated RAD18, as reported previously (Figure 4A) (48). Subunit composition of the purified complexes was determined by western blotting with anti-RAD6 and anti-RAD18 antibodies using RAD6A–(RAD18)2 complex purified from E. coli cells (26) as references. Ratio of signals from HARAD6A and FLAGRAD18 well fitted those from RAD6A–(RAD18)2 complexes (Figure 4A), suggesting the subunit composition to be RAD6A–(RAD18)2. This was further confirmed using partially purified untagged RAD6A–RAD18 complexes generated by conventional column chromatography from RAD6A and RAD18 expressing human cells. We found that RAD6A, RAD18 and mono-ubiquitinated RAD18 all eluted together from a gel filtration column (Figure 4B), similar to the RAD6A–(RAD18)2 complex (Table 1 and Figure 6C). Again, the subunit composition of the eluted fraction corresponded to RAD6A–(RAD18)2 (Figure 4B). These results strongly suggest ternary complex formation in vivo.
Figure 4.
Ternary complex formation of RAD6A and RAD18 in vivo. (A) HARAD6–FLAGRAD18 complexes isolated from human cells were analyzed by SDS–PAGE (lane 1) together with RAD6A–(RAD18)2 isolated from E. coli (lanes 2–4) followed by western blotting probed with anti-RAD18 and anti-RAD6 antibodies. Relative chemiluminescence signals of HARAD6A and FLAGRAD18 (sum of unmodified FLAGRAD18 and Ub-FLAGRAD18) detected with a CCD camera were plotted with those of RAD6A and RAD18 purified from E. coli as references. The theoretical 1:1 ratio of RAD6 and RAD18 is shown as a broken line. (B) Untagged RAD6–RAD18 complexes isolated from human cells were analyzed by SDS–PAGE as (A).
Analysis of UBZ roles in complex formation and ligase activity
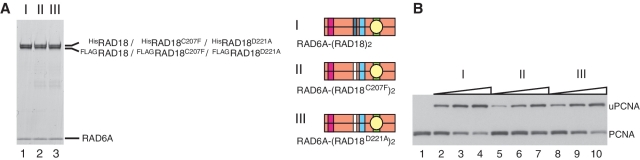
Our system to purify RAD6–RAD18 complex with two different tags is a useful tool for analysis of structure–function relationships in RAD18. First, we applied this system to test whether UBZ is required for dimerization of RAD18 using two loss-of-function mutants, RAD18C207F and RAD18D221A (34,37) (Figure 3B and D). The results demonstrated that these mutants were successfully reconstituted into the complexes, RAD6A–(RAD18C207F)2 and RAD6A–(RAD18D221A)2 (Figure 5A) and their ligase activities were similar to that of wild-type complex (Figure 5B). These results indicate UBZ to be dispensable for complex formation and ligase activity, in line with recent reports (15,39), demonstrating reliability of our system for analysis of the structure and function of RAD6–RAD18 complexes.
Figure 5.
Analysis of ligase activity and complex formation with UBZ mutants of RAD18. (A) Purified complexes (3.7 pmol) were analyzed by SDS–PAGE followed by staining with CBB. I, RAD6A–(HisRAD18–FLAGRAD18); II, RAD6A–(HisRAD18C207F–FLAGRAD18C207F); III, RAD6A–(HisRAD18D221A–FLAGRAD18D221A). Structures are represented schematically, UBZ domains with a mutation being shown in white boxes. (B) Ligase activity of the respective RAD6A–RAD18 complexes. Increasing amounts of the complexes (0.5, 1 and 2 pmol) shown in (A) were subjected to standard assays.
Functional interaction between RAD6 and the RING domains of the two subunits of RAD18 in the complex
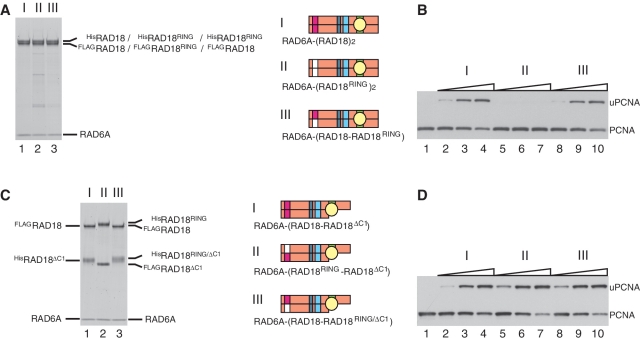
Next, we addressed the question whether the N-terminal part containing a RING domain mediates dimerization. We generated another truncated RAD18 mutant consisting of the 1–115 amino acid residues of RAD18 (hereafter designated as RAD18ΔC3, Figure 3D). When HisRAD18ΔC3 and FLAGRAD18ΔC3 were co-produced with RAD6A in the same E. coli cells, we found the HisRAD18ΔC3-FLAGRAD18ΔC3 dimer to be reconstituted, but that RAD6A did not co-purify with the dimer. The observed elution profile of the dimer from a gel filtration column is shown in Figure 6A. HisRAD18ΔC3 and FLAGRAD18ΔC3, confirmed by western blotting (Figure 6B), co-eluted with an apparent molecular mass of ∼37 kDa, slightly larger than the calculated molecular mass of 29 kDa as a dimer (Figure 6A and B). As a complementary experiment, an N-terminal deletion mutant of RAD18 consisting of 113–495 amino acid residues (hereafter designated as RAD18ΔN1, Figure 3D), was generated. When HisRAD18ΔN1 was coproduced with FLAGRAD18 and RAD6A, HisRAD18ΔN1 co-purified with RAD6A but not with FLAGRAD18 (Figure 6D). As mentioned above, the RAD6A–(HisRAD18–FLAGRAD18) ternary complex eluted at a position corresponding to 490 kDa in gel filtration (Figure 6C). In contrast, the complex of RAD6A–HisRAD18ΔN1 eluted at a position corresponding to 220 kDa, which is much smaller than the ternary complex (Figure 6C). Furthermore the molecular ratio of RAD6A and HisRAD18ΔN1 determined by western blotting, compared with the ternary complex as a reference, was close to 1:1 (Figure 6D). These results suggest that the RAD6A–HisRAD18ΔN1 complex is a dimer composed of one RAD6A and one HisRAD18ΔN1 molecule and imply that HisRAD18ΔN1 neither self-associates nor forms a heterodimer with FLAGRAD18. From these data shown in Figure 6, we conclude that the N-terminal region (1–115) is necessary and sufficient for dimerization, while UBZ and SAP domains are dispensable.
In general, the RING domains of E3s have an essential function in ligase activity by mediating interactions with E2s (1,2). It is of great interest to clarify how the two RING domains interact with one RAD6 subunit in the RAD6A–(RAD18)2 complex. To address this point, we made several mutants in which one or two conserved amino acid residues in the RING domain were replaced as indicated in Figure 3A. We found that RING mutants with C25A, C25S, C25F, I27A, C28S, C27F or F53A/L54A substitutions eluted in void volumes on gel filtration chromatography, suggesting these mutants to form large aggregates due to highly disordered structures by misfolding. Therefore we did not further analyze them. However, we could successfully obtain one mutant complex containing I50A/R51A substitutions in RAD18 (hereafter designated as RAD18RING, see Figures 3A and 7A, lane 2). A previous report suggested, based on the crystal structure of the c-Cbl-UbcH7 complex, that the amino acid residues Ile50 and Arg51 of RAD18 should be located in a predicted RAD6-interacting α-helix (2). Thus, it was expected that I50A/R51A mutations would affect the ligase activity of RAD18. In fact, it was much reduced when compared to that of the wild-type complex (Figure 7B). Then, we reconstituted a hetero complex with the mutant and wild-type, RAD6A–(HisRAD18RING–FLAGRAD18) (Figure 7A, lane 3). Surprisingly, its ligase activity was essentially identical to that of the wild-type (Figure 7B), indicating that an interaction between RAD6 and only one RING domain in the two RAD18 subunits is sufficient for ligase activity. In addition, I50A/R51A mutations were combined with the ΔC1 mutation and two mutant complexes, RAD6A–(HisRAD18RING–FLAGRAD18ΔC1) and RAD6A–(HisRAD18RING/ΔC1–FLAGRAD18) were reconstituted (Figure 7C). Analysis of their ligase activities demonstrated these to be as active as the wild-type (Figure 7D), indicating that one RAD6 molecule in the complex has the potential to interact with either subunit of the RAD18 dimer.
Figure 7.
Analysis of ligase activity and complex formation of a RING mutant of RAD18. (A) Purified complexes (3.7 pmol) were analyzed by SDS–PAGE followed by staining with CBB. I, RAD6A–(HisRAD18–FLAGRAD18); II, RAD6A–(HisRAD18RING–FLAGRAD18RING); III, RAD6A–(HisRAD18RING–FLAGRAD18). Structures are represented schematically, RING domains with a mutation being shown in white boxes. (B) Ligase activities of the respective RAD6A–RAD18 complexes. Increasing amounts of the complexes (0.5, 1 and 2 pmol) shown in (A) were subjected to standard assays. (C) Purified complexes (3.7 pmol) were analyzed by SDS–PAGE followed by staining with CBB. I, RAD6A–(HisRAD18ΔC1–FLAGRAD18); II, RAD6A–(HisRAD18RING–FLAGRAD18ΔC1); III, RAD6A–(HisRAD18RING/ΔC1–FLAGRAD18). (D) Ligase activities of the respective RAD6A–RAD18 complexes. Increasing amounts of the complexes (0.5, 1 and 2 pmol) shown in (C) were subjected to standard assays.
Functional interaction between SAP and R6BD for ligase activity
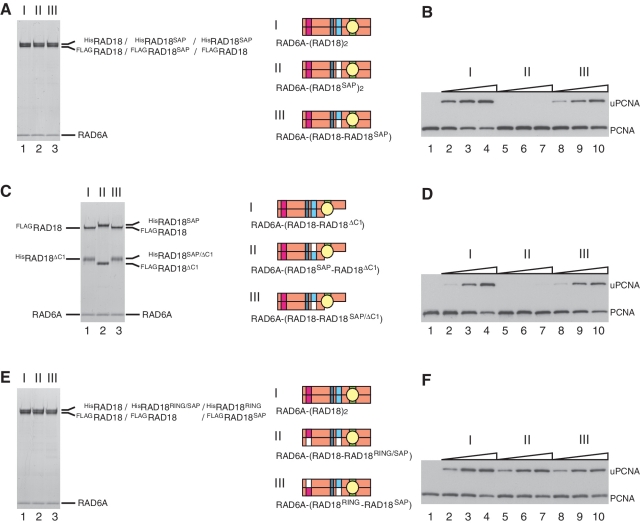
It has been shown that the SAP domain of RAD18 has DNA-binding activity (15,38), which is separable from its essential function for ligase activity (38). When a mutant RAD18 containing L250A/L265A substitutions (hereafter designated as RAD18SAP, see Figure 3D) was successfully reconstituted into the ternary complex, RAD6A–(HisRAD18SAP–FLAGRAD18SAP) (Figure 8A, lane 2), the ligase activity of the mutant complex was reduced to an undetectable level (Figure 8B). However, activity was restored in a hetero complex with the wild-type, RAD6A–(HisRAD18SAP–FLAGRAD18) (Figure 8B), demonstrating that only one of the two SAP domains in the complex is sufficient for the essential SAP function. Next, the SAP mutation was combined with the ΔC1 mutation to reconstitute RAD6A–(HisRAD18SAP–FLAGRAD18ΔC1) and RAD6–(HisRAD18SAP/ΔC1–FLAGRAD18) (Figure 8C). Interestingly, the former had quite reduced ligase activity, but the latter exhibited the wild-type level (Figure 8D), indicating that the active SAP domain should be present on the same RAD18 molecule to which RAD6 binds. In contrast, such functional interaction was not observed between RING and SAP mutants; ligase activities of RAD6A–(HisRAD18RING/SAP–FLAGRAD18) and RAD6–(HisRAD18RING–FLAGRAD18SAP) complexes (Figure 8E) were similar to that of wild-type (Figure 8F).
Figure 8.
Analysis of ligase activity and complex formation of a SAP mutant of RAD18. (A) Purified complexes (3.7 pmol) were analyzed by SDS–PAGE followed by staining with CBB. I, RAD6A–(HisRAD18–FLAGRAD18); II, RAD6A–(HisRAD18SAP–FLAGRAD18SAP); III, RAD6A–(HisRAD18SAP–FLAGRAD18). Structures were represented schematically, SAP domains with a mutation being shown in white boxes. (B) Ligase activities of the respective RAD6A–RAD18 complexes. Increasing amounts of the complexes (0.5, 1 and 2 pmol) shown in (A) were subjected to standard assays. (C) Purified complexes (3.7 pmol) were analyzed by SDS–PAGE followed by staining with CBB. I, RAD6A–(HisRAD18ΔC1–FLAGRAD18); II, RAD6A–(HisRAD18SAP–FLAGRAD18ΔC1); III, RAD6A–(HisRAD18SAP/ΔC1–FLAGRAD18). (D) Ligase activities of the respective RAD6A–RAD18 complexes. Increasing amounts of the complexes (0.5, 1 and 2 pmol) shown in (C) were subjected to standard assays. (E) Purified complexes (3.7 pmol) were analyzed by SDS–PAGE followed by staining with CBB. I, RAD6A–(HisRAD18–FLAGRAD18); II, RAD6A–(HisRAD18RING/SAP–FLAGRAD18); III, RAD6A–(HisRAD18RING–FLAGRAD18SAP). (F) Ligase activities of the respective RAD6A–RAD18 complexes. Increasing amounts of the complexes (0.5, 1 and 2 pmol) shown in (E) were subjected to standard assays.
DISCUSSION
In this study, we established a method to distinguish between the two subunits of RAD18 in the RAD6–(RAD18)2 complex by introducing different tags at the N-termini. This enabled us to purify RAD18 complexes composed of wild–wild, wild–mutant or mutant–mutant subunits and facilitated analysis of structure–function relationships in RAD18.
Previously, it was considered that the RAD6–RAD18 complex could be composed of two RAD6 and two RAD18, since the observations that RAD18 binds to RAD6 and it forms a dimer led to the assumption that each RAD18 molecule in the dimer should bind to RAD6 (15,20,21). However, there is little evidence for such an assignment based on the stoichiometry of RAD6. In this study, we provided hard lines of evidence that the complex is composed of one subunit of RAD6 and two subunits of RAD18, so that the overall structure of the ternary complex should be asymmetric.
The RING domains in E3s have an essential function in ligase activity by mediating interactions with E2s. Zheng et al. (2) have suggested, on the basis of the crystal structure of the c-Cbl-UbcH7 complex, that amino acid residues I50 and R51 in the RING domain of RAD18 are located in a predicted RAD6-interacting α-helix. Consistent with this, a mutant complex containing I50A/R51A substitutions in both RAD18 subunits exhibited severely reduced ligase activity, probably due to an impaired interaction between the altered RING domain and RAD6A. When the mutant subunit was complexed with wild-type RAD18 subunit, ligase activity was restored to the wild-type level, indicating that only one of the two RING domains is necessary for enzyme function. Furthermore, even when the RING mutation was combined with a deletion mutation of R6BD in the same or other RAD18 subunit, the complexes exhibited robust ligase activity. These results imply that RAD6 can bind in either one of the two R6BDs, then interacting with one of the two RING domains in the RAD18 dimer.
Why should the RING domain of RAD18 form a dimer? We found that inactivation of one RING domain does not affect the ligase activity, indicating that the close proximity of two active RING domains in RAD18 is not important for the enzyme function. From our observations that some RING mutants such as C25A, C25S, C25F, I27A, C28S, C28F and F53A/L54A, probably disrupting the RING structure (Figure 3D), appeared to form large aggregates, we suggest that the RING structure is required for stable dimerization, which is in turn necessary for sustaining the functionally active RING structure of RAD18. From the results, we cannot exclude the possibility of retention of some enzyme functions even in aggregates. Indeed, it has been reported that the C28F mutant can fully complement the homologous recombination defects of RAD18-null cells (39).
The SAP domain is a unique eukaryotic module involved in sequence- or structure-specific DNA binding (36,38,49–52). Additionally, in RAD18 it has an essential role in ligase activity, separable from its function in DNA binding (38). We further demonstrated that one of the two SAP domains in the RAD6–(RAD18)2 complex is sufficient for ligase activity. Interestingly, experiments using hetero complexes, in which mutation in the SAP domain was combined with a deletion mutation of R6BD, revealed an interesting relationship between SAP and R6BD. Although one of the two SAP domains and one of the two R6BDs are sufficient for enzyme activity, the active SAP domain should be present on the same RAD18 molecule to which RAD6 binds in the complex. It makes a sharp contrast to the case of RING mutant as described above. A simple explanation is that the SAP domain acts as a hinge connecting the R6BD and RING domain to enable precise juxtaposition between RING and RAD6. This might be very important for ligase activity because a deletion mutant of R6BD hardly supported PCNA ubiquitination in the presence of an excess amount of RAD6A, which should be high enough to detect ligase activities for many other E2–E3 pairs (4–6,9–2). It seems likely that binding affinity between RAD6 and the RING domain of RAD18 is very low. That could be the reason why R6BD is essential for ligase activity of the RAD6–RAD18 complex. The tight interaction between RAD6 and R6BD confers on the complex the ability to monoubiquitinate PCNA, also inhibiting the activity of RAD6 catalyzing ubiquitin chain formation (19).
In the above, we have shown that RAD6 and RAD18 forms a ternary complex, RAD6–(RAD18)2 and demonstrated that one R6BD site is sufficient for the ligase activity. Then, a question that immediately arises is why RAD6 can bind to one of the two R6BD sites, but not to both of them. Although we have no experimental data to answer the question at the moment, such a case is not unprecedented. CHIP (C-terminal of Hsp70 interacting protein), a protein containing a C-terminal Ubox domain (similar to RING) and an N-terminal TPR domain, forms an asymmetric dimer mediated by the Ubox domains, in which only one of the two Ubox domains is available for binding to E2 (Ubc13) because the other Ubox domain is blocked due to interaction with the TPR domain (53). It is tempting to speculate that a segment including R6BD located in a C-terminal portion of the two RAD18 subunits may interact with one of the two RING domains, consequently preventing the R6BD from interacting with RAD6. Thus, one RAD6 molecule can bind to the other R6BD and is positioned closely to another unoccupied RING domain for catalyzing the ligase function. Evidently, further experiments are required to elucidate this interesting possibility.
During the course of finally completing this manuscript, we have learned that Huang and co-workers have recently determined the structure of a RAD18 RING(1–99) dimer, also showing that this binds to RAD6 at a 2:2 ratio, whereas the full-length RAD18 dimer binds only to a single RAD6 molecule (54). Our study was conducted independently of their work.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Materials and Methods, Supplementary References [26,45,55].
FUNDING
Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to Y.M. and K.K.); Grants-in-Aid for Cancer Research from the Ministry of Health, Labour and Welfare (to K.K.). Funding for open access charge: Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Toshiki Tsurimoto (Kyushu University, Fukuoka, Japan) and Dr Tomohiko Ohta (St Marianna University School of Medicine, Kanagawa, Japan) for providing PCNA expression and ubiquitin-encoding plasmids, respectively. The authors would also like to express our appreciation to Dr Haruo Ohmori (Kyoto University, Kyoto, Japan) for his critical reading of manuscripts and valuable suggestions. The authors are grateful to Fumie Okubo, Kazumi Shimamoto and Mai Yoshida for their laboratory assistance.
REFERENCES
- 1.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 2.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 3.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 4.Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- 5.Xia Y, Pao GM, Chen HW, Verma IM, Hunter T. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J. Biol. Chem. 2003;278:5255–5263. doi: 10.1074/jbc.M204591200. [DOI] [PubMed] [Google Scholar]
- 6.Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satijn DP, Otte AP. RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity. Mol. Cell. Biol. 1999;19:57–68. doi: 10.1128/mcb.19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM. RING domain-mediated interaction is a requirement for MDM2's E3 ligase activity. Cancer Res. 2007;67:6026–6030. doi: 10.1158/0008-5472.CAN-07-1313. [DOI] [PubMed] [Google Scholar]
- 9.Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc. Natl Acad. Sci. USA. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 11.Mace PD, Linke K, Feltham R, Schumacher FR, Smith CA, Vaux DL, Silke J, Day CL. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J. Biol. Chem. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- 12.Liew CW, Sun H, Hunter T, Day CL. RING domain dimerization is essential for RNF4 function. Biochem. J. 2010;431:23–29. doi: 10.1042/BJ20100957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 14.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 15.Notenboom V, Hibbert RG, van Rossum-Fikkert SE, Olsen JV, Mann M, Sixma TK. Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res. 2007;35:5819–5830. doi: 10.1093/nar/gkm615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailly V, Prakash S, Prakash L. Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol. Cell. Biol. 1997;17:4536–4543. doi: 10.1128/mcb.17.8.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tateishi S, Sakuraba Y, Masuyama S, Inoue H, Yamaizumi M. Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc. Natl Acad. Sci. USA. 2000;97:7927–7932. doi: 10.1073/pnas.97.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hibbert RG, Huang A, Boelens R, Sixma TK. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc. Natl Acad. Sci. USA. 2011;108:5590–5595. doi: 10.1073/pnas.1017516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyase S, Tateishi S, Watanabe K, Tomita K, Suzuki K, Inoue H, Yamaizumi M. Differential regulation of Rad18 through Rad6-dependent mono- and polyubiquitination. J. Biol. Chem. 2005;280:515–524. doi: 10.1074/jbc.M409219200. [DOI] [PubMed] [Google Scholar]
- 21.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. 2nd edn. Washington, DC: ASM; 2006. DNA repair and mutagenesis. [Google Scholar]
- 23.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 24.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases η and REV1. Proc. Natl Acad. Sci. USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc. Natl Acad. Sci. USA. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda Y, Piao J, Kamiya K. DNA replication-coupled PCNA mono-ubiquitination and polymerase switching in a human in vitro system. J. Mol. Biol. 2010;396:487–500. doi: 10.1016/j.jmb.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Unk I, Hajdu I, Fatyol K, Hurwitz J, Yoon JH, Prakash L, Prakash S, Haracska L. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl Acad. Sci. USA. 2008;105:3768–3773. doi: 10.1073/pnas.0800563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unk I, Hajdu I, Fatyol K, Szakal B, Blastyak A, Bermudez V, Hurwitz J, Prakash L, Prakash S, Haracska L. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl Acad. Sci. USA. 2006;103:18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 30.Wood A, Garg P, Burgers PM. A ubiquitin-binding motif in the translesion DNA polymerase Rev1 mediates its essential functional interaction with ubiquitinated proliferating cell nuclear antigen in response to DNA damage. J. Biol. Chem. 2007;282:20256–20263. doi: 10.1074/jbc.M702366200. [DOI] [PubMed] [Google Scholar]
- 31.Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ. Regulation of polymerase exchange between Polη and Polδ by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc. Natl Acad. Sci. USA. 2008;105:5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Day TA, Palle K, Barkley LR, Kakusho N, Zou Y, Tateishi S, Verreault A, Masai H, Vaziri C. Phosphorylated Rad 18 directs DNA polymerase η to sites of stalled replication. J. Cell. Biol. 2010;191:953–966. doi: 10.1083/jcb.201006043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bish RA, Myers MP. Werner helicase-interacting protein 1 binds polyubiquitin via its zinc finger domain. J. Biol. Chem. 2007;282:23184–23193. doi: 10.1074/jbc.M701042200. [DOI] [PubMed] [Google Scholar]
- 34.Crosetto N, Bienko M, Hibbert RG, Perica T, Ambrogio C, Kensche T, Hofmann K, Sixma TK, Dikic I. Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner. J. Biol. Chem. 2008;283:35173–35185. doi: 10.1074/jbc.M803219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair. 2009;8:544–556. doi: 10.1016/j.dnarep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 37.Nakajima S, Lan L, Kanno S, Usami N, Kobayashi K, Mori M, Shiomi T, Yasui A. Replication-dependent and -independent responses of RAD18 to DNA damage in human cells. J. Biol. Chem. 2006;281:34687–34695. doi: 10.1074/jbc.M605545200. [DOI] [PubMed] [Google Scholar]
- 38.Tsuji Y, Watanabe K, Araki K, Shinohara M, Yamagata Y, Tsurimoto T, Hanaoka F, Yamamura K, Yamaizumi M, Tateishi S. Recognition of forked and single-stranded DNA structures by human RAD18 complexed with RAD6B protein triggers its recruitment to stalled replication forks. Genes Cells. 2008;13:343–354. doi: 10.1111/j.1365-2443.2008.01176.x. [DOI] [PubMed] [Google Scholar]
- 39.Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell. Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukuda K, Morioka H, Imajou S, Ikeda S, Ohtsuka E, Tsurimoto T. Structure-function relationship of the eukaryotic DNA replication factor, proliferating cell nuclear antigen. J. Biol. Chem. 1995;270:22527–22534. doi: 10.1074/jbc.270.38.22527. [DOI] [PubMed] [Google Scholar]
- 41.Masuda Y, Suzuki M, Piao J, Gu Y, Tsurimoto T, Kamiya K. Dynamics of human replication factors in the elongation phase of DNA replication. Nucleic Acids Res. 2007;35:6904–6916. doi: 10.1093/nar/gkm822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomida J, Masuda Y, Hiroaki H, Ishikawa T, Song I, Tsurimoto T, Tateishi S, Shiomi T, Kamei Y, Kim J, et al. DNA damage-induced ubiquitylation of RFC2 subunit of replication factor C complex. J. Biol. Chem. 2008;283:9071–9079. doi: 10.1074/jbc.M709835200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 44.Maki H, Kornberg A. Proofreading by DNA polymerase III of Escherichia coli depends on cooperative interaction of the polymerase and exonuclease subunits. Proc. Natl Acad. Sci. USA. 1987;84:4389–4392. doi: 10.1073/pnas.84.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 46.Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 47.Worthylake DK, Prakash S, Prakash L, Hill CP. Crystal structure of the Saccharomyces cerevisiae ubiquitin-conjugating enzyme Rad6 at 2.6 Å resolution. J. Biol. Chem. 1998;273:6271–6276. doi: 10.1074/jbc.273.11.6271. [DOI] [PubMed] [Google Scholar]
- 48.Yuasa MS, Masutani C, Hirano A, Cohn MA, Yamaizumi M, Nakatani Y, Hanaoka F. A human DNA polymerase η complex containing Rad18, Rad6 and Rev1; proteomic analysis and targeting of the complex to the chromatin-bound fraction of cells undergoing replication fork arrest. Genes Cells. 2006;11:731–744. doi: 10.1111/j.1365-2443.2006.00974.x. [DOI] [PubMed] [Google Scholar]
- 49.Ahn JS, Whitby MC. The role of the SAP motif in promoting Holliday junction binding and resolution by SpCCE1. J. Biol. Chem. 2003;278:29121–29129. doi: 10.1074/jbc.M302314200. [DOI] [PubMed] [Google Scholar]
- 50.Chou CH, Wang J, Knuth MW, Reeves WH. Role of a major autoepitope in forming the DNA binding site of the p70 (Ku) antigen. J. Exp. Med. 1992;175:1677–1684. doi: 10.1084/jem.175.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gohring F, Schwab BL, Nicotera P, Leist M, Fackelmayer FO. The novel SAR-binding domain of scaffold attachment factor A (SAF-A) is a target in apoptotic nuclear breakdown. EMBO J. 1997;16:7361–7371. doi: 10.1093/emboj/16.24.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kipp M, Gohring F, Ostendorp T, van Drunen CM, van Driel R, Przybylski M, Fackelmayer FO. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol. 2000;20:7480–7489. doi: 10.1128/mcb.20.20.7480-7489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation–crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 54.Huang A, Hibbert RG, de Jong RN, Das D, Sixma TK, Boelens R. Symmetry and asymmetry of the RING-RING dimer of Rad18. J. Mol. Biol. 2011;410:424–435. doi: 10.1016/j.jmb.2011.04.051. [DOI] [PubMed] [Google Scholar]
- 55.Gomes XV, Gary SL, Burgers PM. Overproduction in Escherichia coli and characterization of yeast replication factor C lacking the ligase homology domain. J. Biol. Chem. 2000;275:14541–14549. doi: 10.1074/jbc.275.19.14541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.