Abstract
Compared to protein enzymes, our knowledge about how RNA accelerates chemical reactions is rather limited. The crystal structures of a ribozyme that catalyzes Diels–Alder reactions suggest a rich tertiary architecture responsible for catalysis. In this study, we systematically probe the relevance of crystallographically observed ground-state interactions for catalytic function using atomic mutagenesis in combination with various analytical techniques. The largest energetic contribution apparently arises from the precise shape complementarity between transition state and catalytic pocket: A single point mutant that folds correctly into the tertiary structure but lacks one H-bond that normally stabilizes the pocket is completely inactive. In the rate-limiting chemical step, the dienophile is furthermore activated by two weak H-bonds that contribute ∼7–8 kJ/mol to transition state stabilization, as indicated by the 25-fold slower reaction rates of deletion mutants. These H-bonds are also responsible for the tight binding of the Diels–Alder product by the ribozyme that causes product inhibition. For high catalytic activity, the ribozyme requires a fine-tuned balance between rigidity and flexibility that is determined by the combined action of one inter-strand H-bond and one magnesium ion. A sharp 360° turn reminiscent of the T-loop motif observed in tRNA is found to be important for catalytic function.
INTRODUCTION
RNA can fold into 3D scaffolds that generate binding sites and catalytic centers. While natural ribozymes that catalyze the cleavage or ligation of RNA strands have been studied quite thoroughly, RNAs ability to accelerate chemically different reactions—and in particular those involving small molecules—received less attention. Consequently, the nature of RNA active sites evolved for such functions remains poorly understood (1–4).
Carbon–carbon bond formation is a fundamental task in organic synthesis, biochemistry and prebiotic chemistry, and finding ribozymes that catalyze such processes has been a tremendous challenge (5,6). Our laboratory has previously reported the combinatorial selection of ribozymes that catalyze C–C bond formation by Diels–Alder reaction (5), an important reaction type in organic chemistry (7). The RNA molecules were selected to accelerate the cycloaddition reaction of an aromatic anthracene diene tethered to their 5′-ends (in cis reaction) with a biotinylated maleimide dienophile. The majority of the active sequences was found to contain a small secondary structure motif consisting of three helices (shown in cyan, yellow and green, Figure 1a and Supplementary Figure S1), an asymmetric internal loop, and the formally single-stranded 5′-end (shown in red), and a rationally re-designed 49-mer RNA containing these elements accelerated the in cis reaction about 20 000-fold. These ribozymes were later found to catalyze the cycloaddition reaction in a truly bimolecular fashion between dienes and dienophiles free in solution (in trans reaction). Other distinctive features are fast multiple-turnover catalysis and enantioselective bond formation (8).
Figure 1.
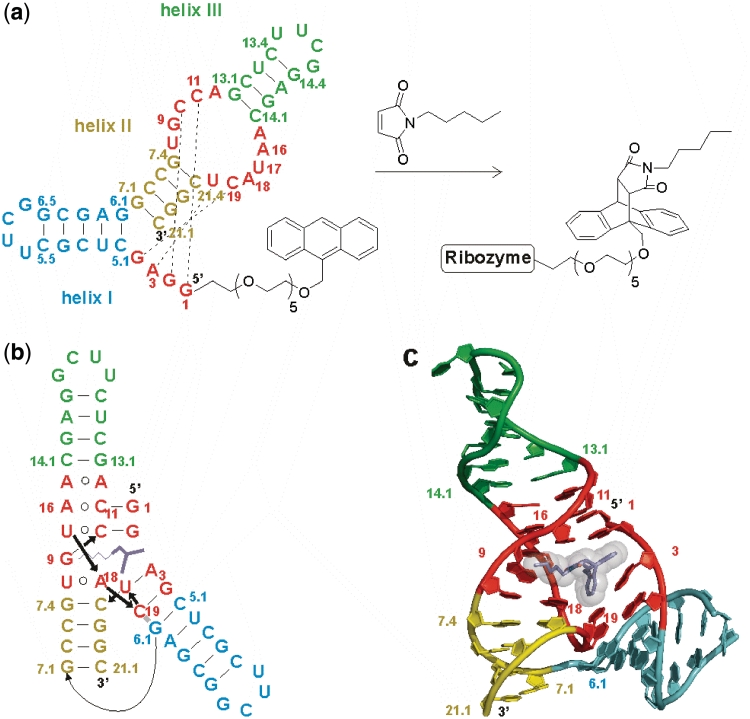
Minimized Diels–Alderase ribozyme. (a) Ribozyme secondary structure with helices I, II and III (cyan, yellow, green), the asymmetric bubble and the conserved 5′-end (both in red), and in-cis Diels–Alder reaction. (b) Tertiary fold and (c) 3D topology in the crystal structure of the ribozyme–product complex. One enantiomer of the Diels–Alder product (dark blue sticks with transparent spheres) is bound into the catalytic pocket of the ribozyme. This manuscript uses the original numbering scheme for this ribozyme (5). A translation map between the different schemes is provided as Supplementary Figure S1.
A first insight into the principles of substrate recognition was obtained by studying the interactions of the ribozyme with a panel of systematically varied substrate analogs (9). These results suggested a major role for hydrophobic and van der Waals interactions, while hydrogen bonding and metal-ion coordination appeared to be less important for catalysis. Mutation analysis and chemical probing identified interactions between nucleotides of the 5′-terminus and the internal asymmetric bubble, and indicated a preformed catalytic pocket (10). These findings could be confirmed by the solution of the crystal structure of this ribozyme in the unbound state and in complex with the Diels–Alder reaction product (Figure 1b and c) (11). The RNA was found to adopt a λ-shaped nested pseudoknot architecture, whose hydrophobic pocket shows precise shape complementarity (according to P1L21, P10L73, P11L9) to the reaction product (shown in dark blue). RNA folding and product binding are dictated by extensive stacking and hydrogen bonding, whereas stereoselection is governed by the shape of the catalytic pocket. The 5′-terminal tetranucleotide segment plays a critical role in shaping both the RNA scaffold and the catalytic pocket. All four residues align through Watson–Crick base pairing with residues on both sides of the asymmetric bubble segment (dotted black lines in Figure 1a), thereby forming the pseudoknot and zippering up the bubble (Figure 1b). The crystal structure suggests three hydrogen bonds between the ribozyme and the Diels–Alder product, involving amino and hydroxyl groups of the former and carbonyl and ether oxygen atoms of the latter (vide infra).
These two crystallized complexes (11), however, represent only two points on the reaction coordinate (empty catalyst and catalyst–product complex), whereas other relevant intermediates (e.g. the ribozyme bound to the substrates or to a transition state analog) could not be crystallized. Therefore, alternative approaches are required for elucidating the principles that govern RNA-substrate interaction and catalytic rate acceleration.
Considerable effort has been devoted recently to understanding the structural dynamics of the Diels–Alderase ribozyme. Single-molecule fluorescence resonance energy transfer (smFRET) and nuclear magnetic resonance (NMR) spectroscopy have revealed that the ribozyme is highly dynamic in aqueous solution at room temperature (12,13). The concentration of divalent metal ions was established as a major determinant of folding and dynamics (12–14). Recent molecular dynamics (MD) simulations revealed the existence of conformations with similar overall architecture but either closed or open catalytic pocket, resulting in either inactive or active catalysts (15).
Based on this static and dynamic structural information, we set out to elucidate the principles that govern RNA–substrate interaction and catalytic rate acceleration. The availability of high-resolution structural information allows to rationally design experiments that support or disprove the relevance of individual structural elements for the catalytic function of the ribozyme.
In this study, we use the tool box of atomic mutagenesis. We add, delete or replace functional groups either in a rational site-directed or in a combinatorial fashion, and determine the enzymatic activity of the mutants. As these assays measure the rate of product formation, they can detect interference of the atomic mutation with any essential step that precedes or is concurrent with this event, from RNA folding to substrate binding to the chemical step of carbon–carbon bond formation. To identify interacting positions, rescue experiments are carried out that involve compensatory double and triple mutations. These investigations are supplemented by chemical probing, inhibition and binding studies, and single-molecule fluorescence spectroscopy. In particular, we focus on the H-bonding network in the catalytic pocket. The results obtained present unprecedented insight into the strategies RNA utilizes to catalyze carbon–carbon bond formation.
MATERIALS AND METHODS
Oligonucleotide synthesis and labeling
Oligonucleotides were synthesized by phosphoramidite chemistry (Expedite 8900 synthesizer, standard protecting groups, standard deprotection) or purchased from CSS Chemical Synthesis Services, Craigavon and IBA, Göttingen. Phosphoramidites were from Proligo, Cruachem and ChemGenes. T7 transcription reactions were performed as described (16). α-Thio-modified nucleoside triphosphate analogues were from Glen Research or IBA. Incorporation of hexa-ethylene glycol (HEG)-tethered anthracene was performed either chemically by using the respective phosphoramidite, or by transcription initiation (17). Oligonucleotides were purified by HPLC and/or PAGE. Oligonucleotides and conjugates were 32P-labeled at their 3′-end with T4 RNA ligase or Klenow fragment of DNA Polymerase I, or 5′-end labeled with polynucleotide kinase, according to standard procedures (18). The 11-mer anthracene conjugates were always synthesized chemically, while RNA 18-mers and 24-mers were either chemically synthesized, or prepared by run-off transcription.
Ribozyme activity assays
Two different gel electrophoretic assays were used. True initial rates of product formation were determined by monitoring the first 15% of conversion. For rapid screening of mutants, semi-quantitative data were obtained by performing single-time point measurements with a standard reaction time of 30 min. A wild-type combination of the unmutated strands was always run in parallel (typical yield: 65% conversion in 30 min) and used as reference. About 0.1 µg/µl dodecyl maltoside was added to avoid unspecific binding in the true initial rate assays. Other reaction conditions were as described (10). All assays were done in triplicate.
The product inhibition kinetics were determined by UV/vis-spectroscopy in an in trans assay that employed the free (i.e. non-tethered) substrates anthracene-HEG (100 µM) and N-pentylmaleimide (500 µM) as well as 28 µM double mutant ribozyme (G9Pu U17dU) in reaction buffer containing 10% ethanol in the presence of increasing concentrations of Diels–Alder products. Second-order rate constants of triple determinations were calculated over the first 5% conversion as k (M−1 h−1). All other conditions were as in reference (9).
Chemical probing
The 5′-32P-labeled RNAs (fmol amounts, 50 000 cpm/sample) were supplemented with 0.15 µg/µl unlabeled tRNA. Reactions were performed in 0.75 mM freshly prepared lead acetate solution, in 40 mM Tris–HCl (pH 7.5), 40 mM NaCl, 0–10 mM MgCl2, 5% ethanol, with or without 1 mM Diels–Alder product at 20°C for 45 min and were terminated by 1:1 mixing with a 7 M urea, 10 mM EDTA solution. Electrophoretic separation was performed on 15% denaturing polyacrylamide gels at 60 W for 3 h. Further reaction conditions were as in reference (10).
Nucleotide analog interference mapping
SP-α-thio modified nucleoside triphosphate analogues (NTPs) were statistically incorporated into in vitro transcribed RNAs using a modification of published protocols (19–21). Anthracene-HEG-GMP initiator nucleotides were incorporated at the 5′-terminus (5). Transcriptions were performed in 40 mM Tris–HCl, pH 8.0, 22 mM MgCl2, 4–7 mM spermidine, 10 mM DTT, 2.4 mg/ml bovine serum albumine, 1 µM double-stranded DNA template, 1 mM initiator nucleotide A-HEG-GMP and 80 U T7-RNA polymerase. While most experiments were conducted with the wild-type sequence, investigation of the role of nucleotide G9 required (for incorporation of 2-aminopurine, purine and 2,6-diaminopurine riboside) the use of a G9A mutant transcription template, while inosine was incorporated using the wild-type template. The two resulting transcript libraries showed largely different reactivities in Diels–Alder reactions and required different conditions (see below). Concentrations of SP-αS-NTPs were adjusted according to the frequency of the respective nucleotide and the length of the ribozyme and had to be optimized for each analog. Transcription mixtures were ethanol precipitated and the re-dissolved pellets applied onto a C18-HPLC reversed phase column. Full length ribozyme conjugates were identified by their spectroscopic properties, isolated and 3′-end-labeled using 5′-32P-pCp and T4 RNA ligase. The resulting Diels–Alderase-HEG-anthracene conjugate libraries containing modified nucleosides (final concentrations 100 nM) were subjected to Diels–Alder reaction with 5 µM biotin maleimide, and the reaction time was adjusted to achieve a library conversion (Cr) of 25–33% (2 min for the wild-type sequence, 30 min for the less reactive G9A mutant). After electrophoretic separation on 20% polyacrylamide gels, (unreacted) substrate and (reacted) product bands were quantified by phosphorimaging, excised, eluted and subjected to iodine cleavage in denaturing loading buffer (5.3 M urea, 34 mM EDTA pH 8.0) using 1/10 sample volume of ethanolic iodine solution (100 mM) for 3 min at room temperature and additionally for 1 min at 65°C. Cleavage patterns were analyzed by phosphorimaging of denaturing 20% PAGE sequencing gels (0.4 mm) using alkaline hydrolysis and RNase T1 digest ladders for sequence assignments. Slight offsets of these reference lanes with respect to the phosphorothioate cleavage lanes are due to the different termini generated in the reactions. Analog interferences and phosphorothioate effects were identified by comparing the densities of substrate and product bands of individual positions, which were also used for quantitative data calculation. Product band densities Dp were multiplied with the respective library conversion values Cr (vide supra), while substrate band densities Ds were multiplied by the factor (1 − Cr). Reaction yields r = Dp*Cr/[Dp*Cr + Ds*(1 − Cr)] of individual point mutants were calculated, taking into account the adjustment of the specific radioactivities of substrate and product lanes on the sequencing gels to equal levels. Relative activity values were calculated by comparison with the wild-type sequence. It should be noted that these values represent semi-quantitative activity data rather than true initial rates, as the samples were analyzed after 25–33% conversion, which was necessary due to general practical requirements of the nucleotide analog interference mapping (NAIM) methodology (19,22).
Single-molecule FRET experiments
Wild-type and U17isoC mutant ribozyme with a Cy3 dye attached to the 5′-end, a Cy5 dye attached to position L2.2 and a 3′-biotin were synthesized by a chemoenzymatic approach as described in reference (23). The wild-type construct was found to have full activity (compared to a ribozyme without modifications), while the U17isoC mutant was found to be inactive (23). smFRET experiments were carried out on a home-build confocal laser scanning microscope with single-molecule sensitivity, essentially as described in reference (24,25). Briefly, probe solutions with freely diffusing RNA molecules at ∼100 pM concentration were placed in the sample cell. Excitation was done with 532 (green) and 633 nm (red) laser lines in alternating fashion at 10 kHz with 70% of duty cycle for the green laser. Detection of fluorescence light was performed synchronously with the excitation in two channels spectrally optimized for the donor (Cy3, 550–610 nm) and acceptor (Cy5, 650–750 nm) emission. FRET efficiency histograms were compiled from selected fluorescent bursts with simultaneously photoactive donor and acceptor, durations between 0.2 and 2 ms, and more than 30 photon counts.
Graphics
Figures were prepared from the PDB data set 1YLS, chains A and B, by using PyMOL (26).
RESULTS
Individual site-specific atomic mutations
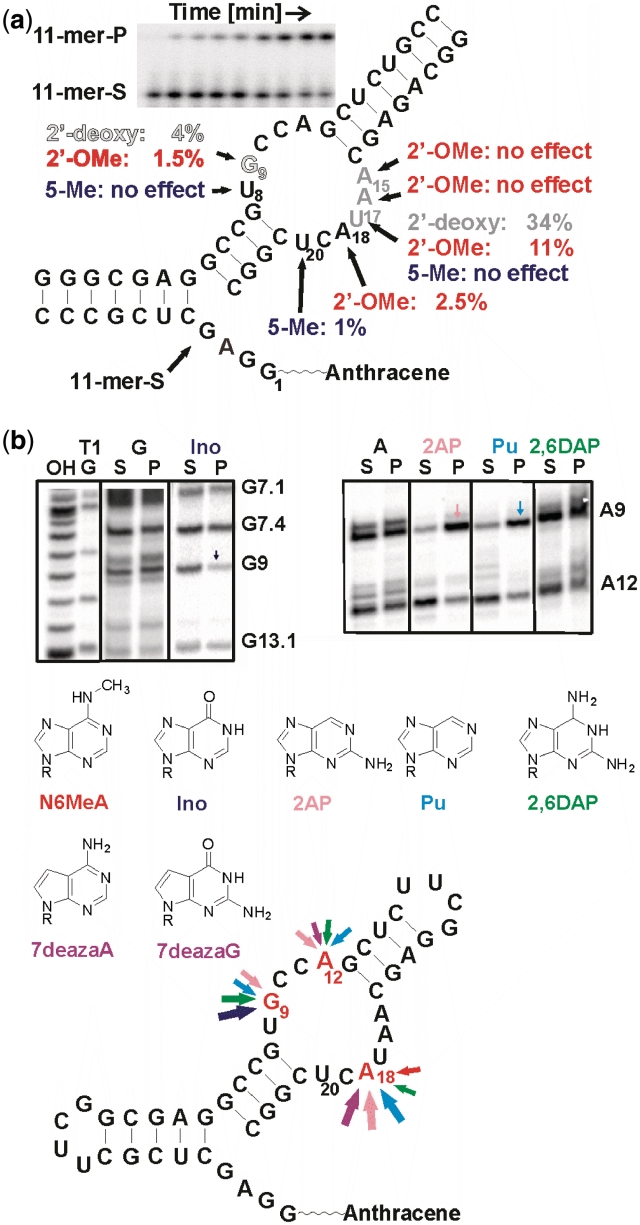
To assess the role of individual functional groups for catalysis, single mutations were introduced into the ribozyme by chemical synthesis, and the influence on the ribozyme-catalyzed reaction estimated using an earlier described tripartite Diels–Alderase assay (10). This ribozyme system of a 24-mer strand, an 18-mer and an anthracene-tether-11-mer spontaneously assembles after mixing, and reactions could be easily monitored by gel electrophoresis (see inset in Figure 2a). Important atomic positions were identified using a rapid screening assay, and only those mutants were selected for further characterization for which the amount of product formed within 30 min was reduced at least by half, compared to the wild-type. For these mutants, apparent rate constants kobs were determined by measuring the initial rate within the first 15% of conversion. Division by kobs, wild-type (0.12 min−1) yielded krel values.
Figure 2.
Single mutation analysis. (a) Individual site-specific mutations. Gray scale represents activity of 2′-deoxy mutants: black—no effect, gray—slight reduction, white w. black hairline—strong effect. Red: 2′-O-methyl substitutions. Blue: 5-methyl substitution. The inset shows a typical assay gel for determination of kobs in the tripartite ribozyme assay. S—substrate, P—product. (b) NAIM. PAGE gels showing interferences at positions 9 and 12. Bottom: summary of nucleotide analog effects. Arrow size corresponds to the magnitude of the effects.
Site-specific 2′-deoxy-substitutions introduced at all nucleotides of the internal bubble and the 5′-GGAG end revealed that not a single 2′-hydroxyl group is absolutely required (Figure 2a). The strongest influence was observed at guanosine G9 (kobs = 0.005 min−1, krel = 4%), and slight activity reductions were seen at adenosines A15 and A16, while for all other positions, the 2′-OH group is apparently not critical. The high residual activity of the U17dU mutant (kobs = 0.04 min−1, krel = 34%) is noteworthy, as the 2′-hydroxyl group of this nucleotide is proposed to be involved in the catalytic mechanism by hydrogen bonding to a maleimide carbonyl oxygen (11).
For selected positions, 2′-O-methyl substitutions were studied, too (red in Figure 2a). Nucleotide G9 tolerated this substitution even worse than the deoxy-modification (krel= 1.5%). While A15 and A16 were insensitive to 2′-O-methylation, the U17 and A18 mutants showed reduced activities with krel values of 11% and 2.5%, respectively.
For the three uridine positions inside the internal bubble, the addition of a methyl group to the 5-position was additionally investigated by incorporating thymidine (blue in Figure 2a). This modification was found to be detrimental at U20 (kobs = 0.001 min−1, krel = 1%), while it has no significant influence at U8 and U17.
Combinatorial atomic mutations
Atomic mutagenesis can also be carried out in a combinatorial fashion (NAIM) (19–21). While this technique was successfully applied to RNA-processing ribozymes for which the change in molecular size due to self-cleavage is a convenient selection marker, the investigation of the Diels–Alder reaction required the development of a modified approach. This involved the preparation and purification of RNA-tether-anthracene conjugates (17) with statistically incorporated α-thio-nucleoside triphosphates (α-thio-NTPs), their 3′-terminal 32P-labeling, reaction with biotin maleimide, and the separation of reacted and unreacted fractions by denaturing PAGE, followed by iodine cleavage at the thiophosphate group and gel analysis (Supplementary Figure S2).
The role of the non-bridging pro-RP-phosphate oxygen atoms was studied by using the parental α-thio-NTPs (i.e. those nucleotides that do not carry additional modifications). While no phosphorothioate interferences were observed for the purine nucleotides, weak but significant interferences were found in the pyrimidine series primarily at positions C10, C11, C19, U8, U17 and U20 (Supplementary Figure S3). According to the crystal structure, all these positions are candidates for metal ion coordination (11). Using modified α-thio-NTPs, rather strong interferences were observed for inosine, purine riboside and 2-aminopurine riboside substitutions at position G9, a nucleotide proposed to make a mechanistically important H-bond to the transition state (Figure 2b). Another strong interference was seen at position A18, in particular for 7-deaza-adenosine, 2-aminopurine riboside and purine riboside, while weaker interferences were seen for N6-methyl-adenosine and 2,6-diaminopurine riboside, suggesting an importance of the Hoogsteen face of this nucleotide. Weak interferences are also seen for nucleotide A/G 12 at the interface of helix III and the internal loop for 7-deaza-adenosine, 2-aminopurine riboside, purine riboside and 2,6-diaminopurine riboside (Figure 2b).
Site-specific double mutations
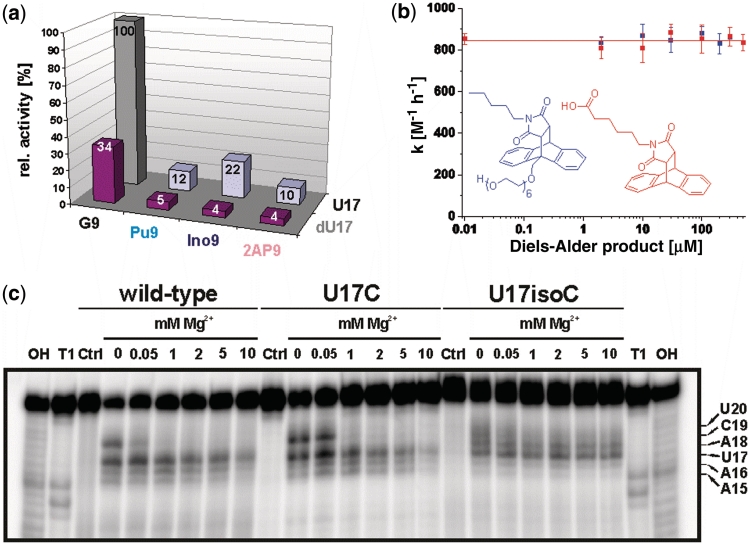
To quantitatively determine the contribution of the two proposed H-bonds between Diels–Alder transition state and ribozyme (2-NH2 of G9 and 2′-OH of U17, both with the same maleimide carbonyl oxygen), we combined individual RNA strands with chemical modifications at the G9 and U17 positions and determined initial rates in the tripartite assay. These measurements revealed krel values of 10% (2-aminopurine), 12% (purine) and 22% (inosine) for the single-mutants in position 9 and 4–5% for the double mutants that carried the U17dU substitution (Figure 3a), thereby showing that both proposed H-bonds indeed contribute to catalytic rate acceleration in an additive manner.
Figure 3.
Investigation of mutations: (a) Relative catalytic activity of the differently combined modifications in positions 9 and 17. Gray column represents the wild-type. (b) Catalytic activity of the U17dU G9Pu double mutant in the presence of two different Diels–Alder products. (c) Lead probing PAGE gel of 5′-32P-labeled wild-type, U17C and U17isoC mutant at different Mg2+ ion concentrations (0–10 mM). 'OH' and 'T1' correspond to alkaline hydrolysis and RNase T1 ladder, 'Ctrl' to control incubation (absence of lead ions). For precise assignment of the bands, compare Figure 6 in reference (10).
Product inhibition studies
The Diels–Alderase is known to be strongly product-inhibited (IC50 ∼10–20 µM) (9), and these two above-mentioned H-bonds are likely to contribute. To study their relevance in product binding, a 49-mer double mutant was synthesized that carries the U17dU and the G9Pu substitutions in one strand, and its catalytic activity towards the free (i.e. non-tethered) substrates determined in the presence of increasing amounts of two different Diels–Alder products. No effect on the reaction rate could be observed up to the solubility limits of these compounds (200 and 500 µM, respectively, Figure 3b), supporting the hypothesis that the product normally makes critical interactions with the 2-NH2 of G9 and the 2′-OH of U17.
Site-specific rescue experiments involving modified nucleotides
To probe the relevance for catalysis of other secondary and tertiary interactions observed in the crystal structure by activity restoration (rescues), site-specifically substituted double and triple mutants were chemically synthesized and assayed for catalytic activity. These chemically modified ribozyme variants involved 2′-deoxyguanosine, 2′-methoxyguanosine, 7-deazaguanosine, 8-oxo-2′-deoxyguanosine, isoguanosine and isocytidine substitutions. The results are summarized in Table 1 and discussed in a structural context below. The (putative) structures of modified base pairs and triples are shown in Supplementary Figure S4. One of the most interesting results is the dependence of catalytic activity on the presence of a single H-bond (4-NH2 group of C10 versus carbonyl oxygen O2 of U17).
Table 1.
Catalytic activities of various single, double and triple mutants of the Diels–Alderase ribozyme
| 5′-end | Upper bulge | Lower bulge | kobs (min−1)a,b | krel (%) |
|---|---|---|---|---|
| U8–A18 reverse Hoogsteen base pair | ||||
| U8C | 0.0023 | 1.9c | ||
| U8iC | 0.001 | 0.8 | ||
| A18G | 0 | 0 | ||
| A18deazaG | 0 | 0 | ||
| A18-8OxodG | 0.0029 | 2.4 | ||
| U8C | A18G | 0 | 0 | |
| U8C | A18deazaG | 0 | 0 | |
| U8C | A18-8OxodG | 0.0032 | 2.6 | |
| U8iC | A18G | 0 | ||
| U8iC | A18deazaG | 0 | ||
| U8iC | A18-8OxodG | 0.0002 | 0.2 | |
| Pseudoknot base pairs G1–C11 and G2–C10 | ||||
| G1iG | ∼0.05 | ∼40 | ||
| C11iC | 0.0004 | 0.3 | ||
| G1iG | C11iC | 0.0037 | 3.1 | |
| G2iG | 0.01 | 8 | ||
| C10iC | 0.001 | 0.8 | ||
| G2iG | C10iC | 0.0004 | 0.3 | |
| Base triple G2–C10–U17 | ||||
| U17C | 0.04 | 33 | ||
| U17iC | 0 | 0 | ||
| G2iG | C10iC | U17iC | 0 | 0 |
| G2iG | C10iC | U17C | 0 | 0 |
| Spine junction G9–U17 | ||||
| G9dG9 | 0.005 | 4 | ||
| G9-2′OMeG | 0.0018 | 1.5 | ||
| U17C | 0.04 | 33 | ||
| G9dG9 | U17C | 0.0018 | 1.5 | |
| G9-2'OMeG | U17C | 0 | 0 | |
aInitial rates were determined in the tripartite gel-shift assay over the first 15% of conversion. The initial rate of the wild-type sequence combination was defined as 100%.
b‘0’ denotes a kobs < 0.0001 min−1 and a krel < 0.1%.
cThe U8 position was earlier referred to as ‘absolutely conserved’ (10). This assignment has to be changed into ‘highly conserved’, as the U8C mutant shows significant residual (1.9% wild-type) activity.
Chemical probing
To elucidate the reason for the functional relevance of this interaction, lead probing experiments were carried out with the U17 wild-type, a 33% active U17C mutant and the completely inactive U17isoC mutant. Lead probing detects both, specific metal ion coordination and general accessibility of the phosphodiester bonds (27). While the overall probing patterns are very similar (indicating similar overall structures), the lower half of the asymmetric bubble shows significant differences. Nucleotide A18 (the direct neighbor of U17) is completely protected from lead cleavage in the wild-type and U17C mutants, while it is efficiently cleaved in the isoC mutant (Figure 3c). While wild-type and U17C mutant are stabilized against cleavage by increasing Mg2+ ion concentration, the probing pattern of the U17isoC mutant is not influenced by divalent cations. Another striking difference is the alteration of the probing patterns upon addition of Diels–Alder product (Supplementary Figure S5). In contrast to wild-type and U17C mutant, the U17isoC mutant exhibits no changes at all, indicating a strongly reduced affinity for the reaction product.
smFRET spectroscopy
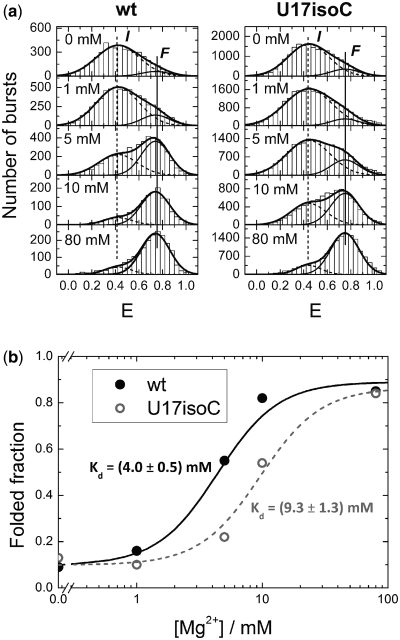
To further elucidate the role of this particular H-bond, variants of the wild-type and the U17isoC mutant were synthesized, labeled with a pair of dyes (Cy3 at the 5′-end and Cy5 in the upper tetraloop) (Supplementary Figure S6) and investigated by smFRET spectroscopy on freely diffusing molecules in solution (24), probing conformational states by varying the concentration of Mg2+ ions (Figure 4). For both constructs, a similar behavior was observed: Two conformations could be distinguished with nearly identical average FRET intensities E of 0.42 and 0.74 (wild-type) and 0.44 and 0.75 (mutant). According to our previous work, these conformations can be assigned to intermediate (I) and folded (F) states (12). For both constructs, the fractional population of the F state increases with the Mg2+ ion concentration at the expense of the I state in a similar fashion with midpoints of 4.0 and 9.3 mM, respectively. The essentially identical E-values and the similar shapes of the distributions indicate that the distances between the dyes are the same and, thus, suggest similar 3D structures.
Figure 4.
Histograms of smFRET efficiency values, E, measured on freely diffusing wild-type ribozyme (left) and U17isoC mutant (right) molecules in buffer solutions at five different Mg2+ concentrations. Dashed and solid thin lines represent best-fit model distributions for the I and F states, respectively; the solid thick line gives the sum of the distributions. It should be noted that, in comparison to our previous work (12), the dye positions were changed to ensure full catalytic activity of the wild-type construct.
DISCUSSION
Architectural principles: the ‘spine’ and the stabilization of the cross-strand junction
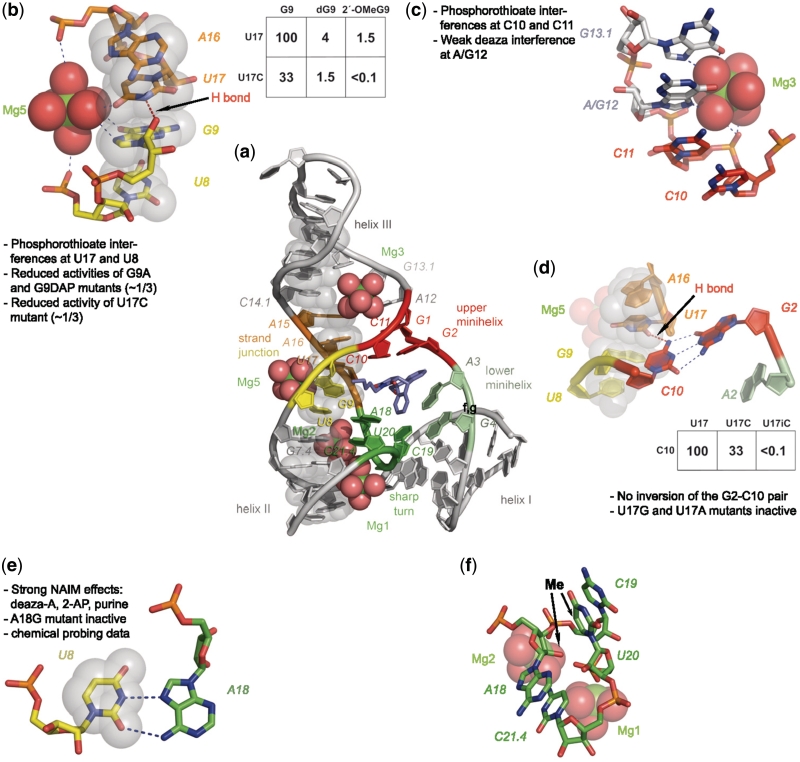
The data obtained in this study allow to more precisely define structural elements and to assign function. While X-ray crystallography showed that extensive stacking is an important principle of this ribozyme's structure formation (11), the atomic mutagenesis data presented here identify the extended cross-strand stack that runs through the whole molecule from the bottom of helix II through the catalytic pocket to the top of helix III (semi-transparent gray spheres in Figure 5a, for a stereo picture see Supplementary Figure S7) as arguably the single most important and most sensitive structural element. We call this element the ‘spine’, and it shapes and reinforces the overall architecture of the catalyst. The junction point of the cross-strand stack is located between nucleotides G9 and U17 (Figure 5b). Nucleotide G9 makes—according to the crystal structure—a number of non-standard interactions, one of them being a hydrogen bond proposed between its 2′-OH group and ring nitrogen HN3 of nucleotide U17 (11). This position is found to be sensitive to 2′-deoxy substitution (Table 1 and Figure 2a, krel = 4%), and placement of two H-bond acceptors opposite each other completely abolishes catalytic activity (G9-2′OMeG versus U17C, krel < 0.1%), thereby strongly supporting the importance of this particular interaction.
Figure 5.
Visualization of structural features of the Diels–Alderase ribozyme. (a) Overview of the structure. Nucleotides belonging to the ‘spine’ are represented as semi-transparent gray spheres through all panels of this figure. (b) Cross-strand junction stabilized by magnesium ion Mg5 and one H-bond. Inset: krel values of selected atomic mutants. (c) Coordination of magnesium ion Mg3. (d) Importance of the U17–C10 H-bond with atomic mutagenesis data. (e) The reverse Hoogsteen base pair U8–A18 with supporting data. (f) Sharp turn involving nucleotides 18–21.4. The two arrows indicate positions where the addition of a CH3 group interferes with activity.
The crystallographic data localize a Mg2+ ion (#5) in the direct vicinity of the junction point. The analysis of the activity data indicates a stabilizing role of this metal ion. It is apparently coordinated to a 5′-phosphate oxygen of U8 (as suggested by the phosphorothioate interference at U8) and carbonyl oxygen O6 of nucleobase G9 on one side of the junction point, as well as with carbonyl oxygen O4 of U17 on the other side. Replacement of either carbonyl oxygen atom by an amino group reduces ribozyme activity by a factor of 3 (Table 1 and Figure 5a), presumably due to attenuated interaction with the hydrated Mg2+ ion. We interpret these data as indicating a synergistic stabilization of the cross-strand junction by the combined action of the G9-U17 H-bond and Mg #5. Interference with the H-bond apparently ‘breaks the spine’, thereby causing loss of function, while an attenuated interaction with the metal ion has a less dramatic effect.
The formation of the catalytic pocket: the ‘roof’
The spine is involved in numerous critical interactions: In the central part, it holds the upper pseudoknot minihelix G1–C11 and G2–C10, which—in the proper orientation—can be viewed as the ‘roof’ of the catalytic pocket. This minihelix seen in the crystal had so far only been supported by indirect evidence from chemical probing, as all mutations in this part (including pair-wise complementary ones) resulted in inactive constructs (10). Here, a partial rescue observed with the isomeric G1isoG—C11isoC double mutant (3.1% activity versus 0.3% activity for the C11isoC single mutant, Table 1) provides some support for this interaction in solution, whereas no rescue was seen for the G2–C10 interaction (28).
Attachment of the roof to the spine is apparently accomplished by a single hydrogen bond, namely between the exocyclic 4-NH2 group of C10 (of the G2–C10 base pair) and carbonyl oxygen O2 of U17 (the nucleotide already found important for the stabilization of the cross-strand junction). In agreement with a critical role of this interaction, incorporation of isoC at position 17 in which the critical carbonyl oxygen O2 is replaced by a NH2 group yields a completely inactive ribozyme (krel < 0.1%, Table 1, Figure 5d). A triple rescue experiment for this tertiary interaction is conceivable but not trivial, as the interacting nucleotides constitute the active site and make direct contacts with the substrates. The inactivity of the isosteric triple mutant U17isoC-C10isoC-G2isoG (krel < 0.1%, Table 1 and Supplementary Figure S4) is, therefore, not surprising.
Further insight into the role of this particular interaction was obtained by chemical probing (Figure 3c) and smFRET spectroscopy (Figure 4) of the U17isoC mutant. Both techniques indicate a global structure very similar to the wild-type ribozyme, but with a different state of the catalytic pocket. The lower half of the bulge becomes more accessible to lead ions, and both methods fail to detect any binding to the Diels–Alder product.
Intriguingly, MD simulations recently established the existence of Diels–Alderase ribozyme conformations with a closed pocket (15). These species still contain the global nested pseudoknot fold, but the register of the upper pseudoknot minihelix is shifted (G2:C11 and G1:A12 instead of G2:C10 and G1:C11), the stabilizing interaction with U17 is missing, and nucleotide G2 stacks directly on A3, thereby closing the catalytic pocket. The U17isoC mutant might therefore be an RNA that exists in the closed pocket state only, is catalytically incompetent, and cannot bind the reaction product.
In the wild-type ribozyme, the critical C10-U17 hydrogen bond is apparently the only stabilizing inter-strand interaction in this section and needs precise positioning for fine-tuned interaction. If perturbed, the catalytic pocket may either collapse, or the upper minihelix may fall off the spine, thereby removing the roof and parts of the back wall from the catalytic pocket. The possibility to disassemble the catalytic pocket by breaking a single hydrogen bond may be important for the multiple-turnover behavior of this catalyst (8,9), and also for the possibility of a ‘back door’ reaction pathway that requires substrate passage through a narrow orifice (29). Such a frequent breakdown and re-formation of the tertiary structure would be consistent with the structural transitions between ‘folded’ and ‘intermediate’ conformations on the 100 ms timescale, which were observed by both single-molecule (12) and NMR spectroscopy (13).
The interface between catalytic pocket and helix III
Following the spine toward the top, the next structural element is the interface between catalytic pocket and helix III. According to the crystal structure (11), a magnesium ion (#3) is localized in this region, where the sugar–phosphate backbone of the counter-strand shows deviations from a standard helical conformation. The observed phosphorothioate interferences are in agreement with metal ion binding to the 5′-phosphate of C11, and the weak nucleotide analog effects at position A12 (7-deazaadenine, 2-aminopurine, purine) support the importance of its Hoogsteen face, which points toward the bound ion (Figure 5c). Likely, metal ion coordination at this site helps positioning the upper pseudoknot minihelix for proper interaction with its partners.
The central scaffold element: helix II, the sharp turn and Mg ions #1 and 2
From the cross-strand junction towards the bottom, the spine holds a very unusual element that is critical for aligning helices I and II in a special kind of three-way junction (Figure 5a). Neighboring nucleotides G7.4 and U8 form stacked base pairs; a standard Watson–Crick pair G7.4-C21.4 and a reverse Hoogsteen pair U8–A18, thereby formally extending helix II by one base pair. In a sharp turn of the counter-strand, the intervening nucleotides C19 and U20 are pushed outwards and rotated, allowing them to form—with their Watson–Crick faces—the lower pseudoknot minihelix that stacks upon helix I and forms the bottom of the catalytic pocket. Thus, while the first and the last member of this four-nucleotide stretch form one helix, the second and third one are involved in another one. While the phosphate backbone of the counter-strand makes only a ∼180° turn, bases A18 and C21.4 stack upon each other and enclose a full 360° segment (Figure 5f). The importance of the sharp turn is confirmed here by two findings: While a 2′-deoxy-substitution is tolerated well at nucleotide A18, incorporation of a 2′-OMe group causes a rather large reduction in activity (Figure 2, krel ∼ 2.5%). Strikingly, the addition of a methyl group to carbon atom C5 of nucleotide U20 leads to inactivation of comparable magnitude (Figure 2, krel= 1%). These two phenomena seem to be connected: in the crystal structure, the 2′-O atom of A18 and C5 of U20 are only 3.3 Å apart and point (roughly) towards each other (see arrows in Figure 5f). Thus, the addition of a methyl group at either side of this hydrophobic compartment has a similarly detrimental effect.
The U8–A18 reverse Hoogsteen base pair (Figure 5e) has been supported by independent data. Both nucleotides are highly conserved, and chemical probing indicated that the N1 position (Watson–Crick face) gets exposed on tertiary structure formation, while N7 (Hoogsteen side) becomes protected (10). An A18G substitution mutant is completely inactive, in agreement with the different Hoogsteen pairing schemes for the two purine nucleotides (10). The removal of the N7 ring nitrogen (deazaA) or the exocyclic amino group (2-aminopurine, purine) lead to reduced activities in NAIM experiments. The complete or near-complete inactivity of all double mutants designed to probe this interaction (Table 1) is likely due to direct interaction of this base pair with the substrates.
In the crystal structure, two Mg2+ ions (#1 and #2) are observed right next to this sharp turn, bound centrally in the major grove of helix II, and at the same time coordinate to the opposed U19 and C20 of the pseudoknot minihelix (Figure 5a and f). A significant phosphorothioate interference was observed here for C19 (Supplementary Figure S3), likely due to direct coordination of its pro-RP phosphate to magnesium ion #2. The pro-RP phosphate oxygen of nucleotide U20 is located 4.0–4.5 Å away from both Mg1 and Mg2, which is a typical distance for outer sphere interactions and may explain the weaker interference. The dimeric metal ion site was also supported by EPR-spectroscopy that identified—by a distinct coupling pattern—two metal ions in close proximity (∼7 Å distance) (14), and by MD simulations (15).
The hydrogen bonding network inside the catalytic pocket
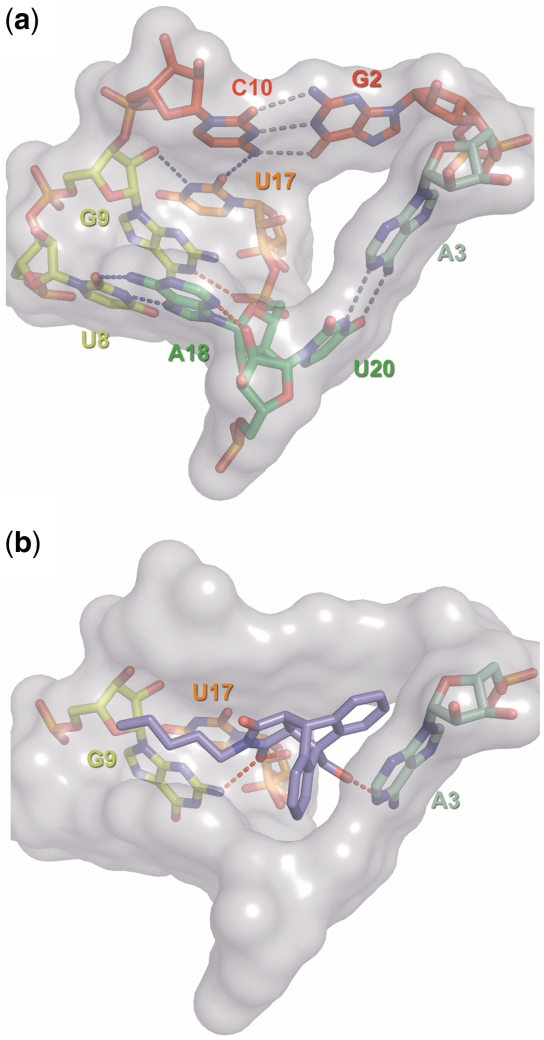
The catalytic center is formed by the central spine section (including the junction), the roof, the bottom, and the reverse Hoogsteen adenosine, and a total of 8 nt make contact with the reaction product (Figure 6). The pocket is held together by an intricate hydrogen bonding network that looks like those seen in typical enzyme active sites. Conventional and atomic mutagenesis was used to probe the relevance of each interaction. The two Watson–Crick standard type interactions (gray dotted lines) are found important, but conservation levels differ. The bottom A-U pair tolerates pair-wise complementary substitutions (10), while for the roof G–C base pair the G2isoG (krel = 8%) and the C10isoC exchanges (krel = 0.8%) are the only mutations that do not completely abolish catalytic activity. This may be due to the different functions; the bottom apparently provides primarily a platform for diene stacking, while the roof has to interact with both substrates. Several non-standard interactions are found to be critically important for function (blue dotted lines), in particular those involving either junction nucleotide U17 or the reverse Hoogsteen pair U8–A18. Other interactions that are conceivable on the basis of the crystallographic information are found to be not essential for catalysis (red dotted lines). The hydrogen bond between ring nitrogen N3 of nucleotide A18 and the 2′-OH group of U20 is not essential, since the U20dU mutant shows high catalytic activity (Figure 2a), and the interaction between ring nitrogen HN1 of G9 and the 5′-phosphate group of A18 is also not critically important, as G9 mutants with an unprotonated N1 (purine and diaminopurine, Figures 2 and 3) are found to be active.
Figure 6.
Surface representation of the catalytic pocket. (a) Empty catalytic pocket with stick representations of the nucleotides. Standard Watson–Crick type interactions are indicated by gray dotted lines, blue indicates non-standard interaction and red highlights interactions for which no experimental evidence was found. (b) RNA–product interactions. Only those nucleotides are shown that were proposed to interact with the Diels–Alder product (red dotted lines).
RNA–substrate interactions and mechanistic considerations
Two of the three hydrogen bonds between RNA and bound cycloaddition product observed in the crystal structure involve a carbonyl oxygen (arising from the maleimide substrate) of the reaction product and have the exocyclic 2-NH2 group of guanosine G9 and the 2′-OH group of U17 as partners. Such hydrogen bonds could indeed be mechanistically meaningful, as a dienophile's reactivity in a Diels–Alder reaction is known to be enhanced by the withdrawal of electron density via hydrogen bonds (30), a strategy also observed in catalytic Diels–Alderase antibodies (31,32).
The atomic mutagenesis data presented here support this view (Figure 3a). While removal of the 2′-OH group of U17 causes only a 3-fold reduction in activity, destruction of the H-bond to the 2-NH2 group of G9 by incorporation of inosine and purine results in a 5- to 8-fold decrease. The respective double mutants have residual activities of 4–5% of the wild-type, demonstrating an additive effect of the two interactions. This additivity suggests that the ribozyme combines the energy of two interactions of moderate strength to make a significant contribution to catalysis (33). The measured rates suggest that these two hydrogen bonds together contribute ∼7–8 kJ/mol to transition state stabilization. On the other hand, such a residual activity represents a ∼600-fold rate acceleration with respect to the uncatalyzed background (corresponding to a ΔΔG# of ∼16 kJ/mol). Thus, the larger part of the energetic contribution comes from other interactions. The crystal structures indicated an excellent shape complementarity between the catalytic pocket and both, the Diels–Alder product and the transition state of the reaction. The magnitude of the equilibrium dissociation constant of the RNA-product complex (9,25) (∼10 µM, corresponding to a ΔG of ∼30 kJ/mol) suggests that noncovalent interactions between the transition state and the catalytic pocket can easily provide the required amount of activation energy. Our finding that the point mutant U17isoC folds into the pseudoknot shape but cannot stabilize the appropriately shaped catalytic pocket and is completely inactive highlights the importance of shape complementarity.
These two H-bonds between RNA and Diels–Alder product are also found to be responsible for the high affinity of this ribozyme for the Diels–Alder product and the associated strong product inhibition (9). In contrast to the wild-type ribozyme, the G9Pu U17dU double mutant was not even slightly inhibited by two different Diels–Alder products up to their respective solubility limits. This finding is not surprising, considering the strong similarity of reaction product and transition state.
The third hydrogen bond proposed in the RNA-product complex between the 6-NH2 group of adenosine A3 and the first ether oxygen of the polyethylene glycol tether attached to the reaction product is not critical either, since both partners of this interaction were previously shown to be variable (9,10,29).
The T-loop motif and its role in the catalysis of Diels–Alder reactions
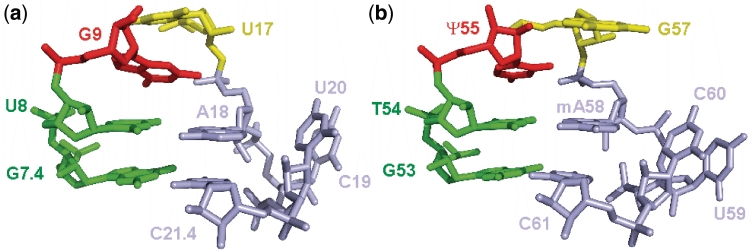
Careful analysis of the structural features of the Diels–Alderase ribozyme revealed the presence of an important tertiary structure motif that is well known in a different context. The sharp 4 base-pair turn, with the bottom base pair being a Watson–Crick and the top base pair a reverse Hoogsteen, is known as the UA_handle which is, e.g. part of the T-loop signature found in tRNA (34). There, it directly interacts with the D-loop and thereby makes an essential contribution to the formation of the L-shaped tertiary structure. The nucleotides involved in its formation are among the most conserved ones in the whole tRNA structure, and the U54-A58 reverse Hoogsteen interaction was identified as one of the key structural aspects of tRNA functionality (35). The striking structural similarities between the respective sections of the Diels–Alderase ribozyme and tRNAPhe (from yeast, PDB Data set 1EHZ) are highlighted in Figure 7 (for more details, see Supplementary Figure S8). Several occurrences of this T-loop motif and the UA_handle submotif have also been reported in ribosomal and other large RNAs (34,36,37). Thus, rather than inventing a new structural solution for the problem of catalyzing a Diels–Alder reaction, in vitro evolution has recycled a motif that natural evolution has created a long time ago for completely different purposes, providing yet another example for the modular character of RNA architecture (38–40).
Figure 7.
Comparison of the structures of the Diels–Alderase sharp turn (panel a) and the T-loop of tRNAPhe (b).
The occurrence of the T-loop motif in the Diels–Alderase ribozyme makes an interesting connection to a report about the catalysis of Diels–Alder reactions by native tRNA (tRNAPhe from yeast) under high pressure (41). In that study, a different set of substrates (6-furfuryl-adenine as diene and maleic anhydride as dienophile) was used. The authors concluded from pressure-dependent lead probing and CD spectroscopy that high pressure disrupts the T-loop–D-loop interaction, and proposed that this opened tRNA forms a scaffold to which the diene is docked by stacking interaction. Considering the structural parallels with the Diels–Alderase ribozyme, this suggestion appears conceivable, as the left half of the catalytic center would already be in place. The adenine part of the diene substrate could furthermore be sandwiched between the G57 (yellow) and reverse Hoogsteen A58 (blue) in the space that is normally occupied by G18 from the D-loop, thereby placing the furfuryl ring roughly at the same position (relative to the T-loop motif) as the anthracene ring system in the Diels–Alderase ribozyme. How the dienophile substrate is positioned and how the catalytic site is formed remains unclear, however.
CONCLUSIONS AND OUTLOOK
While the X-ray crystal structures of the Diels–Alderase provided a first detailed glimpse at the structural principles governing the architecture of this remarkable ribozyme, the atomic mutagenesis studies presented here allow the classification of structural elements and the assignment of function. The roles of several individual hydrogen bonds and of four magnesium ions for the stability and catalytic function of the ribozyme could be unraveled. The critical importance of the central hydrogen bonding network was established by experimental data. Moreover, new insight was obtained regarding the mechanistic role of hydrogen bonds between catalyst and substrates, and the integrity of the catalytic pocket. The loss of a single H-bond on the surface of the catalytic pocket leads to complete inactivation. The base triplet G2–C10–U17 and, in particular, the acceptor–donor pair 2O-U17 versus 4NH2-C10, are of extraordinary importance for the formation of the active fold. A sharp 360° turn reminiscent of the T-loop motif observed in tRNA is found to be crucially important for catalytic function, although its precise role is not clear yet. Future work will address some of the open questions regarding the structure and mechanism of this ribozyme, such as the order of substrate binding and the role of conformational changes during the catalytic cycle, and the molecular details about how this ribozymes folds into its nested pseudoknot tertiary architecture.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1–S8.
FUNDING
Deutsche Forschungsgemeinschaft (DFG Ja 794/3 and CFN); BioFuture program of the German Federal Government (BEO 0311861); Human Frontiers Science Program (RGP 76/2003); Fonds der Chemischen Industrie, and the Volkswagen Foundation (VW-I/82549). Funding for open access charge: Institutional Budget.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 2.Lilley DM. Structure, folding and mechanisms of ribozymes. Curr. Opin. Struct. Biol. 2005;15:313–323. doi: 10.1016/j.sbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Pitt JN, Ferre-D'Amare AR. How RNA closes a Diel. Nat. Struct. Mol. Biol. 2005;12:206–208. doi: 10.1038/nsmb0305-206. [DOI] [PubMed] [Google Scholar]
- 4.Scott WG. Ribozymes. Curr. Opin. Struct. Biol. 2007;17:280–286. doi: 10.1016/j.sbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Seelig B, Jäschke A. A small catalytic RNA motif with Diels-Alderase activity. Chem. Biol. 1999;6:167–176. doi: 10.1016/S1074-5521(99)89008-5. [DOI] [PubMed] [Google Scholar]
- 6.Tarasow TM, Tarasow SL, Eaton BE. RNA-catalysed carbon-carbon bond formation. Nature. 1997;389:54–57. doi: 10.1038/37950. [DOI] [PubMed] [Google Scholar]
- 7.Nicolaou KC, Snyder SA, Montagnon T, Vassilikogiannakis GE. The Diels-Alder reaction in total synthesis. Angew. Chem. Int. Ed. 2002;41:1668–1698. doi: 10.1002/1521-3773(20020517)41:10<1668::aid-anie1668>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 8.Seelig B, Keiper S, Stuhlmann F, Jäschke A. Enantioselective ribozyme catalysis of a bimolecular cycloaddition reaction. Angew. Chem. Int. Ed. Engl. 2000;39:4576–4579. [PubMed] [Google Scholar]
- 9.Stuhlmann F, Jäschke A. Characterization of an RNA active site: interactions between a Diels-Alderase ribozyme and its substrates and products. J. Am. Chem. Soc. 2002;124:3238–3244. doi: 10.1021/ja0167405. [DOI] [PubMed] [Google Scholar]
- 10.Keiper S, Bebenroth D, Seelig B, Westhof E, Jäschke A. Architecture of a Diels-Alderase ribozyme with a preformed catalytic pocket. Chem. Biol. 2004;11:1217–1227. doi: 10.1016/j.chembiol.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 11.Serganov A, Keiper S, Malinina L, Tereshko V, Skripkin E, Höbartner C, Polonskaia A, Phan AT, Wombacher R, Micura R, et al. Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation. Nat. Struct. Mol. Biol. 2005;12:218–224. doi: 10.1038/nsmb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobitski AY, Nierth A, Helm M, Jäschke A, Nienhaus GU. Mg2+-dependent folding of a Diels-Alderase ribozyme probed by single-molecule FRET analysis. Nucleic Acids Res. 2007;35:2047–2059. doi: 10.1093/nar/gkm072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manoharan V, Fürtig B, Jäschke A, Schwalbe H. Metal-induced folding of Diels-Alderase ribozymes studied by static and time-resolved NMR spectroscopy. J. Am. Chem. Soc. 2009;131:6261–6270. doi: 10.1021/ja900244x. [DOI] [PubMed] [Google Scholar]
- 14.Kisseleva N, Kraut S, Jäschke A, Schiemann O. Characterizing multiple metal ion binding sites within a ribozyme by Cadmium-induced EPR silencing. HFSP J. 2007;1:127–136. doi: 10.2976/1.2756332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berezniak T, Zahran M, Imhof P, Jäschke A, Smith JC. Magnesium-dependent active-site conformational selection in the Diels-Alderase ribozyme. J. Am. Chem. Soc. 2010;132:12587–12596. doi: 10.1021/ja101370e. [DOI] [PubMed] [Google Scholar]
- 16.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seelig B, Jäschke A. Ternary conjugates of guanosine monophosphate as initiator nucleotides for the enzymatic synthesis of 5'-modified RNAs. Bioconjug. Chem. 1999;10:371–378. doi: 10.1021/bc980085h. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 19.Ryder SP, Ortoleva-Donnelly L, Kosek AB, Strobel SA. Chemical probing of RNA by nucleotide analog interference mapping. Methods Enzymol. 2000;317:92–109. doi: 10.1016/s0076-6879(00)17008-9. [DOI] [PubMed] [Google Scholar]
- 20.Ryder SP, Strobel SA. Nucleotide analog interference mapping. Methods. 1999;18:38–50. doi: 10.1006/meth.1999.0755. [DOI] [PubMed] [Google Scholar]
- 21.Strobel SA, Shetty K. Defining the chemical groups essential for Tetrahymena group I intron function by nucleotide analog interference mapping. Proc. Natl Acad. Sci. USA. 1997;94:2903–2908. doi: 10.1073/pnas.94.7.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oyelere AK, Kardon JR, Strobel SA. pK(a) perturbation in genomic Hepatitis Delta Virus ribozyme catalysis evidenced by nucleotide analogue interference mapping. Biochemistry. 2002;41:3667–3675. doi: 10.1021/bi011816v. [DOI] [PubMed] [Google Scholar]
- 23.Nierth A, Singer M, Jäschke A. Efficient photoactivation of a Diels-Alderase ribozyme. Chem. Commun. (Camb) 2010;46:7975–7977. doi: 10.1039/c0cc03162c. [DOI] [PubMed] [Google Scholar]
- 24.Kobitski AY, Hengesbach M, Helm M, Nienhaus GU. Sculpting an RNA conformational energy landscape by a methyl group modification–a single-molecule FRET study. Angew. Chem. Int. Ed. Engl. 2008;47:4326–4330. doi: 10.1002/anie.200705675. [DOI] [PubMed] [Google Scholar]
- 25.Nierth A, Kobitski AY, Nienhaus GU, Jäschke A. Anthracene-BODIPY dyads as fluorescent sensors for biocatalytic Diels-Alder reactions. J. Am. Chem. Soc. 2010;132:2646–2654. doi: 10.1021/ja9084397. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WL. 2002. The PyMOL Molecular Graphics System, Version 1.1beta4, Schrödinger, LLC. http://www.pymol.org (30 September 2011, date last accessed) [Google Scholar]
- 27.Lindell M, Brannvall M, Wagner EG, Kirsebom LA. Lead(II) cleavage analysis of RNase P RNA in vivo. RNA. 2005;11:1348–1354. doi: 10.1261/rna.2590605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, Kierzek R, Turner DH. Stability and structure of RNA duplexes containing isoguanosine and isocytidine. J. Am. Chem. Soc. 2001;123:1267–1274. doi: 10.1021/ja002623i. [DOI] [PubMed] [Google Scholar]
- 29.Wombacher R, Keiper S, Suhm S, Serganov A, Patel DJ, Jäschke A. Control of stereoselectivity in an enzymatic reaction by backdoor access. Angew. Chem. Int. Ed. Engl. 2006;45:2469–2472. doi: 10.1002/anie.200503280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodward RB, Hoffmann R. Conservation of orbital symmetry. Angew. Chem. Int. Ed. Engl. 1969;8:781–853. [Google Scholar]
- 31.Heine A, Stura EA, Yli-Kauhaluoma JT, Gao C, Deng Q, Beno BR, Houk KN, Janda KD, Wilson IA. An antibody exo Diels-Alderase inhibitor complex at 1.95 angstrom resolution. Science. 1998;279:1934–1940. doi: 10.1126/science.279.5358.1934. [DOI] [PubMed] [Google Scholar]
- 32.Romesberg FE, Spiller B, Schultz PG, Stevens RC. Immunological origins of binding and catalysis in a Diels-Alderase antibody. Science. 1998;279:1929–1933. doi: 10.1126/science.279.5358.1929. [DOI] [PubMed] [Google Scholar]
- 33.Shan SO, Herschlag D. Energetic effects of multiple hydrogen bonds. Implications for enzymatic catalysis. J. Am. Chem. Soc. 1996;24:5515–5517. [Google Scholar]
- 34.Jaeger L, Verzemnieks EJ, Geary C. The UA_handle: a versatile submotif in stable RNA architectures. Nucleic Acids Res. 2009;37:215–230. doi: 10.1093/nar/gkn911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zagryadskaya EI, Doyon FR, Steinberg SV. Importance of the reverse Hoogsteen base pair 54-58 for tRNA function. Nucleic Acids Res. 2003;31:3946–3953. doi: 10.1093/nar/gkg448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krasilnikov AS, Mondragon A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA. 2003;9:640–643. doi: 10.1261/rna.2202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagaswamy U, Fox GE. Frequent occurrence of the T-loop RNA folding motif in ribosomal RNAs. RNA. 2002;8:1112–1119. doi: 10.1017/s135583820202006x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 39.Jaeger L, Westhof E, Leontis NB. TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001;29:455–463. doi: 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mielcarek M, Barciszewska MZ, Salanski P, Stobiecki M, Jurczak J, Barciszewski J. Native transfer RNA catalyzes Diels-Alder reaction. Biochem. Biophys. Res. Commun. 2002;294:145–148. doi: 10.1016/S0006-291X(02)00452-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.