Abstract
HBP1 is a sequence-specific DNA-binding transcription factor with many important biological roles. It activates or represses the expression of some specific genes during cell growth and differentiation. Previous studies have exhibited that HBP1 binds to p16INK4A promoter and activates p16INK4A expression. We found that trichostatin A (TSA), an inhibitor of HDAC (histone deacetylase), induces p16INK4A expression in an HBP1-dependent manner. This result was drawn from a transactivation experiment by measuring relative luciferase activities of p16INK4A promoter with HBP1-binding site in comparison with that of the wild-type p16INK4A promoter by transient cotransfection with HBP1 into HEK293T cells and 2BS cells. HBP1 acetylation after TSA treatment was confirmed by immunoprecipitation assay. Our data showed that HBP1 interacted with histone acetyltransferase p300 and CREB-binding protein (CBP) and also recruited p300/CBP to p16INK4A promoter. HBP1 was acetylated by p300/CBP in two regions: repression domain (K297/305/307) and P domain (K171/419). Acetylation of Repression domain was not required for HBP1 transactivation on p16INK4A. However, luciferase assay and western blotting results indicate that acetylation of P domain, especially K419 acetylation is essential for HBP1 transactivation on p16INK4A. As assayed by SA-beta-gal staining, the acetylation of HBP1 at K419 enhanced HBP1-induced premature senescence in 2BS cells. In addition, HDAC4 repressed HBP1-induced premature senescence through permanently deacetylating HBP1. We conclude that our data suggest that HBP1 acetylation at K419 plays an important role in HBP1-induced p16INK4A expression.
INTRODUCTION
HBP1 is homologous to the sequence-specific high mobility group (HMG) family of transcription factors (1,2). We have previously isolated HBP1 as a retinoblastoma (RB) partner and have determined that it functions as a proliferation regulator by inhibiting oncogenic pathways as a transcriptional repressor (3–8). Recently, the HBP1 transcriptional repressor has been reported as a putative substrate for the p38 MAPK in cell-cycle arrest (9). Mechanistically, p38 MAPK-mediated phosphorylation of the HBP1 leads to increased protein stability and G1 arrest. A specific p38 MAPK phosphorylation site (serine 401) has been identified in the HBP1 protein. Moreover, our previous work demonstrated that HBP1 is necessary for premature senescence induced by Ras-p38 MAPK (10). Furthermore, HBP1 itself also induces premature senescence through upregulating p16INK4A expression in primary cells (18). p16INK4A knockdown can abolish HBP1- and Ras-induced premature senescence. Together, our previous data support a model in which HBP1 is a downstream effector of Ras, and p38 MAPK, and provide new insights into the signaling mechanisms that lead to premature senescence. All of the data indicate that HBP1 modulation is a complex process, and the biological consequences of HBP1 activation induced by certain stimuli may depend on HBP1 post-translational modification at multiple sites. There is no report about the roles of the acetylated HBP1. Investigating whether or not HBP1 is acetylated by acetyltransferase and if the acetylated HBP1 increases its DNA binding as well as downstream transcriptional activity are the objectives of this report.
Histone deacetylase (HDAC) inhibitors have been extensively studied in basic biological research to gain an understanding of basic chromatin structure and transcriptional control and have recently been introduced as potential clinical treatment for cancer (11–15). Generally, HDAC inhibitors induce accumulation of hyperacetylated nucleosome core histones and cause transcriptional activation of genes (11,23). In addition, HDAC inhibitors are reported to increase acetylation of non-histone proteins (16,17). The common laboratory HDAC inhibitor trichostatin A (TSA) is a potential candidate for the study of HBP1 acetylation.
In this study, normal human lung fibroblast 2BS cells were treated with TSA to test changes of HBP1 acetylation. When assayed for relative luciferase activity using mutagenized p16INK4A promoters and transfection of wild-type HBP1 into 2BS cells, TSA-induced p16INK4A expression was partially attributed to HBP1 acetylation. Furthermore, HBP1 interacted with both histone acetyltransferase p300 and CREB-binding protein (CBP). HBP1 was acetylated by p300/CBP at specific sites: the Repression domain (K297/305/307) and the P domain (K171/419). The acetylation of K419 within the P domain is essential for HBP1 transcriptional activation on p16INK4A by increasing its DNA binding to the p16INK4A promoter.
MATERIALS AND METHODS
Cell culture, transfection and treatment
Human lung fibroblasts 2BS (27,28), Human embryonic kidney HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 µg/ml streptomycin, at 37°C in 5% CO2. All the plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. The cells were collected at 24 to 48 h after transfection for further analysis. The HDAC inhibitor Trichostatin A (TSA) (Sigma) was dissolved in ethanol and added into culture medium at 1 or 2 µM for 12 to 24 h. The control cells were treated with an equal volume of ethanol for the same time periods as mentioned above.
Expression and knockdown plasmids
The overexpression vectors of HBP1 and its point mutants K171R, K419R, K171R/K419R were constructed in the pcDNA3.1 background. The point mutations were introduced at positions 171aa, 419aa, 171aa and 419aa by changing Lysine (K, AAA or AAG) into Arginine (R, AGA or AGG) with overlap PCR. All the plasmids were transfected into cells using Lipofectamine 2000 for transient expression. To obtain stable expression cell lines, cells were infected using lentiviral gene expression system. To prepare lentiviral particles, HEK293T cells were transfected with the three packaging plasmids of pLP1, pLP2 and pLP3 and one expression plasmid of pITA inserted with the gene of interest. The supernatant was harvested and added to the culture medium of target cells for 8–12 h. Then the cells were selected by puromycin and these cells were further used in later experiments. The short hairpin RNA (shRNA) plasmid targeting 19 residues for human HBP1 or HDAC4 was constructed in the pSuper-retro background: HBP1 knockdown (GATCCCCACTGTGAGTGCCACTTCTCTTCAAGAGAGAGAAGTGGCACTCACAGTTTTTTGGAAA), HDAC4 knockdown (GATCCCCGTCGGGCCAGTGGTCACTGTTCAAGAGACAGTGACCACTGGCCCGACTTTTTGGAAA). The underlined sequences represent the hairpin (10). The plasmids were transfected into cells by using Lipofectamine 2000 according to the manufacturer's instruction. For the p300/CBP knockdown, cells were transfected with p300 siRNA (AACAGAGCAGUCCUGGAUUAG) and CBP siRNA (UAGUAACUCUGGCCAUAGC) using oligofectamine reagent (Invitrogen) according to the manufacturer's instructions.
RT–PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) following the manufacturer's protocol. One microgram of RNA was analyzed by reverse transcription (RT)–PCR with the Access RT–PCR Kit (Promega). The DNA sequences of the human HBP1 primers (10) were 5′-ATCATCTCCTGTACACATCATAGC-3′ and 5′-CATAGAAAGGGTGGTCCAGCTTAC-3′. These primers resulted in an RT–PCR product of 523 bp. The DNA sequences of the human p16INK4A primers were 5′-GCCGGCGGCGGGGAGCAGCATGG-3′ and 5′-CAGCATTCGAGAGATCTGTACGC-3′. These primers resulted in an RT–PCR product of 379 bp. The DNA sequences of the human GAPDH primers were 5′-CGAGTCAACGGATTTGGTGGTAT-3′ and 5′-AGCCTTCTCCATGGTGAAGAC-3′. These primers resulted in an RT–PCR product of 320 bp. GAPDH was applied as internal control for normalizing the RT–PCR results.
Immunoblotting and antibody
The cells were collected and lysed in cell lysis buffer (50 mM Tris–HCl, pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EGTA, 1 mM Na3VO4, 1 mM NaF) containing protease inhibitors: 1 mM PMSF (Sigma), 1 μg/ml Leupeptin (Sigma) and 1 μg/ml Pepstatin A (Sigma). After the insoluble part of the lysates was cleared by centrifugation, protein concentrations were determined by the BCA Protein Assay Kit (Pierce). Forty micrograms of proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred onto a nitrocellulose (NC) membrane. The primary antibodies used for western blot analysis were against HBP1 (N-20X, Santa Cruz), HBP1 (ab83402, Abcam), p16INK4A (C-20, Santa Cruz), p38 (A-12, Santa Cruz), HA.11 antibody (Convance), Flag (Sigma), p300 (sc-585, Santa Cruz), CBP (sc-369, Santa Cruz), PCAF (sc-8999, Santa Cruz), GST (M071-3, MBL) and Anti-Multi Ubiquitin (D058-3, MBL) and Acetylated-Lysine Monoclonal Antibody (#9681, CST).
Immunoprecipitation
Cells were lysed in cell lysis buffer containing protease inhibitors and incubated on ice for 20 min. Extracts were cleared by centrifugation at 12 000 rpm for 10 min at 4°C. Supernatants were subjected to immunoprecipitation with the specific antibodies by rocking for 1 h at 4°C followed by the addition of 40 μl of a 50% slurry of protein A agarose beads (Sigma) and further incubation for 1 h. Precipitated complexes were pelleted, washed three times with cell lysis buffer and boiled in 2× protein loading buffer for 5 min. The supernatants were transferred to new tubes for further western blot analysis.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assay was performed using the Chromatin Immunoprecipitation Assay Kit (Upstate) following the manufacturer's instruction with slight modification. For the p16INK4A promoter, the PCR product to be amplified resides at the position of −272 to −699 bp. The sequences of PCR primers were 5′-CCTTCCAATGACTCCCTC-3′ and 5′-AACCTTCCTAACTGCCAAA-3′. The HBP1-binding site is located in the region from −426 to −433 bp of the p16INK4A promoter (11).
Purification of recombinant proteins
The glutathione S-transferase (GST) fusion protein expression vectors of p53, HBP1 and its mutants DelHMG, DelRepression, Repression, HMG, P, P1, P2, P3, P2-K144, P2-K160, P2-K 171, P3-K416, P3-K419 were inserted into the basic pGEX-4T-1 vector at the BamHI and EcoRI sites. The vectors were transformed into Escherichia coli BL21. Expression of fusion proteins were induced by 0.5–1 mM of isopropyl-β-d-thiogalactoside (IPTG) at 20°C to 25°C overnight. The purification of GST fusion proteins was performed using Glutathione Sepharose 4B (GE Healthcare) as the manufacturer's instruction with slight modification. The purification efficiency and protein concentration were identified using silver staining as described previously (34). The purified recombination proteins were stored at −80°C for further in vitro protein acetylation assay. GST and p53 (29) fusion protein were used as negative and positive controls, respectively.
In vitro protein acetylation assay
The HAT domain of p300/CBP protein was purchased from Upstate company (#14-418, Upstate). The protein acetylation assay was performed as previously described with slight modification (30). In the standard assay, a 50 μl reaction mixture contained 20 μg of purified GST fusion protein, 0.5 μg of HAT of p300/CBP protein, 4 μg of acetyl-CoA (Ac-CoA) (Sigma) and 10 μl of 5× HAT assay buffer (250 mM Tris–HCl, pH 8.0, 50% glycerol, 0.5 mM EDTA, 5 mM dithiothreitol). The contents were mixed gently and placed in a 30°C shaking incubator for 4 h. Fifty microliters of 2× protein loading buffer was added into the reaction and boiled for 5 min. The reaction products were separated by SDS–PAGE and immunoblotted with the acetylated-lysine antibody (#9681, CST).
Reporter gene assay
Cells were transfected with the above-mentioned plasmids using Lipofectamine 2000 (Invitrogen). Cell lysates were prepared with the Dual Luciferase Reporter Assay Kit (Promega) as the manufacturer's instruction at 36–48 h post-transfection. The normal promoter plasmid pGL3-NP was constructed by inserting part of the normal p16INK4A promoter (length of 658 bp from transcriptional origin) into pGL3-basic, and the mutant promoter plasmid pGL3-MP was constructed by inserting part of the mutated p16INK4A promoter (point mutation at HBP1-binding site: change gggggTaggggg to gggggAaggggg). Changing T at position of −430 bp to A results in the HBP1 abortive binding to p16INK4A promoter. The Firefly Luciferase activity measurements were normalized to Renilla Luciferase activity for the same sample. The luciferase assay was performed on three biological replicates and each replicate was measured at least three times.
Senescence-associated-β-gal staining
Cells were washed twice with PBS, fixed for 3–5 min in 2% formaldehyde/0.2% glutaraldehyde or 3% formaldehyde at room temperature, and washed again with PBS. The cells were then incubated overnight at 37°C without CO2 in fresh senescence-associated-beta-gal (SA-β-Gal) stain solution (31) consisting of 1 mg/ml 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal), 40 mM citric acid–sodium phosphate, pH 6.0, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl and 2 mM MgCl2. At least 300 cells were counted in randomly chosen fields.
RESULTS
HBP1-induced p16INK4A expression is enhanced by TSA treatment
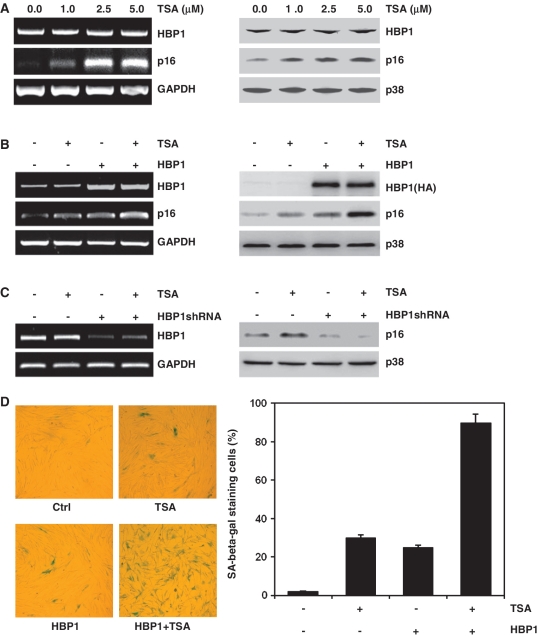
First, we found that HDAC inhibitor TSA increased p16INK4A mRNA and protein levels significantly in a dose-dependent manner in 2BS cells, but had no effect on transcription factor HBP1 expression, as shown in Figure 1A. As previously reported, HBP1 upregulated p16INK4A expression through targeting to the p16INK4A promoter (18). This role of HBP1 was enhanced by TSA treatment (Figure 1B). TSA was not able to induce p16INK4A expression if HBP1 was knocked-down with shRNA (Figure 1C). With senescence-associated β-galactosidase staining (SA-β-gal staining), a senescence biomarker, Figure 1D shows that both HBP1 and TSA increased the percentage of cells for SA-β-gal staining. Furthermore, HBP1-induced premature senescence was enhanced by TSA treatment. All of the data indicate that HBP1 induced p16INK4A expression and that premature senescence was enhanced by TSA. TSA-induced p16INK4A expression depends on HBP1.
Figure 1.
HBP1-induced p16INK4A expression was enhanced by TSA treatment. (A) 2BS cells were treated with TSA at different concentration (0, 1, 2.5, 5 μM) for 18 h. The mRNA levels of p16INK4A and HBP1 were determined by RT–PCR (left panel). The protein levels of HBP1 and p16INK4A were determined by western blot (right panel). (B) 2BS cells were stably infected with pITA-HBP1. After selection for 3 days, cells were treated with TSA at 1 μM for 18 h. mRNA (left panel) and protein (right panel) were extracted for analysis. (C) 2BS cells were transfected with pHBP1shRNA or pSuper.retro (as control). After selection for 3 days, cells were treated with TSA at 1 μM for 18 h. The left panel shows HBP1 knockdown efficiency determined by RT–PCR and the right panel shows the protein level of p16INK4A measured by western blot. (D) Young 2BS cells (PD20) were transfected with pITA-HBP1. After selected with puromycin for 3 days, cells were treated with TSA. Then the cells were stained for SA-β-gal (left panel). The percentage of cells positive for SA-β-gal in 2BS cells is shown in right panel. At least 300 cells were counted for each sample.
TSA enhances the transcriptional activity of HBP1 through increasing DNA binding of HBP1 to the p16INK4A promoter
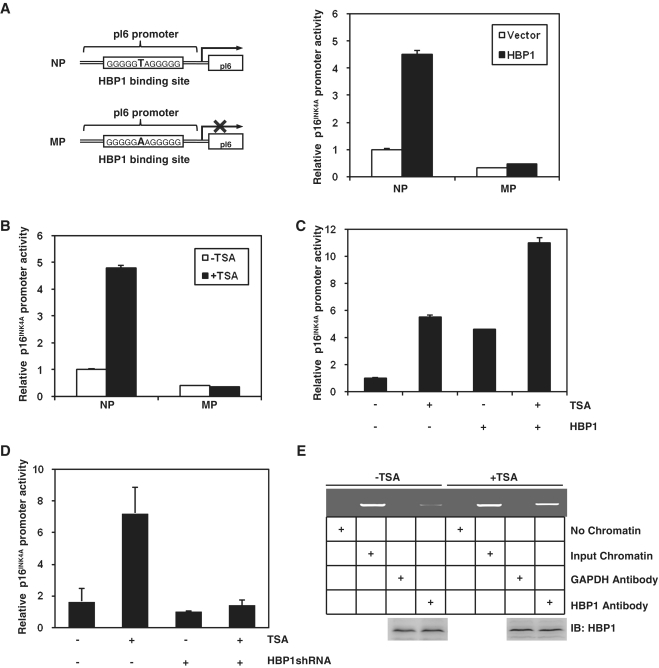
Here, we asked whether TSA increased the transcriptional activity of HBP1 on p16INK4A. We performed overlap PCR to construct a mutant promoter in the background of the native p16INK4A promoter (NP), designated as MP (point mutation at position −430 in HBP1-binding site: change T to A) (Figure 2A). 2BS cells were then transfected with HBP1 and luciferase reporters with the native p16INK4A promoter (NP) or the mutant p16INK4A promoter (MP). In the luciferase assay, HBP1 failed to activate MP, whereas it increased NP activity significantly as previously reported. The result indicates that the integrity of binding site is indispensible for HBP1 to enhance p16INK4A promoter activity in vivo. As shown in Figure 2B, TSA also enhanced the activity of the p16INK4A promoter (NP), but it had no effect on MP, which indicates that TSA-induced p16INK4A expression needs HBP1 binding. The transcriptional activity of p16INK4A promoter in TSA-treated cells was ∼10-fold higher than that in control cells. Furthermore, TSA enhanced the transcriptional activity of HBP1 on p16INK4A (Figure 2C). But TSA lost its transcriptional activation ability at the p16INK4A promoter when HBP1 was knocked-down with shRNA (Figure 2D), which means that TSA-induced p16INK4A expression requires HBP1. We have reported that HBP1 regulates p16INK4A expression through binding to the p16INK4A promoter. In this study, we used the ChIP assay to test the change of DNA binding of HBP1 to the p16INK4A promoter after TSA treatment. As shown in Figure 2E, the binding increased after TSA treatment, consistent with previous result showing that p16INK4A expression increased in TSA-treated cells. All the results indicate that TSA enhances the transcriptional activity of HBP1 through increasing DNA binding of HBP1 to the p16INK4A promoter.
Figure 2.
TSA increased HBP1 transactivation on p16INK4A by enhancing the DNA binding of HBP1 to the promoter region of p16INK4A. (A) The integrity of binding site is indispensible for HBP1 enhancing p16INK4A promoter. Schematic diagrams of the native p16INK4A promoter (NP) and its mutant promoter (MP) (left panel). The mutant promoter was constructed by changing one point (T to A) in HBP1-binding site in the background of native p16INK4A promoter. Luciferase assay was performed in cells transfected with NP, or MP and HBP1 to test p16INK4A mutant promoter (right panel). (B) HBP1-binding site is required for activating TSA-induced p16INK4A expression. Thirty hours after transfection with either pGL3-NP or pGL3-MP, 2BS cells were treated with TSA at 1 μM for 18 h. Relative luciferase activity was measured after TSA treatment. (C) The transactivation of HBP1 on p16INK4A is enhanced by TSA. Thirty hours after transfection with HBP1, cells were treated with TSA at 1 μM for 18 h. Then the cells were harvested for luciferase assay. (D) TSA loses its transactivation ability at the p16INK4A promoter when HBP1 was knocked-down with shRNA. Thirty hours after transfection with HBP1shRNA, cells were treated with TSA at 1 μM for 18 h. Then the cells were harvested for luciferase assay. (E) TSA enhances the DNA binding of HBP1 to the promoter region of p16INK4A in vivo. ChIP assay with antibody against HBP1 was performed in 2BS cells with or without TSA treatment at 1 μM for 18 h. Lanes with input chromatin were positive controls (lanes 2, 6). Lanes without chromatin or GAPDH antibody were used for negative controls (lanes 1, 3, 5, 7). At the bottom, anti-HBP1 western immunoblotting (IB) for HBP1 protein expression is shown.
HBP1 is an acetylated protein
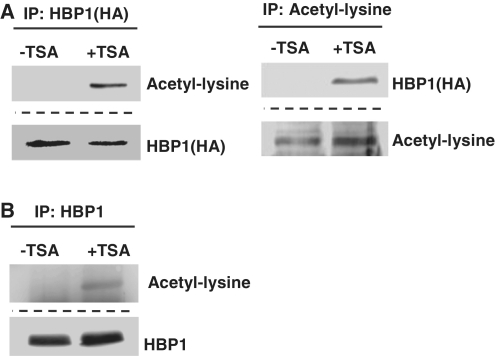
Figure 1A shows that HBP1 mRNA level was not obviously changed upon TSA treatment in 2BS cells. However, TSA was not able to induce p16INK4A expression if HBP1 was knocked-down with shRNA (Figure 1C). The data imply that a modification in HBP1 induced by TSA may lead to HBP1 transcriptional activation of p16INK4A. To determine whether HBP1 is acetylated in the cells, 2BS cells were transfected with a HA-HBP1 expression vector and then treated with TSA. Subsequently, an acetyl-lysine antibody was used for detecting HBP1 acetylation in the TSA-treated 2BS cells. By performing IP with anti-HA and then probing with acetyl-lysine antibody, or IP with acetyl-lysine antibody and then probing with anti-HA, exogenous HBP1 acetylation was confirmed in the TSA-treated cells (Figure 3A). In addition, we also tested the endogenous HBP1 acetylation. As shown in Figure 3B, with HBP1 antibody to substitute for anti-HA, by performing IP with anti-HBP1 and then probing with acetyl-lysine antibody, endogenous HBP1 was also acetylated within cells. In summary, the data allowed us to conclude that HBP1 was indeed acetylated within the cells upon TSA treatment.
Figure 3.
HBP1 was acetylated within the cells. (A) Exogenous HBP1 was acetylated within the cells. 2BS cells transfected with HBP1(HA-tagged) were treated with TSA at 1 μM for 18 h. IP assay was carried out by using HA antibody and followed by western blot with acetyl-lysine antibody (left). IP assay was carried out by using acetyl-lysine antibody and followed by western blot with HA antibody (right). (B) Endogenous HBP1 was acetylated within the cells. 2BS cells were treated with TSA at 1 μM for 18 h. IP assay was carried out by using HBP1 antibody and followed by western blot with acetyl-lysine antibody.
HBP1 interacts with p300/CBP and recruits p300/CBP onto the p16INK4A promoter
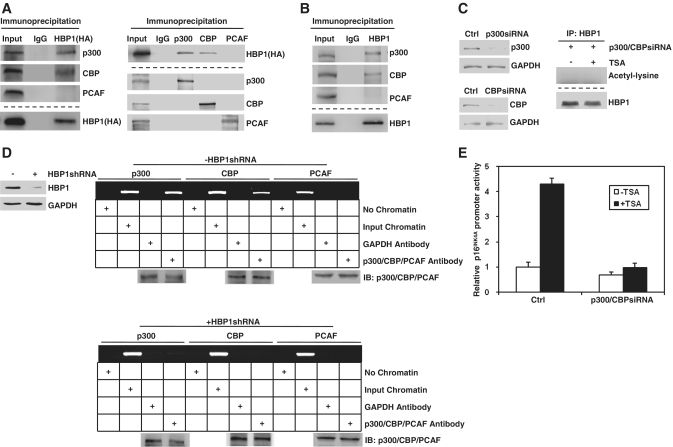
Given that HBP1 is an acetylated protein, then which enzyme can acetylate it? To answer the question, we tested the physical association between HBP1 and p300/CBP or PCAF by co-immunoprecipitation analysis with nuclear extracts of HEK293T cells ectopically expressing HA-tagged full-length HBP1 since p300/CBP and PCAF have been shown to acetylate transcription factors in addition to their histone substrates (36). Western blot analysis showed the presence of HA-HBP1 in the IP of p300 or CBP but not that of PCAF. The reciprocal analysis also showed the presence of p300 and CBP in the HA-HBP1 IP (Figure 4A). As shown in Figure 4B, we further confirmed this interaction between endogenous HBP1 and p300/CBP. When p300/CBP knockdown with siRNA, HBP1 cannot be acetylated even though cells were treated with TSA, which indicates that p300/CBP acetylates HBP1 through directly binding (Figure 4C). Transcriptional activation by a transcription factor involves the recruitment of various coactivators to the specific binding site on DNA, which then directly or indirectly interact with components of the basal transcription machinery. As p300/CBP is known to function as a coactivator connecting sequence-specific transcription factors with the basal transcription machinery, it is possible that HBP1 could recruit p300/CBP onto p16INK4A promoter. This hypothesis was supported by the ChIP assay results shown in Figure 4D, in which p300/CBP, but not PCAF, were recruited to the p16INK4A promoter. The recruitment of p300/CBP is abolished in the absence of HBP1. All of the data indicates that HBP1 interacts with p300/CBP and is acetylated by p300/CBP. As shown in Figure 4E, TSA lost its transactivation on p16INK4A when p300/CBP are knocked-down, which means that the function of TSA is to inhibit the HDAC activity and allow the p300/CBP-dependent acetylation to become dominant.
Figure 4.
HBP1 interacted with p300/CBP and recruits p300/CBP onto the promoter of p16INK4A. (A) Exogenous HBP1 interacts with p300/CBP. HEK293T cells transfected with HA-HBP1 were subjected to immunoprecipitation with anti-HA antibody. Immunoprecipitated complexes were used for western blot analysis with anti- p300, anti-CBP or anti-PCAF antibody, respectively (left). The interaction of HBP1 and p300/CBP was further confirmed by reciprocal analysis. IP was performed with anti-p300, anti-CBP or anti-PCAF antibody respectively and followed with western blot with anti-HA antibody (right). The bottom lanes were for determining IP efficiency. (B) Endogenous HBP1 interacted with p300/CBP. HEK293T cells were subjected to immunoprecipitation with anti-HBP1 antibody. Immunoprecipitated complexes were used for western blot analysis with anti-p300, anti-CBP or anti-PCAF antibody respectively. The bottom lanes were for determining IP efficiency. (C) Endogenous HBP1 cannot be acetylated when p300/CBP knockdown. HEK293T cells were transfected with p300siRNA and CBPsiRNA. The protein levels of p300 and CBP were determined by western blot (left panel). Then the cells were subjected to immunoprecipitation with anti-HBP1 antibody with or without TSA treatment. Immunoprecipitated complexes were used for western blot analysis with acetyl-lysine antibody (Right panel). (D) HBP1 is required for p300/CBP recruitment onto p16INK4A promoter when TSA was applied. HEK293T cells were transfected with HBP1shRNA. The protein level of HBP1 was determined by western blot (left panel). ChIP assay with anti-p300, anti-CBP and anti-PCAF antibody was performed in HEK293T cells after treated with TSA at 2.5 μM for 18 h with or without HBP1shRNA transfection. (E) TSA loses its transactivation ability at the p16INK4A promoter when p300/CBP knockdown. Thirty hours after transfection with p300/CBP siRNA, cells were treated with TSA at 2.5 μM for 18 h. Then the cells were harvested for luciferase assay.
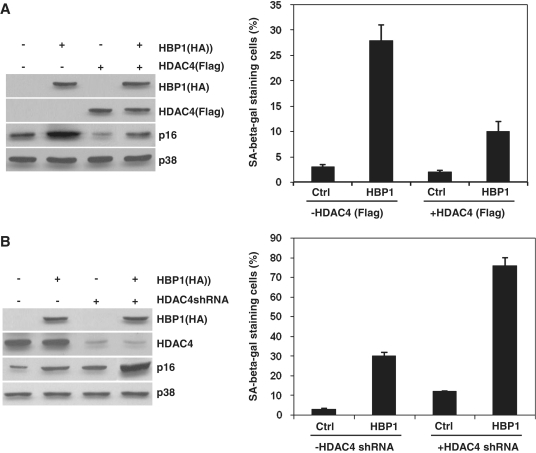
HBP1-induced premature senescence is repressed by HDAC4
HBP1 and TSA cooperate to induce senescence, meaning that, despite the acetylation of HBP1 by p300/CBP, a HDAC permanently deacetylates HBP1 and prevents the induction of senescence. It is very important to identify the HDAC involved. We previously reported that HDAC4, a class II HDAC binds with HBP1 within a region of amino acids 220–250 (35). Our next work is to test whether HDAC4 influences HBP1-induced premature senescence. As shown in Figure 5, with western blot and SA-β-gal staining assay, exogenous HDAC4 represses HBP1-induced p16INK4A expression, thus represses HBP1-induced premature senescence. While HDAC4 knockdown highly enhances HBP1-induced p16INK4A expression, thus enhances HBP1-induced premature senescence.
Figure 5.
HDAC4 represses HBP1 ability to induce senescence. (A) 2BS cells were stably infected with pITA-HBP1. After selection for 3 days, cells were transfected with pITA-HDAC4. The protein levels of HBP1 (HA), HDAC4 (Flag) and p16INK4A were measured by western blot (left panel). Then the cells were stained for SA-β-gal. The percentage of cells positive for SA-β-gal in 2BS cells is shown in right panel. At least 300 cells were counted for each sample. (B) 2BS cells were stably infected with pITA-HBP1. After selection for 3 days, cells were transfected with pSR-HDAC4shRNA. The protein levels of HBP1 (HA), HDAC4 and p16INK4A were measured by western blot (left panel). Then the cells were stained for SA-β-gal. The percentage of cells positive for SA-β-gal in 2BS cells is shown in right panel. At least 300 cells were counted for each sample.
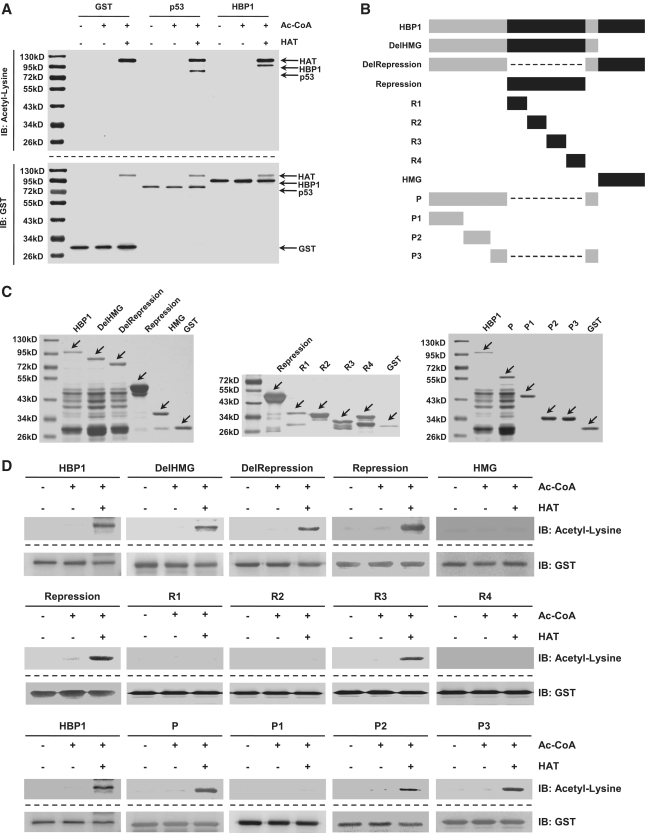
Wild-type HBP1, Repression domain and P domain of HBP1 are acetylated by p300/CBP in vitro
Acetylation has been shown to regulate the activity of many transcription factors, including sequence-specific DNA-binding factor p53 (17). To determine whether p300/CBP acetylate HBP1, we performed an in vitro acetylation reaction using GST-tagged HAT (a catalytic subunit of p300/CBP) purified from transformed E. coli as an enzyme. HBP1 and p53 (as positive control) were similarly purified from E. coli as GST fusion proteins and used as substrates. As shown in Figure 6A, GST-HAT efficiently acetylated GST-HBP1, as well as GST-p53. GST alone was not acetylated by HAT, showing that the in vitro acetylation was specific to HBP1. Our next step was to identify the specific lysines acetylated by p300/CBP. As the HBP1 amino acid sequence contains 31 lysines, our strategy was to design three mutants, containing either the C-terminal DNA-binding domain (HMG box binding), middle region (the Repression domain), or the regions outside of the HMG box binding and the Repression domains (designated as P domain). In addition, we designed two deletion mutants lacking either the HMG box (designated as DelHMG) or Repression domain (designated as DelRepression) (Figure 6B) for further confirmation. We then performed the same GST-IP/anti-acetyl-lysine western blot with each HBP1 mutants as was done with wild-type HBP1 as shown in Figure 6A. All of the mutants were efficiently overexpressed and purified in cells (Figure 6C). Acetylation was still detectable in the Repression domain and P domain as well as DelHMG and DelRepression mutants, whereas acetylation in the HMG box was absent (Figure 6D), suggesting that the Repression domain and the P domain contain the acetylation target. As the Repression domain and the P domain contain 11 and 8 lysines respectively, we produced several more deletion mutants. As shown in Figure 6B, the Repression domain was divided into four short fragments (R1, R2, R3, R4), and the P domain was divided into three short fragments (P1, P2, P3). Then we performed the same GST-IP/anti-acetyl-lysine western blot with each HBP1 mutant as was done previously with wild-type HBP1. Acetylation was still detectable in R3, P2 and P3 (Figure 6D), suggesting that these three domains contain the acetylation targets.
Figure 6.
Wild-type HBP1 and mutants were acetylated by HAT of p300/CBP in vitro. (A) HBP1 was acetylated by HAT in vitro. At the top, purified GST-tagged HBP1 protein was incubated with HAT in the presence of Ac-CoA. The reaction products were separated by SDS–PAGE and immunoblotted with the acetyl-lysine antibody. At the bottom, the same nitrocellulose membrane as used in the in vitro acetylation assay described above was stripped and immunoblotted with anti-GST antibody. GST and GST-tagged p53 proteins were used for negative and positive controls, respectively. (B) Schematic diagram of wild-type HBP1 and associated mutants. (C) Expression of wild-type HBP1 and associated mutants proteins in E. coli. GST-tagged proteins were immunoprecipited with GST, then ran on SDS–PAGE and stained with silver. (D) HBP1 and some mutants (Repression domain, P domain) were acetylated by HAT in vitro. GST-tagged proteins from experiments shown in (B) were incubated with Ac-CoA and HAT in 30°C for 4 h. The reaction products were separated by SDS–PAGE and immunoblotted with the acetyl-lysine antibody. GST was as loading control.
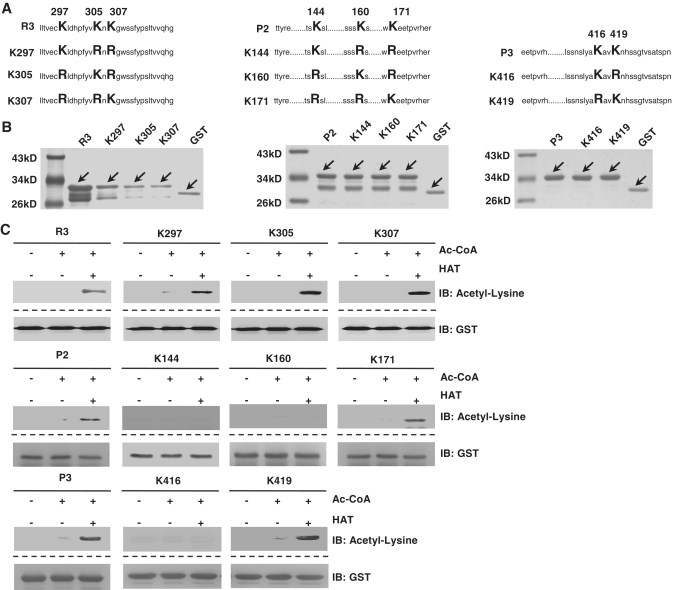
The K297, K305, K307 of the Repression domain and K171, K419 of the P domain are acetylated by p300/CBP in vitro
There are only three lysines in R3, three lysines in P2, and two lysines in P3. To determine which lysine can be acetylated by p300/CBP, we performed overlap-PCR to mutate each lysine to arginine, individually as well as simultaneously (Figure 7A). All the mutants were efficiently overexpressed in cells (Figure 7B). We then performed the GST-IP/acetyl-lysine western blot with each mutant. We found that K297, K305, K307 in the Repression domain and K171, K419 in the P domain can be acetylated by p300/CBP (Figure 7C), suggesting p300/CBP targets specific lysines on HBP1 for acetylation.
Figure 7.
The K297, K305, K307 of Repression domain and K171, K419 of P domain were acetylated by HAT of p300/CBP in vitro. (A) Schematic diagram of associated mutants. (B) Expression of associated mutants proteins in E. coli. GST-tagged proteins were immunoprecipited with GST, then ran on SDS–PAGE and stained with silver. (C) The K297, K305, K307 of Repression domain and K171, K419 of P domain are acetylated by HAT of p300/CBP in vitro. GST-tagged proteins from experiments shown in panel A were incubated with Ac-CoA and HAT in 30°C for 4 h. The reaction products were separated by SDS–PAGE and immunoblotted with the acetyl-lysine antibody. GST was as loading control.
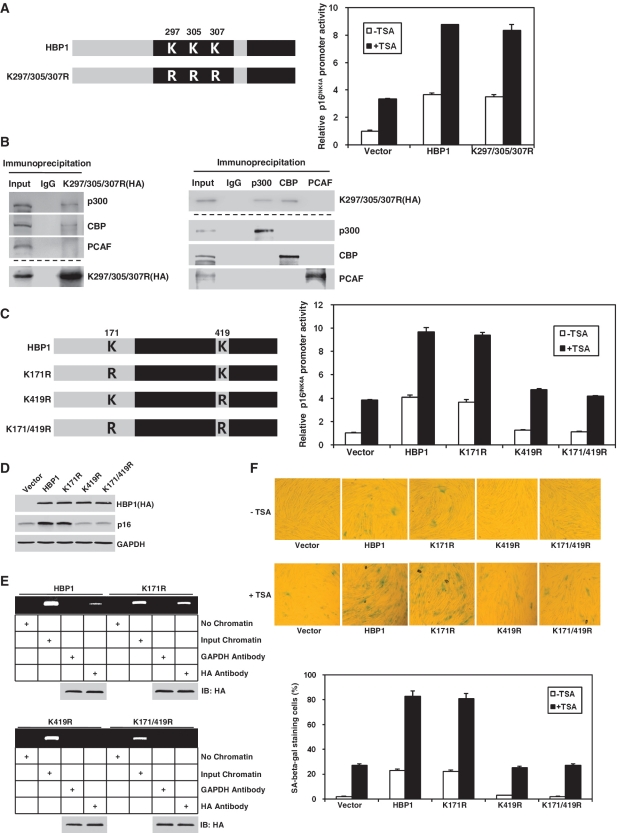
Acetylation of HBP1 at K419 is required for HBP1-induced premature senescence
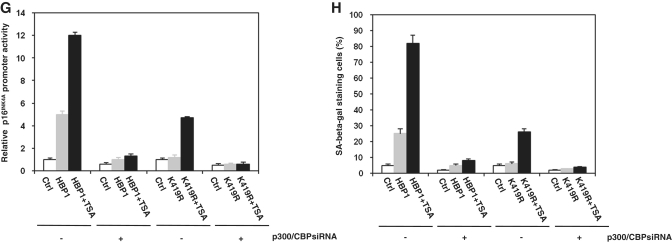
Given that acetylation of a transcription factor often has an effect on its transcriptional activity, we decided to investigate the ability of the lysine mutant HBP1 in activating p16INK4A. Cotransfection of a p16INK4A promoter-luciferase construct with wild-type HBP1 into HEK293T cells resulted in an ∼4-fold increase in the activation of the reporter (Figure 8A). Acetylation-deficient mutant K297/305/307R of the Repression domain maintained same result after cotransfection with p16INK4A promoter–luciferase construct and still responded to TSA, suggesting that acetylation of Repression domain has no effect on HBP1 transactivation on p16INK4A. Furthermore, we tested the binding of the mutant and p300/CBP with IP. As shown in Figure 8B, exogenous K297/305/307R (HA-tag) still binds to p300/CBP, but not PCAF. The result was consistent with our previous data that the Repression domain is not required for HBP1-induced p16INK4A expression. K171R of the P domain had no effect on HBP1 transactivation either (Figure 8C). However, HBP1-K419R was only able to activate the reporter to ∼30% level of the wild-type HBP1. The cells transfected with K419R or K171/419R have almost same response to TSA compared with the control cells. As shown in Figure 8D, we found that overexpression of the wild-type HBP1 resulted in an ∼10-fold induction of p16INK4A protein. Overexpression of mutant K171R resulted in almost the same level of p16INK4A protein induction as that of wild-type HBP1, whereas overexpressing K419R or K171/419R produced no induction compared with control vector. These results suggest that acetylation of HBP1 at K419 is important for its transactivation on p16INK4A. Furthermore, we tested the influence of acetylation on the binding of HBP1 onto p16INK4A promoter with the ChIP assay. As shown in Figure 8E, the binding of HBP1 onto p16INK4A promoter was abrogated as HBP1 was mutated at K419R or K171R/419R, while no difference was observed when only mutated at K171R. This demonstrates that HBP1 acetylation at K419 is required for the binding of HBP1 onto the p16INK4A promoter. To further detect the functional importance of K419 acetylation in transcriptional activation, we transfected wild-type and mutant HBP1 into human normal fibroblasts 2BS cells. After 3 days selection, we treated the cells with or without TSA, then fixed the cells and stained with SA-β-gal. As shown in Figure 8F, both wild-type HBP1 and K171R increased the percentage of cells for SA-β-gal staining, whereas K419R and K171/419R did not change the percentage. The cells transfected with K419R or K171/419R have almost same response to TSA compared with the control cells, suggesting that acetylation of HBP1 at K419 is required for HBP1-induced premature senescence. All these data indicate that acetylation of HBP1 at K419 regulates HBP1-induced premature senescence through enhancing the binding of HBP1 to the p16INK4A promoter. The cells transfected with K419R are still slightly responsive to TSA because of endogenous HBP1. To further investigate the impact of acetylation on HBP1-induced transactivation on p16INK4A and premature senescence, we induced p300/CBPsiRNA into the cells transfected with wild-type HBP1 or K419R. As shown in Figure 8G, 8H, with luciferase assay and SA-β-gal staining, the cells transfected with either wild-type HBP1 or K419R are not responsive to TSA, suggesting that acetylation at K419 is essential for the HBP1 functions in the activation of p16INK4A and for the activation of premature senescence.
Figure 8.
The acetylation of HBP1 at K419 was indispensible for HBP1-induced premature senescence. (A) HBP1 acetylation at K297/305/307 was not required for transactivation on p16INK4A. HEK293T cells were cotransfected with HBP1 or point mutants K297/305/307R and pGL3-p16INK4A promoter for 48 h. After transfection, cells were treated with TSA at 2.5 μM for 18 h. Then the cells were harvested for luciferase assay. (B) K297/305/307 was not required for interaction of HBP1 and p300/CBP. The whole cell extracts were subjected to immunoprecipitation with anti-HA antibody. Immunoprecipitated complexes were used for western blot analysis with anti-p300, anti-CBP or anti-PCAF antibody respectively. The bottom lanes were for determining IP efficiency. The interaction of HBP1 and p300/CBP was further confirmed by reciprocal analysis. (C) HBP1 acetylation at K419 was required for transactivation on p16INK4A. HEK293T cells were transfected with pGL3-p16INK4A promoter and HBP1 or mutants for 48 h. After transfection, cells were treated with TSA at 2.5 μM for 18 h. Then the cells were harvested for luciferase assay. (D) HBP1 acetylation at K419 was required for HBP1-induced p16INK4A expression. p16INK4A protein level in 2BS cells transfected with HBP1 and mutants for 5-7 days was determined by western blot. (E) HBP1 acetylation at K419 was required for the binding of HBP1 onto p16INK4A promoter. ChIP assay with antibody against HA was performed in HEK293T cells transfected with HBP1 or mutants. (F) HBP1 acetylation at K419 is required for HBP1-induced premature senescence. 2BS cells transfected with HBP1 or mutants with or without TSA treatment were stained for SA-β-gal (up panel). The percentage of cells positive for SA-β-gal is shown in low panel. At least 300 cells were counted for each sample. (G) p300/CBP knockdown attenuates the HBP1-induced p16INK4A gene activity. 2BS cells were stably infected with pITA-HBP1. After selection for 3 days, cells were transfected with p300/CBP siRNA and pGL3-p16INK4A promoter. After transfection, cells were treated with TSA at 1 μM for 18 h. Then the cells were harvested for luciferase assay. (H) p300/CBP knockdown attenuates the HBP1-induced senescence. 2BS cells were stably infected with pITA-HBP1. After selection for 3 days, cells were transfected with p300/CBP siRNA. After transfection, cells were treated with TSA at 1 μM for 18 h. Then the cells were stained for SA-β-gal.
DISCUSSION
Histone acetylation regulates many cellular processes, including nucleosome assembly, chromatin compaction, heterochromatin formation and gene transcription. Hyperacetylated histones have long been known to associate with activated genes, and some studies further suggest a causal relationship between histone acetylation and gene activation. It is thought that addition of acetyl groups to the lysine residues of histone tails facilitates access of transcription factors to DNA by disrupting higher-order packaging of the chromatin and by recruiting bromodomain factors, which recognize acetyl-lysine motifs (32,33). The bromodomain, found in chromatin-associated proteins and histone acetyltransferase, functions as the sole protein module known to bind acetyl-lysine motifs. Recently, acetylation of transcription factors has been drawing more attention. Acetylation has been shown to be a key regulatory mechanism for the activity of many transcription factors (24–26). Although several other intensely studied transcription factors have been shown to be acetylated by p300 (19–22), the acetylation of HBP1 has not been reported previously. We had reported that HBP1 participates in Ras-induced premature senescence through targeting p16INK4A, but whether the acetylation of HBP1 impacted p16INK4A expression remained unknown. In this study, we used the HDAC inhibitor TSA to treat cells, and found that TSA led to increased p16INK4A expression. Moreover, TSA-induced p16INK4A expression depended on HBP1 (Figure 1C), suggesting that HBP1 is a key regulator for TSA-induced p16INK4A expression. Although no change in the HBP1 mRNA level was detected after TSA treatment, the p16INK4A mRNA and protein levels increased significantly in 2BS cells. We proposed that post-translational modifications of HBP1, especially acetylation, might play a role in regulating p16INK4A expression. This hypothesis was supported by evidence that TSA enhances the transcriptional activity of HBP1 through increased DNA binding of HBP1 onto p16INK4A promoter. Furthermore, we found that HBP1 was acetylated within the cells, as after immunoprecipitation of HA-HBP1 from HEK293T cells, we could detect HBP1 acetylation with an acetyl-lysine antibody.
Since HBP1 is an acetylated protein, which acetyltransferase can acetylate it? With IP-western blot and ChIP assay, we identified that HBP1 interacted with p300/CBP and recruited p300/CBP onto p16INK4A promoter. In addition, HBP1 was acetylated by p300/CBP in vitro. By making deletions and site mutations of full-length HBP1, we identified the acetylated lysines at Lys-297, 305, 307 in the Repression domain and Lys-171, 419 in the P domain. All five lysines were acetylated by p300/CBP in vitro, and they were not acetylated when mutated to arginine, which indicates that these are specific acetylation sites.
The acetylation of HBP1 appears to be important in HBP1-mediated transactivation of p16INK4A, as mutation of Lys-419 in HBP1 sequence resulted in decreased activation of a p16INK4A reporter and significantly blocked the HBP1-mediated induction of the p16INK4A protein. Acetylation of HBP1 at Lys-419 is also indispensable for HBP1-induced premature senescence because HBP1 lost the ability to induce senescence when Lys-419 was mutated to arginine. However, acetylation of the other lysines (Lys-297, 305, 307 and Lys-171) had no effect on HBP1-mediated transactivation on p16INK4A. The results are in concert with our previous report that the Repression domain (Lys-297, 305, 307 are in the Repression domain) is not required for HBP1-induced p16INK4A expression. HBP1 is a dual transcription factor. It has many targets: Cyclin D1, c-myc, p47 phox, and p16INK4A, etc. The identification of the diverse set of targets, either being activated or repressed by HBP1, suggests that HBP1 has multiple mechanisms for affecting G1 progression and for suppressing oncogenic pathways. Although it was known that the Repression domain of HBP1 is required for its suppression function, the underlying mechanism for such function was unclear. Our results suggest that acetylation of Repression domain may be important for HBP1-induced repression. We identified that HDAC4 represses HBP1-induced p16INK4A expression, thus represses HBP1-induced premature senescence. While HDAC4 knockdown highly enhances HBP1-induced p16INK4A expression, thus enhances HBP1-induced premature senescence. The function of HDAC4 may be through directly binding to the Repression domain, because there is binding site within the domain. More experiments are needed to test this hypothesis and to answer the question of whether HBP1 interacts with HDACs directly or possibly through other corepressors.
In this study, we observed that HBP1 interacted with, and was acetylated by, the HAT protein p300/CBP. This observation is physiologically relevant, as we found that HBP1 is acetylated in vivo as well. Furthermore, we found that HBP1 recruited p300/CBP onto p16INK4A promoter and enhanced HBP1 transactivation on p16INK4A. The recruitment of p300/CBP is abolished in the absence of HBP1. Our data indicate that HBP1 interacts with p300/CBP and is acetylated by p300/CBP. The function of TSA is to inhibit the HDAC activity and allow the p300/CBP-dependent acetylation to become dominant. Therefore, TSA would lose its transactivation on p16INK4A when p300/CBP are knocked-down.
Actually, HBP1 has dual function in transcription. Most of our previous works focused on its transcriptional repression and regarded HBP1 as a novel transcriptional repressor in differentiation (1–3,6,9). Only several papers reported that HBP1 is a transcriptional activator in myeloid differentiation and transformation (10,18). The acetylation of HBP1 at Lys-419 was required for transactivation of p16INK4A and for the activation of premature senescence. Might K419 acetylation (or non-acetylation) determine whether HBP1 functions as an activator or repressor? Is K419R mutant functional for transcriptional repression? To answer the questions, we cotransfected with a Wnt promoter-luciferase construct and wild-type HBP1 or K419R mutant into HEK293T cells. With luciferase assay, we found that both HBP1 and K419R repressed activation of the reporter (data not shown). The result indicates that the acetylation of K419 does not switch HBP1 from a transcriptional repressor to a transcriptional activator, except for its transactivation for p16INK4A. Other modifications or co-repressors might be involved.
Our previous study linked HBP1 with p16INK4A. The present study describes that acetylation by p300/CBP enhances the effect of HBP1 on the regulation of p16INK4A, thereby impacts on the process of replicative senescence. The expression of p16INK4A in replicative senescence is hugely elevated. Although the mechanisms underlying is not fully understood, both transcriptional and post-transcriptional regulation are considered important. Factors reported to be involved in the transcriptional regulation of p16INK4A include CREG1, SP1 and HBP1, etc. The contribution of HBP1- p16INK4A regulatory process to the elevation of p16INK4A in replicative senescence is not easy to be evaluated. However, our study suggests that the HBP1- p16INK4A regulatory process is at least partly responsible for the regulation of p16INK4A. Overall, the molecular mechanisms of HBP1-mediated gene expression will have important implications within several fields, including senescence biology, cancer biology and stem cell biology.
FUNDING
National Basic Research Programs of China (No. 2007CB507400); National Natural Science Foundation of China (No. 81170320). Funding for open access charge: National Natural Science Foundation of China (No. 81170320).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Wengong Wang for helpful discussion on the article and Dr Amy Yee for plasmid and antibodies
REFERENCES
- 1.Berasi SP, Xiu M, Yee AS, Paulson KE. HBP1 repression of the p47phox gene: cell cycle regulation via the NADPH oxidase. Mol. Cell. Biol. 2004;24:3011–3024. doi: 10.1128/MCB.24.7.3011-3024.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paulson KE, Rieger-Christ K, McDevitt MA, Kuperwasser C, Kim J, Unanue VE, Zhang X, Hu M, Ruthazer R, Berasi SP, et al. Alterations of the HBP1 transcriptional repressor are associated with invasive breast cancer. Cancer Res. 2007;67:6136–6145. doi: 10.1158/0008-5472.CAN-07-0567. [DOI] [PubMed] [Google Scholar]
- 3.Lavender P, Vandel L, Bannister AJ, Kouzarides T. The HMG-box transcription factor HBP1 is targeted by the pocket proteins and E1A. Oncogene. 1997;14:2721–2728. doi: 10.1038/sj.onc.1201243. [DOI] [PubMed] [Google Scholar]
- 4.Lemercier C, Duncliffe K, Boibessot I, Zhang H, Verdel A, Angelov D, Khochbin S. Involvement of retinoblastoma protein and HBP1 in histone H1(0) gene expression. Mol. Cell. Biol. 2000;20:6627–6637. doi: 10.1128/mcb.20.18.6627-6637.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shih HH, Tevosian SG, Yee AS. Regulation of differentiation by HBP1, a target of the retinoblastoma protein. Mol. Cell. Biol. 1998;18:4732–4743. doi: 10.1128/mcb.18.8.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shih HH, Xiu M, Berasi SP, Sampson EM, Leiter A, Paulson KE, Yee AS. HMG box transcriptional repressor HBP1 maintains a proliferation barrier in differentiated liver tissue. Mol. Cell. Biol. 2001;21:5723–5732. doi: 10.1128/MCB.21.17.5723-5732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith JM, Bowles J, Wilson M, Koopman P. HMG box transcription factor gene Hbp1 is expressed in germ cells of the developing mouse testis. Dev. Dyn. 2004;230:366–370. doi: 10.1002/dvdy.20053. [DOI] [PubMed] [Google Scholar]
- 8.Tevosian SG, Shih HH, Mendelson KG, Sheppard KA, Paulson KE, Yee AS. HBP1: a HMG box transcriptional repressor that is targeted by the retinoblastoma family. Genes Dev. 1997;11:383–396. doi: 10.1101/gad.11.3.383. [DOI] [PubMed] [Google Scholar]
- 9.Xiu M, Kim J, Sampson E, Huang CY, Davis RJ, Paulson KE, Yee AS. The transcriptional repressor HBP1 is a target of the p38 mitogen-activated protein kinase pathway in cell cycle regulation. Mol. Cell. Biol. 2003;23:8890–8901. doi: 10.1128/MCB.23.23.8890-8901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Kim J, Ruthazer R, McDevitt MA, Wazer DE, Paulson KE, Yee AS. The HBP1 transcriptional repressor participates in RAS-induced premature senescence. Mol. Cell. Biol. 2006;26:8252–8266. doi: 10.1128/MCB.00604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat. Rev. Drug Discov. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 12.Marks PA, Richon VM, Breslow R, Rifkind RA. Histone deacetylase inhibitors as new cancer drugs. Curr. Opin. Oncol. 2001;13:477–483. doi: 10.1097/00001622-200111000-00010. [DOI] [PubMed] [Google Scholar]
- 13.Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J. Natl Cancer Inst. 2000;92:1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 14.Yu X, Guo ZS, Marcu MG, Neckers L, Nguyen DM, Chen GA, Schrump DS. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J. Natl Cancer Inst. 2002;94:504–513. doi: 10.1093/jnci/94.7.504. [DOI] [PubMed] [Google Scholar]
- 15.Zhu WG, Otterson GA. The interaction of histone deacetylase inhibitors and DNA methyltransferase inhibitors in the treatment of human cancer cells. Curr. Med. Chem. Anti-Cancer Agents. 2003;3:187–199. doi: 10.2174/1568011033482440. [DOI] [PubMed] [Google Scholar]
- 16.Chan HM, Kristic-Demonacos M, Smith L, Demonacos C, La Thangue NB. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat. Cell Biol. 2001;3:667–674. doi: 10.1038/35083062. [DOI] [PubMed] [Google Scholar]
- 17.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 18.Li HK, Wang WB, Liu XW, Paulson KE, Yee AS, Zhang XW. Transcriptional factor HBP1 targets p16INK4A, upregulating its expression and consequently is involved in Ras-induced premature senescence. Oncogene. 2010;29:5083–5094. doi: 10.1038/onc.2010.252. [DOI] [PubMed] [Google Scholar]
- 19.Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J. Biol. Chem. 2007;282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- 20.Lee CW, Ferreon JC, Ferreon AC, Arai M, Wright PE. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc. Natl Acad. Sci. USA. 2010;107:19290–19295. doi: 10.1073/pnas.1013078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang FM, Chen YJ, Ouyang HJ. Regulation of unfolded protein response modulator XBP1s by acetylation and deacetylation. Biochem. J. 2010;433:245–252. doi: 10.1042/BJ20101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M, Partridge NC. Parathyroid hormone activation of matrix metalloproteinase-13 transcription requires the histone acetyltransferase activity of p300 and PCAF and p300-dependent acetylation of PCAF. J. Biol. Chem. 2010;285:38014–38022. doi: 10.1074/jbc.M110.142141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, Botrugno OA, Parazzoli D, Oldani A, Minucci S, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471:74–79. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 25.Kawai Y, Garduño L, Theodore M, Yang J, Arinze IJ. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related Factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J. Biol. Chem. 2011;286:7629–7640. doi: 10.1074/jbc.M110.208173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh SK, Wilczynska KM, Grzybowski A, Yester J, Osrah B, Bryan L, Wright S, Griswold-Prenner I, Kordula T. The unique transcriptional activation domain of nuclear factor-I-x3 is critical to specifically induce marker gene expression in astrocytes. J. Biol. Chem. 2011;286:7315–7326. doi: 10.1074/jbc.M110.152421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan J, Zhang Z, Tong T. Senescence delay of human diploid fibroblast induced by anti-sense p16INK4a expression. J. Biol.Chem. 2001;276:48325–48331. doi: 10.1074/jbc.M104814200. [DOI] [PubMed] [Google Scholar]
- 29.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 30.Costanzo A, Merlo P, Pediconi N, Fulco M, Sartorelli V, Cole PA, Fontemaggi G, Fanciulli M, Schiltz L, Blandino G. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell. 2002;9:175–186. doi: 10.1016/s1097-2765(02)00431-8. [DOI] [PubMed] [Google Scholar]
- 31.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 33.Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- 34.Yan JX, Wait R, Berkelman T, Harry RA, Westbrook JA, Wheeler CH, Dunn MJ. A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis. 2000;21:3666–3672. doi: 10.1002/1522-2683(200011)21:17<3666::AID-ELPS3666>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Yee AS, Paulson EK, McDevitt MA, Rieger-Christ K, Summerhayes I, Berasi SP, Kim J, Huang CY, Zhang X. The HBP1 transcriptional repressor and the p38 MAP kinase: unlikely partners in G1 regulation and tumor suppression. Gene. 2004;336:1–13. doi: 10.1016/j.gene.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]