Abstract
Mammalian tissues display a remarkable complexity of splicing patterns. Nevertheless, only few examples of tissue-specific splicing regulators are known. Herein, we characterize a novel splicing regulator named RBM11, which contains an RNA Recognition Motif (RRM) at the amino terminus and a region lacking known homology at the carboxyl terminus. RBM11 is selectively expressed in brain, cerebellum and testis, and to a lower extent in kidney. RBM11 mRNA levels fluctuate in a developmentally regulated manner, peaking perinatally in brain and cerebellum, and at puberty in testis, in concomitance with differentiation events occurring in neurons and germ cells. Deletion analysis indicated that the RRM of RBM11 is required for RNA binding, whereas the carboxyl terminal region permits nuclear localization and homodimerization. RBM11 is localized in the nucleoplasm and enriched in SRSF2-containing splicing speckles. Transcription inhibition/release experiments and exposure of cells to stress revealed a dynamic movement of RBM11 between nucleoplasm and speckles, suggesting that its localization is affected by the transcriptional status of the cell. Splicing assays revealed a role for RBM11 in the modulation of alternative splicing. In particular, RBM11 affected the choice of alternative 5′ splice sites in BCL-X by binding to specific sequences in exon 2 and antagonizing the SR protein SRSF1. Thus, our findings identify RBM11 as a novel tissue-specific splicing factor with potential implication in the regulation of alternative splicing during neuron and germ cell differentiation.
INTRODUCTION
The multi-exon nature of genes greatly expands the coding potential of eukaryotic genomes, by allowing production of multiple mRNA variants from each gene through differential assortment of exons (1,2). This process, known as alternative splicing (AS), is operated by the spliceosome, and modulated by the interaction between trans-acting factors and cis-regulatory sequences within the regulated exons and/or the flanking introns (3). The best characterized trans-acting factors are the heterogeneous nuclear ribonucleoproteins (hnRNPs) and the serine–arginine rich (SR) RNA-binding protein (RBP) families, which typically act antagonistically to regulate exon recognition (3–5).
Tissues and organs display remarkable differences in terms of AS frequency and specificity (1,2,6). This tissue-specific repertoire of splicing variants can be achieved, at least in part, through modulation of the expression of ubiquitous trans-acting factors. Indeed, cells expressing variable amounts of SR proteins or hnRNPs are expected to differentially regulate a given splicing event, due to the opposite effects they usually exert on regulated exons (3–5). Furthermore, these RBPs can regulate their own expression and that of other splicing factors in specific cell contexts. The autoregulation of hnRNP I, also known as polypyrimidine tract-binding protein (PTB), provides an excellent example. PTB binds its own pre-mRNA and triggers the skipping of exon 11, thus creating a premature termination codon (PTC), which targets the transcript to the nonsense-mediated decay pathway (NMD) (7). The SRSF1 gene is also subject to extensive AS leading to production of six different variants, one being the full-length variant while the others are retained into the nucleus or targeted to NMD (8). SRSF1 enhances the production of the nuclear-retained splice variants, causing its own downregulation (8). In addition, Sam68, a ubiquitous splicing factor, promotes the retention of a cryptic intron in SRSF1 3′-UTR, thus preventing degradation by NMD of the full-length mRNA (9).
Tissue-specific splicing factors provide an additional layer of complexity, particularly in organs characterized by highly differentiated cell types like brain and testis. For instance, the neuron-specific NOVA proteins play an essential role in neurogenesis (10,11), likely due to regulation of AS in genes important for synaptogenesis (10). Tissue-specific splicing factors might also cooperate with ubiquitous proteins to regulate neuron-specific AS. The FOX family comprises three members (FOX-1–3) that are alternatively spliced to yield multiple protein variants (1,12). FOX-1 and FOX-2 are expressed in brain and muscle, whereas FOX-3 is restricted to brain. However, not all neurons express all FOX proteins and splicing of at least one neuron-specific exon specifically correlates only with FOX-3 expression (12). Notably, FOX-3 strictly requires the interaction with the PTB-associated splicing factor (PSF) to regulate this exon (12), thus enrolling a ubiquitous factor in a neuron-specific AS event. Splicing reprogramming in neurons is also regulated by the switch occurring from PTB to the neuron-specific nPTB, which are expressed in a mutually exclusive fashion in developing brain (7). Gene silencing experiments showed that PTB and nPTB modulate splicing changes of different sets of alternative exons during neurogenesis (7), which may underlie neural cell differentiation.
Germ cell differentiation is another dynamic process possibly guided by tissue-specific splicing factors and characterized by extensive AS (13). Two male germ cell-specific members of the RNA-binding motif (RBM) protein family, RBMY and hnRNPG-T (13), were shown to regulate testis-specific exons (14,15). RBMY and hnRNPG-T interact with two other RBPs highly expressed in testis, SLM-2 and Sam68 (13). SLM-2 expression is restricted to neurons and germ cells (16), while Sam68 is present in most tissues (17) but it is essential for male fertility (18). Sam68 is expressed in transcriptionally active male germ cells (18–20), where it promotes AS (20) and translation of target mRNAs (18).
Given the relatively small number of tissue-specific splicing regulators known, it is likely that additional RBPs are involved in tissue-specific AS. In the present work, we have studied the expression and function of RBM11, a previously uncharacterized RNA Recognition Motif (RRM) protein. The human gene maps on Chromosome 21 (21–23), whereas the mouse counterpart is located on the homologous Chromosome 16. Due to its genomic localization, which potentially links RBM11 to the Down syndrome, the gene has been included in expression studies, which suggested a restricted pattern of expression (21–23). These reports documented expression in brain and testis (21), or in brain, testis and spleen (23) or uniquely in testis (22). Beside these partially divergent results, no direct studies on either human or mouse RBM11 have been performed to date. Herein, we provide evidence that RBM11 is expressed in selected tissues in a developmentally regulated fashion. RBM11 colocalizes with splicing speckles, binds RNA, and modulates splicing events. These results suggest that RBM11 is a novel tissue-specific splicing factor with potential implication in regulation of AS in brain and testis.
MATERIALS AND METHODS
Plasmid constructs
The human RBM11 cDNA (accession no. NM_144770) was amplified by RT–PCR from Hek293T RNA, using the Finnzyme Phusion DNA Polymerase, and cloned into the HindIII–BamHI restriction sites of p3XFlag (Sigma-Aldrich), the XhoI–SalI restriction sites of pEGFP-C1 (Clontech) and the BamHI–XhoI restriction sites of pGEX-4T1 (GE Healthcare). The cDNAs encoding the RRM or ΔRRM portion of RBM11 were amplified by PCR using Flag-RBM11 as template and cloned as above. Mouse Rbm11 cDNA (accession no. NM_198302) was amplified from 30 die post partum (dpp)-old mouse testis RNA, as described above for human RBM11, and cloned into HindIII–BamHI restriction sites of p3XFlag (Sigma-Aldrich). cDNAs were sequenced by Cycle Sequencing (BMR Genomics). The BCL-X (24–26), E1A (27), CD44 (28) and SMN2 (29) minigenes were previously described. All PCR primers used are listed in Supplementary Table S1.
Cell cultures, transfections, treatments and in vivo splicing assay
Germ cells were purified by elutriation from juvenile or adult testis as described (19). Hek293T and HeLa cells were maintained in Dulbecco's modified Eagle Medium (DMEM; Gibco BRL) supplemented with 10% fetal bovine serum (LONZA), penicillin and streptomycin. Transfections were performed with 1 µg of the indicated RBM11 constructs, ±0.5 µg of the indicated minigene, using Lipofectamine 2000 (Invitrogen). RNAi transfections were performed as described (29) with RBM11 and SRSF1 siRNAs (Supplementary Table S1). Twenty-four hours after transfection, cells were collected for RNA or biochemical analyses and processed as described (24). Total RNA was isolated using cold TRIzol reagent (Invitrogen) and resuspended in RNase-free water (Sigma-Aldrich). For confocal analysis, cells were seeded on cover slips and treated with or without 1 µM mitoxantrone (MTX, Sigma-Aldrich) for 2 h, 0.5 mM arsenite for 30 min or 50 µM cisplatin (Sigma-Aldrich) for 1 h. Cells were then fixed and stained for immunofluorescence analysis. For transcription inhibition/release experiments, HeLa cells were incubated for 6 h with 75 µM 5,6-Dichlorobenzimidazole 1-β-d-ribofuranoside (DRB, Sigma-Aldrich), rinsed and let recover in fresh medium for the indicated time.
Immunofluorescence analysis
HeLa cells were fixed and stained for immunofluorescence analysis as previously described (28). The primary antibodies used were: mouse anti-SRSF2 and anti-Flag (1:500; Sigma-Aldrich), goat anti-TIA-1 (1:200; Santa Cruz Biotechnology), mouse anti-RNAP II H5 IgM (1:200; Abcam). The secondary antibodies used were: Alexa Fluor 488 goat anti-mouse (1:500; Invitrogen) and cy3-coniugated donkey anti-mouse IgM (1:500; Jackson Immunoresearch). Confocal analyses were performed using a Leica confocal microscope and a Plan-Neofluar HCX 40.0×/1.25 oil UV objective and acquired using IAS AF Lite software (Leica Microsystems). Images were saved as TIFF files; Photoshop (Adobe) was used for composing the panels.
In vitro RNA synthesis, biotin–RNA pull down and in vitro UV crosslink
Templates for RNA synthesis were generated using PCR products amplified with forward primers containing the T7 promoter sequence upstream of the exon 2 sequence of either wild-type or mutated BCL-X minigenes. Reverse primers were complementary to either the sequence in exon 2 or intron 2. Amplified bands were gel-purified and used as templates (1 µg) for in vitro RNA transcription with T7 polymerase (MAXIScript kit, Ambion) and Biotin RNA Labeling Mix (Roche). RNA was purified by phenol–chloroform and diluted in RNAse-free water (Sigma-Aldrich). Biotin–RNA/streptavidin pull-down experiments were performed using nuclear extracts from Hek293T cells transfected with wild-type or mutant Flag-RBM11 plasmids. In vitro UV crosslink was performed as described (29) using BCL-X exon 2 RNA transcribed in vitro from wild X2.13 or ΔB2 plasmids in the presence of 30 µCi GTP and purified on Sephadex G50. RNA was incubated for 10 min in the presence of GST or GST-RBM11 (200 ng). After RNase treatment, samples were resolved by 10% SDS–PAGE and analyzed by autoradiography.
UV crosslink RNA–protein immunoprecipitation (CLIP)
Hek293T cells transfected with Flag-RBM11 alone or with BCL-X minigene plasmids were washed with PBS and UV-irradiated (400 mJ). Cells were collected in lysis buffer: 50 mM Tris pH 8, 100 mM NaCl, 1% NP40, 1 mM MgCl2, 0.1 mM CaCl2, 0.5 mM NaVO4, 1 mM dithiothreitol, protease inhibitor cocktail, RNase inhibitor (New England Biolabs) and incubated 10 min on ice. Extracts were then incubated with RNase-free DNase (10 units/100 µg) for 30 min at 37°C, and centrifuged at 15 000g for 10 min at 4°C. An amount of 50 µg of extract (RNA input) was treated with Proteinase K (50 µg) for 30 min at 37°C before RNA purification. The rest of the extract was pre-cleared for 1 h on protein A–Sepharose beads and for 1 h on protein A/G-Sepharose beads with rabbit IgG (2 µg/mg of extract), 0.25 µg/µl tRNA and 0.01% BSA. Extracts were then incubated as for the second pre-clearing but in the presence of 1 µg mouse anti-Flag IgGs for 2 h at 4°C. After three washes at room temperature in lysis buffer supplemented with 1 M NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS and 1 mM EDTA, and two washes in lysis buffer, beads were resuspended in 100 µl of lysis buffer. An aliquot (10%) was kept for western blot analysis and the rest incubated with 50 µg Proteinase K at 37°C for 20 min. RNA was isolated by standard procedures and used for real time PCR analyses.
RT–PCR and real-time PCR analyses
Total RNA (0.2–1 µg) from transfected cells or tissues was used for RT–PCR using M–MLV reverse transcriptase (Invitrogen). A total of 5% of the RT reaction was used as template and PCR products were analyzed by ethidium bromide agarose gel. Quantitative real-time PCR (qPCR) was performed using LightCycler 480 SYBR Green I Master and the LightCycler 480 System (Roche), according to manufacturer's instructions. For crosslink immunoprecipitation (CLIP) analyses, each sample was normalized with respect to its input. BCL-X RNA associated with RBM11 is represented as fold enrichment relative to samples transfected with empty vectors. All primers used are listed in the Supplementary Table S1.
RESULTS
RBM11 is expressed in a cell- and tissue-specific pattern in mouse and human
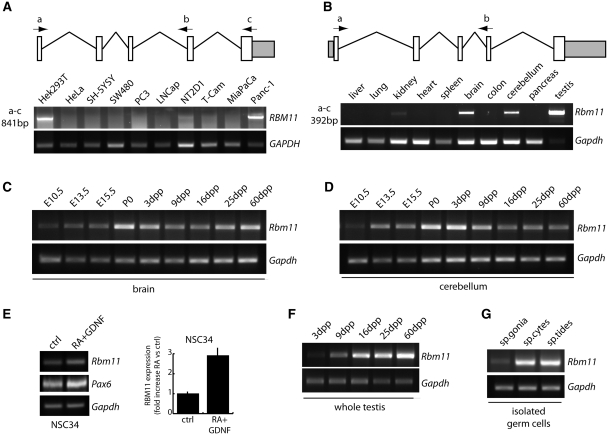
RBM11 expression was analyzed in a panel of cell lines derived from various human tissues by RT–PCR. Screening with primers that amplify the whole open reading frame (a–c in Figure 1A) showed high expression levels only in Hek293T kidney cells and Panc-1 pancreatic adenocarcinoma cells, while weak expression was observed in NT2D1 testicular embryonic carcinoma cells (Figure 1A). By using an internal set of primers that amplify a smaller fragment (a–b in Figure 1A) or quantitative real-time PCR (qPCR) (Supplementary Figure S1A and B), a faint signal was also detected in SH-5YSY neuroblastoma cells but not in HeLa cells. These results confirmed a restricted pattern of expression for human RBM11 (21–23).
Figure 1.
RBM11 is expressed in a cell- and tissue-specific pattern in mouse and human. (A and B) Schematic representation of human RBM11 and mouse Rbm11, with the position of the PCR primers used. (A) RT–PCR analysis (35 cycles) of RBM11 expression in the indicated human cell lines. GAPDH was used as internal standard. (B–D, F and G) RT–PCR analysis of Rbm11 expression in the mouse. Total RNA (1 μg) from 30 dpp mouse tissues (B), fetal and post-natal brain (C) and cerebellum (D), post-natal testis (F) and purified germ cells (G) were collected at the indicated ages or stages of differentiation was used for RT–PCR analyses of Rbm11 expression (28 cycles). Gapdh was used as an internal standard. (E) Conventional (left panels) and qPCR of Rbm11 expression in the mouse neuronal cell line NSC34 under basal conditions and after neuronal differentiation induced by retinoic acid and glial derived neurotrophic factor (RA + GDNF). Induction of Pax6 was analyzed as marker of neuronal differentiation.
Analysis of expression of the highly homologous mouse gene in tissues obtained from 30 dpp mice showed high levels only in brain, cerebellum and testis, which yielded the strongest signal, whereas barely detectable levels were observed in kidney (Figure 1B). Quantitative analysis by qPCR yielded similar results (Supplementary Figure S1C), thus indicating that RBM11 displays a restricted pattern of expression also in the mouse. To investigate whether Rbm11 mRNA expression varies during development, we selected the organs expressing high levels of the gene. RT–PCR analyses starting from Day 10.5 of embryonic development (E10.5) until adulthood showed that Rbm11 expression increased during the fetal life in brain (Figure 1C) and cerebellum (Figure 1D), reaching a peak perinatally (P0-3 dpp). At this time dendrite maturation and synaptic connections are refined (30,31), suggesting a correlation between Rbm11 expression and cell differentiation events. Indeed, we also found that its expression could be induced by triggering differentiation of both mouse (NSC34; Figure 1E) and human (SH-5YSY; Supplementary Figure S1D) neuronal cell lines in culture. In testis, Rbm11 expression increased rapidly from 3 to 16 dpp (Figure 1F), concomitantly with the onset of germ cell meiotic differentiation (32), and then remained high at least until 60 dpp, when testis is largely populated by post-meiotic spermatids (32). This trend was confirmed by analysis of purified germ cells, which showed low Rbm11 expression in mitotic spermatogonia and high expression in meiotic spermatocytes and post-meiotic spermatids (Figure 1G). Collectively, these observations suggest that Rbm11 is expressed in a tissue-specific manner and displays maximal levels in concomitance with cell differentiation events in brain, cerebellum and testis.
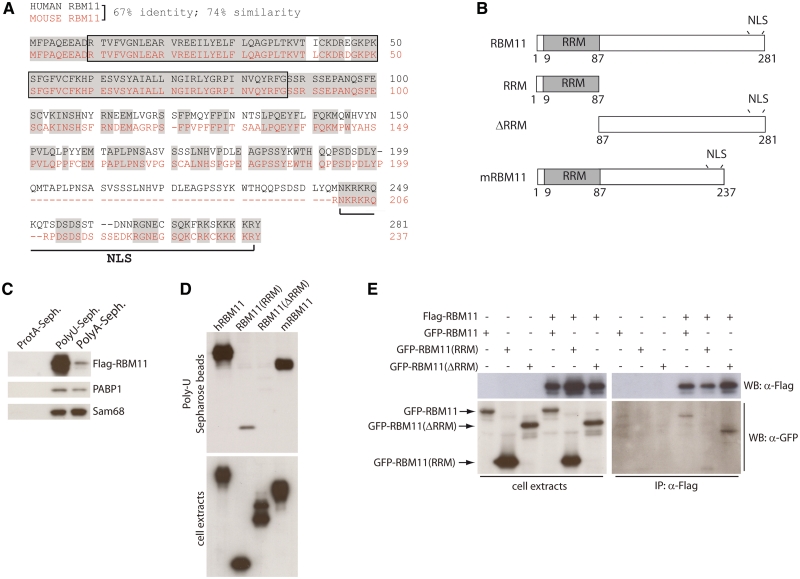
RBM11 protein binds RNA and homodimerizes
Human and murine RBM11 protein sequences show a high degree of homology, with 67% identity and 74% similarity (Figure 2A and B). The main difference is represented by the lack of a stretch of residues within the carboxyl terminal region in murine RBM11, whereas the predicted RRM presents only two conservative substitutions (Figure 2A). Due to the presence of an RRM, RBM11 is annotated as a putative RBP. The RRM is one of the most abundant protein domains in eukaryotes, which characterizes RBPs involved in all steps of RNA processing (33). However, although most RRMs bind RNA, some of these domains are exclusively involved in protein–protein interactions (33). To test whether RBM11 can bind RNA in vitro, we performed pull-down assays with synthetic, homopolimeric RNA. Human Flag-RBM11 expressed in Hek293T cells efficiently bound RNA, with marked selectivity for polyU RNA (Figure 2C). In contrast, two other RBPs, the polyA-binding protein 1 (PABP1) and Sam68, bound with similar efficiency to both polyU and polyA RNAs (Figure 2C). Strong binding to polyU RNA was also observed for murine RBM11 (mRBM11 in Figure 2B and D). Deletion of the RRM (ΔRRM) in human RBM11 completely abolished binding to polyU RNA, whereas the isolated RRM could still bind, albeit with reduced efficiency (Figure 2B and D).
Figure 2.
RBM11 binds synthetic homopolymeric RNA and homodimerizes. (A) Alignment of the human (black) and mouse (red) RBM11 protein sequences. Conserved residues are highlighted in gray; dashed line represents sequences absent in mouse RBM11. The percentage of identity and similarity is reported. The boxed sequence represents the RRM, putative nuclear localization signals are underlined. (B) Schematic representation of RBM11 full length and deletion mutants used in the study. (C, D) Extracts from Hek293T cells transfected with either Flag-RBM11 (C) or with human Flag-RBM11 (hRBM11), Flag-RBM11(RRM), Flag-RBM11(ΔRRM) and mouse Flag-RBM11 (mRBM11) (D), were incubated with polyA- or polyU-RNA sepharose beads. Bound proteins were analyzed by western blot with anti-Flag, anti Sam68 or anti-PABP1 antibodies. (E) Nuclear extracts from Hek293T cells transfected with the indicated plasmids were incubated with ANTI-FLAG® M2 Affinity Agarose Gel beads and bound complexes were analyzed by western Blot with anti-Flag or anti-GFP antibodies.
Several RRM-containing proteins homodimerize, and in some cases this is required for RNA-binding efficiency (33). To test whether RBM11 homodimerizes, Hek293T cells were cotransfected with Flag- and GFP-tagged full-length or mutant RBM11 (Figure 2B). We observed that GFP-RBM11 was efficiently coimmunoprecipitated with Flag-RBM11. Deletion of the RRM in GFP-RBM11(ΔRRM) did not impair this interaction, whereas GFP-RBM11(RRM) was unable to associate with Flag-RBM11 (Figure 2E). These experiments suggest that RBM11 requires the RRM for RNA binding and the C-terminal region for homodimerization.
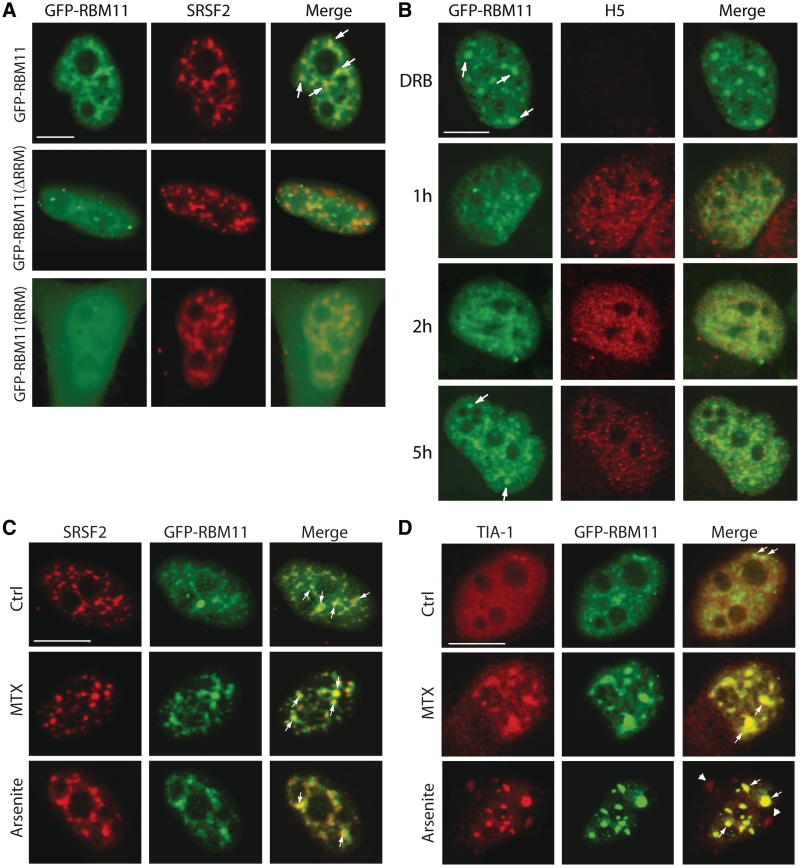
RBM11 colocalizes with splicing speckles in the nucleus
Alignment of the RRM of human RBM11 revealed highest homology with that of proteins involved in pre-mRNA splicing (RBM7, SF3B4, RBMY and hnRNP M), cleavage and polyadenylation (CSTF2), and mRNA translation (RBM3) (Supplementary Figure S2A). To gain insight into the function of RBM11, we analysed its subcellular localization in HeLa cells. GFP-RBM11 and GFP-RBM11(ΔRRM) were exclusively localized in the nucleus, whereas GFP-RBM11(RRM) was equally distributed throughout the cell (Supplementary Figure S2B). In line with this result, analysis of the primary sequence revealed the presence of lysine/arginine-rich potential nuclear localization signals (NLS) in the carboxyl terminus of human and mouse RBM11 (Figure 2A). Confocal analysis showed an enrichment of GFP-RBM11 (Figure 3A) and Flag-RBM11 (Supplementary Figure S2C) in granular structures within the nucleoplasm. Costaining with the SR protein SRSF2 (SC35) (Figure 3A) or the U2AF65 splicing factor (Supplementary Figure S2D) suggested that these structures are splicing speckles. Nonetheless, depletion of RBM11 by RNAi, like depletion of SRSF1, did not affect speckle assembly (Supplementary Figure S2E), indicating that it is not an essential structural component. GFP-RBM11(ΔRRM) localized in the nucleus but did not accumulate in SRSF2 speckles (Figure 3A), nor did the nuclear pool of GFP-RBM11(RRM), which showed a diffused distribution (Figure 3A). These results suggest that the full-length protein is required for association with splicing speckles and that RBM11 might be implicated in splicing regulation.
Figure 3.
Subcellular localization of RBM11. (A) Subcellular localization analysis of wild-type and mutant RBM11 protein. HeLa cells were transfected with the indicated constructs and analyzed by confocal microscopy. Green signal shows GFP fluorescence and red signal shows SFRS2 staining. The full-length RBM11 accumulates in nuclear granules that partially colocalize with SFRS2 (arrows in the merge panels). (B) Confocal microscopy analysis of HeLa cells transfected with GFP-RBM11 and treated for 6 h with DRB (top panels), before release for 1, 2 or 5 h. Cells were stained with the H5 RNAPII antibody, which specifically detects the phosphorylated serine2 protein. GFP signal is in green. White arrows indicate accumulation of RBM11 in speckles in cells treated with DRB. (C, D) HeLa cells were transfected with the GFP-RBM11 plasmid and treated without (Ctrl) or with 1 µM MTX (2 h) or 0.5 mM Arsenite (30 min). Cells were stained with antibodies for SFRS2 (C) or TIA-1 (D) and analyzed by confocal microscopy. The merge panels show the colocalization between RBM11 and SC35 or TIA-1 upon stress in nuclear granules (arrows). Arrowheads indicate localization of TIA-1 in cytoplasmic stress granules. Scale bar = 10 µm.
Analyses of nascent chromatin-associated RNAs have indicated that splicing of most exons occurs cotranscriptionally (34,35). In line with this notion, inhibition of transcription causes accumulation of several SR proteins, including SRSF2, in nuclear speckles (36). To investigate whether RBM11 localization was affected by transcription, we transiently inhibited transcription in HeLa cells. Incubation with the transcription inhibitor DRB suppressed serine 2 phosphorylation of the RNA polymerase II (RNAPII) (H5 staining in Figure 3B) and inhibited transcription, as monitored by bromo-uridine (BrU) incorporation (Supplementary Figure S3A). Under these conditions, RBM11 accumulated in nuclear speckles (Figure 3B). Release of the cells from DRB inhibition caused a sharp increase in RNAPII phosphorylation and redistribution of RBM11 from the speckles to the nucleoplasm. A tight correlation was observed between serine 2 phosphorylation, which marks the RNAPII involved in transcript elongation (37), and diffused localization of RBM11 in the nucleoplasm. Both processes peaked at 1–2 h after DRB release and were partially attenuated by 5 h, when RBM11 started to accumulate again in nuclear speckles (Figure 3B). These observations suggest that, similarly to other splicing regulators, RBM11 is cotranscriptionally recruited to nascent pre-mRNAs.
The localization of RBM11 is influenced by genotoxic and oxidative stresses
Gene transcription is modulated at various steps by RBPs during the cellular response to stress (38,39). To investigate whether the localization of RBM11 is affected by stress, we treated HeLa cells with the DNA topoisomerase II inhibitor mitoxantrone (MTX) (Figure 3C and D) or the alkylating agent cisplatin (Supplementary Figure S3B). Both genotoxic drugs caused accumulation of GFP-RBM11 in nuclear speckles, together with SRSF2 (Figure 3C and Supplementary Figure S3B) and the stress–response RBP TIA-1 (Figure 3D). A similar relocalization was observed after treatment with arsenite to induce oxidative stress (Figure 3C and D). This accumulation of RBPs in nuclear stress granules may reflect a redistribution of transcriptionally active foci within the nucleus in response to genotoxic and oxidative stress (28,38). Notably, while TIA-1 relocalized also in cytoplasmic stress granules after oxidative stress, RBM11, like SRSF2, remained exclusively in the nucleus of cells treated with arsenite (Figure 3D). The relocalization of RBM11 after stress was not a general response of all RBPs, as hnRNP A1 did not accumulate in nuclear granules in these cells (Supplementary Figure S3C).
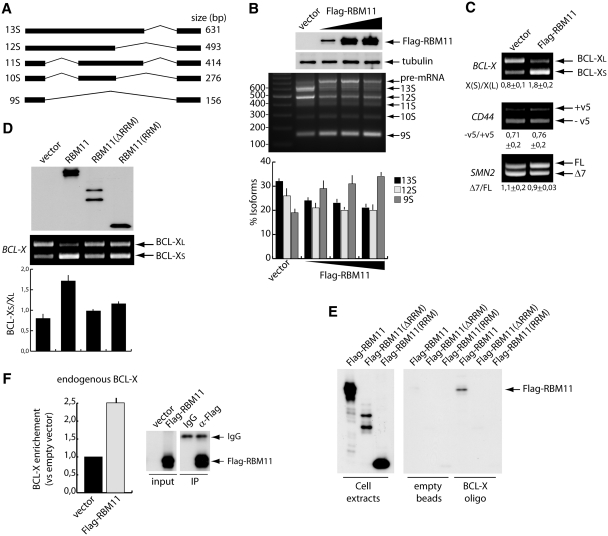
RBM11 is a novel splicing regulator
The presence of an RRM domain is a common feature of splicing factors of the hnRNP and SR protein families (3–5). Given its nuclear localization and association with splicing speckles, we asked if RBM11 displays splicing activity. To address this issue, we performed in vivo splicing assays using the E1A minigene, a commonly used splicing target that contains several 5′ and 3′ alternative splice sites (27) (Figure 4A). RBM11 expression caused a dose-dependent effect on E1A splicing in HeLa cells. In particular, we observed a decrease in the usage of the proximal 5′ splice sites (13S and 12S variants) and a concomitant increase in the choice of the distal 5′ splice site (9S) and of the unspliced pre-mRNA (Figure 4B). In contrast, no change in expression of the 3′ alternatively spliced variants 11S and 10S was observed (Figure 4B). Importantly, RBM11(RRM) and RBM11(ΔRRM) only weakly affected E1A splicing (Supplementary Figure S4A), suggesting that the RRM is required but not sufficient to promote the selection of the distal 5′ splice site in E1A. To further test the splicing activity of RBM11, we used minigenes that recapitulate alternative 5′ splice site choice (BCL-X), variant exon inclusion (CD44) or exon skipping (SMN2). RBM11 strongly enhanced splicing of BCL-XS in Hek293T (Figure 4C) and HeLa cells (Supplementary Figure S4B and C), whereas it exerted negligible effects on CD44 and SMN2 splicing.
Figure 4.
RBM11 regulates AS. (A) Schematic representation of the E1A-derived splicing variants. (B) RT–PCR analysis of E1A splicing in HeLa cells transfected with either empty vector or with increasing amount of Flag-RBM11. Splicing products are indicated on the right of the panel. The bar graph below the PCR panel shows the quantification of the major E1A mRNA variants: light gray bars, dark gray bars, and black bars represent 9S, 12S and 13S variants, respectively. Quantification of the 11S and 10S mRNA isoforms, which showed no change in the splicing assays, was omitted in the graph. Data represent mean ± SD of three experiments. Western blot analysis of RBM11 expression and tubulin as loading control are shown above the PCR panel. (C) RT–PCR analysis of splicing assay in Hek293T cells cotransfected with either empty vector or Flag-RBM11 and the indicated minigene. The ratio of the splicing variants (mean ± SD of three experiments) is shown below each image. (D) RT–PCR analysis of the splicing assays in HEK293T cells cotransfected with the indicated plasmids and the BCL-X minigene. The bar graph represents the BCL–XS/XL ratio (mean ± SD of three experiments). Western blot analysis of wild-type and mutant RBM11 expression is shown above the PCR panel. (E) Western blot analysis of the biotin–RNA pull-down experiment with BCL-X exon-2 RNA and extracts of cells expressing wild-type or mutant Flag-RBM11. Empty streptavidin beads were used as control (right panel). The left panel shows the western blot analysis of the extracts used for the pull-down experiment. (F) UV RNA/protein CLIP of Flag-RBM11. Associated BCL-X RNA was quantified by qRT–PCR. Bar graph (left panel) shows fold enrichment relative to the sample transfected with empty vector. Right panels show western blot analysis of the immunoprecipitated proteins.
RBM11 modulates BCL-X splicing in a sequence specific manner
We further investigated the effect of RBM11 on BCL-X, which represents a functionally relevant splicing event. The BCL-X gene encodes two splice variants, through the choice of alternative 5′ splice sites in exon 2. The long variant, named BCL-XL, displays anti-apoptotic properties, whereas the short BCL-XS variant promotes apoptosis (24). Thus, modulation of BCL-X splicing affects cell survival. We observed that, like for E1A, the BCL-X splicing activity of RBM11 deletion mutants was compromised (Figure 4D). To test whether RBM11 can directly modulate this splicing event, we performed a pull-down assay using biotinylated exon 2 RNA. RBM11 efficiently bound to BCL-X RNA, whereas neither RBM11(ΔRRM) nor the RRM alone interacted (Figure 4E), suggesting that the full-length protein was required for both RNA binding and splicing. Importantly, interaction of RBM11 with BCL-X RNA also occurred in vivo, as indicated by enrichment of endogenous BCL-X mRNA in UV CLIP assays of Flag-RBM11 in Hek293T cells (Figure 4F).
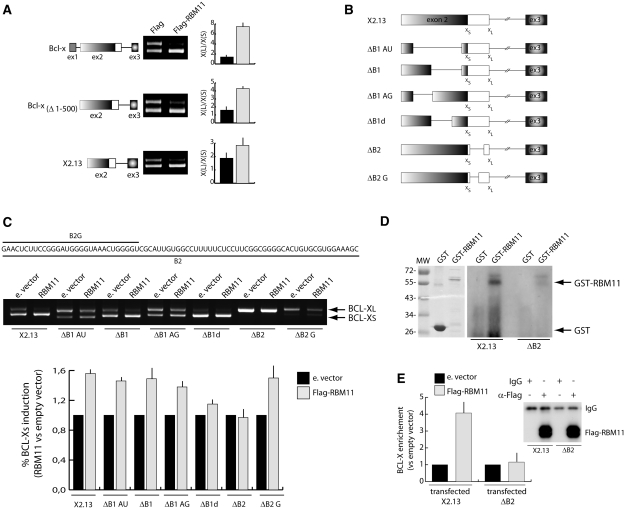
We next asked if specific sequences in BCL-X were required for the effect of RBM11 on splicing. Sequential deletions of the BCL-X minigene showed that RBM11 still exerted an effect when exon 1 and the first half of exon 2 were eliminated (Figure 5A). Thus, we used a set of plasmids containing internal exon 2 deletions (25,26) (Figure 5B). Deletions upstream of the BCL-XS 5′ splice site affected splicing of the minigene, either by reducing or increasing the Xs/XL ratio, but did not abolish the effect of RBM11 (Figure 5C). In contrast, the ΔB2 deletion downstream of the BCL-XS 5′ splice site completely abolished the ability of RBM11 to induce BCL-XS splicing (Figure 5C), whereas deletion of a shorter sequence in the ΔB2G plasmid did not. These results suggest that RBM11 affects BCL-X splicing by directly binding to a sequence downstream of the distal 5′ splice site (Figure 5B and C). Indeed, in vitro UV crosslink experiments showed that GST-RBM11 was crosslinked to wild-type exon 2 RNA (X2.13), whereas binding was almost abolished when the B2 sequence was deleted (Figure 5D). Furthermore, CLIP experiments in Hek293T cells confirmed that RBM11 was efficiently crosslinked in vivo to wild-type BCL-X mRNA, but not to the ΔB2 deletion mutant (Figure 5E). These experiments suggest that RBM11 directly interacts with its splicing target.
Figure 5.
RBM11 binds BCL-X RNA and affects its AS in a sequence-dependent manner. (A) RT–PCR analysis of the in vivo splicing assays in Hek293T cells transfected with Flag-RBM11 and the wild-type BCL-X minigene (top panel) or its 5′-deletion derivatives illustrated on the left side (middle and bottom panel). Bar graphs show the ratio of BCL-XS/XL and represent the mean ± SD of three experiments. (B) Schematic representation of the X2.13 minigene and its mutants. (C) RT–PCR analysis of the in vivo splicing assay. In Hek293T cells cotransfected with either Flag-RBM11 or empty vector as control and the indicated minigene. The bar graph below the PCR panel represents the ratio of BCL–XS/XL (mean ± SD of three experiments). (D) UV-crosslink experiment was performed by incubating 32P-labeled BCL-X exon 2 RNA derived from the indicated minigene with GST or GST-RBM11. The reaction mixtures were UV-irradiated and cross-linked proteins were analyzed by SDS–PAGE and autoradiography. Coomassie staining of the purified proteins used for the experiment is shown on the left. (E) CLIP assay of Flag-RBM11 in Hek293T cells transfected with wild-type (X2.13) or mutated (ΔB2) BCL-X minigenes. Associated BCL-X RNA in the anti-Flag immunoprecipitates was quantified by qRT–PCR (Figure 4). Western blot analysis of the immunoprecipitated proteins is shown on the right.
RBM11 antagonizes SRSF1 in the modulation of BCL-X splicing
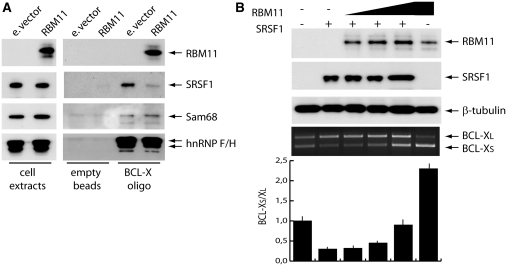
Splicing of the anti-apoptotic BCL-XL variant is promoted by SRSF1 (24). To test whether RBM11 negatively affected SRSF1 activity, we performed a pull-down experiment with nuclear extracts of mock- or RBM11-transfected Hek293T cells. SRSF1 efficiently bound exon 2 RNA when nuclear extracts from mock-transfected cells were used (Figure 6A). However, this interaction was strongly decreased when RBM11 was upregulated and bound to exon 2 RNA. Competition with SRSF1 binding appeared specific, as expression of RBM11 did not affect binding of other regulators of BCL-XS splicing like Sam68 and hnRNP F/H (24,25) (Figure 6A).
Figure 6.
RBM11 antagonizes SRSF1-mediated BCL-X splicing. (A) Western blot analysis of the RNA pull-down experiment performed using biotinylated BCL-X exon-2 RNA and extracts of control or Flag-RBM11 expressing Hek293T cells. Empty streptavidin beads were used as control (right panel). (B) RT–PCR analysis of the in vivo splicing assays of the BCL-X minigene in Hek293T cells transfected with Flag-SRSF1 and increasing amounts of Flag-RBM11, either alone or in combination, as described (top panel). Bar graph shows the BCL–XS/XL ratio and represents the mean ± SD of three experiments. Western blot analysis of RBM11, SRSF1 and tubulin as loading control is shown above the PCR panel.
Next, we tested whether RBM11 competes with SRSF1 for BCL-X splicing also in live cells. As expected, up-regulation of SRSF1 enhanced splicing of BCL-XL in Hek293T cells (Figure 6B). Importantly, coexpression of increasing amounts of RBM11 reverted this effect (Figure 6B). In contrast, the RBM11 deletion mutants that are defective in splicing activity and exon 2 binding did not efficiently counteract SRSF1 activity (Supplementary Figure S5). These results suggest that RBM11 promotes splicing of BCL-XS by competing with recruitment of SRSF1 to exon 2.
DISCUSSION
Our work characterizes the RRM-containing protein RBM11 as a novel tissue-specific splicing factor. RBM11 is selectively expressed in few human cell lines, an evidence that confirmed its restricted pattern of expression in human tissues (21–23). Expression of the mouse Rbm11 gene is also restricted to few tissues, where it is regulated in a developmental manner. In brain and cerebellum, its levels increased during embryogenesis (E10.5–15.5), when neurogenesis and neural cell migration occur, and peaked perinatally (P0–3 dpp), when the vast array of cell types composing the nervous system appears and the mature cytoarchitecture of these organs takes shape (30,31). In the testis, Rbm11 expression levels increased rapidly from 3 to 16 dpp, when mitotic spermatogonia differentiate into meiotic spermatocytes (32). Thus, Rbm11 expression seems to correlate with developmental stages when important cell differentiation events occur, suggesting that it might play a role in the differentiation program of germ cells and neuronal subtypes. Brain and testis have been annotated as the tissues expressing the highest levels of splicing factors and AS events (6,40,41), which could contribute to the generation of proteomic diversity in highly specialized cells, like neurons and germ cells. Nevertheless, only a limited number of brain- and testis-specific splicing factors have been described so far (1,10–16), raising the possibility that additional, uncharacterized RBPs participate to regulation of splicing in these tissues. Although the lack of valid antibodies to confirm RBM11 protein expression may limit the interpretation of our results, the apparently specific expression of RBM11 in brain, cerebellum and testis reinforces the notion that neurons and germ cells express a unique repertoire of splicing factors.
Mouse and human RBM11 are characterized by the presence of a single RRM at the amino terminus. Beside the presence of NLS sequences at the C-terminus, no other known functional domains were identified in the remaining two-thirds of the protein. The RRM is one of the most abundant protein domains in eukaryotes, and characterizes RBPs involved in multiple aspects of RNA metabolism (33). Nevertheless, not all RRMs can directly bind RNA. In some cases, as for the exon junction protein Y14, the RRM is involved in protein–protein interactions (33). In proteins containing multiple RRMs, like hnRNP A1, portions of one RRM can contact another RRM, thereby promoting cooperative binding to RNA (33). Since no information on the functional role of RBM11 was available, it was relevant to investigate the biochemical properties of the protein. We found that RBM11 binds RNA and that this interaction requires its RRM. However, although the RRM was sufficient to bind RNA, its interaction with homopolymeric polyU or with BCL-X exon 2 RNA was much weaker than that of full-length RBM11. As multiple RRMs can allow higher affinity or cooperative binding to RNA (33), one possibility is that RBM11 needs to dimerize or to interact with other proteins to bind RNA with high affinity. Our experiments indicate that RBM11 homodimerizes in live cells, and that the C-terminal region downstream of the RRM is necessary for this interaction. Thus, we propose that RBM11 requires the RRM for the interaction with RNA and the C-terminal region for homodimerization.
RRM-containing proteins have been involved in basically all the steps of RNA processing and metabolism, from cotranscriptional splicing to RNA decay (33). Several observations point to a role of RBM11 in the regulation of splicing. Live imaging and confocal microscopy indicated that RBM11 is exclusively localized in the nucleus. The protein is enriched in nuclear granules that are costained with antibodies for SRSF2, suggesting that these structures are splicing speckles. Furthermore, similarly to other splicing regulators, RBM11 localization is dynamically modulated in response to an altered transcriptional status of the cell. For instance, inhibition of transcription caused accumulation of RBM11 in the nuclear speckles, as observed for SRSF2 and other splicing factors (36). However, upon release from the DRB block, RBM11 rapidly diffused in the nucleoplasm concomitantly with increased serine 2 phosphorylation of RNAPII, which marks the transcriptional elongation phase (37). This redistribution suggests recruitment of RBM11 to nascent transcripts in the nucleoplasm and its involvement in cotranscriptional RNA processing events. Notably, RNAPII accumulates in splicing speckles upon transcription inhibition as well (36). Moreover, transcriptionally active (BrU positive) foci that form after exposure of cells to genotoxic stress also accumulate serine 2-phosphorylated RNAPII and splicing factors (28). RBM11 behaves in the same manner and colocalize with SRSF2 and TIA-1 in these nuclear foci after genotoxic and oxidative stress, but not in TIA-1-containing cytoplasmic stress granules, suggesting that its function is restricted to the nucleus. Thus, we propose that RBM11 is mainly involved in cotranscriptional RNA processing events together with other splicing factors.
Our study shows that RBM11 can modulate AS events. By using a panel of previously characterized minigenes, we found that it selectively affects splicing of E1A and BCL-X, but not that of CD44 and SMN2. This selectivity might indicate a sequence-specific role of RBM11 in splicing. Other observations support this hypothesis. First, we found that, unlike Sam68 and the RRM-containing protein PABP1, RBM11 displays high selectivity for polyU homopolymeric RNA in vitro. Moreover, crosslink experiments in vitro and in live cells showed that RBM11 directly binds BCL-X RNA. Up-regulation of RBM11 strongly induced the BCL-XS variant. However, neither the RRM alone nor RBM11(ΔRRM) could recapitulate the effect of the full-length protein. Since neither deletion mutant efficiently bound BCL-X exon 2, these results suggest that RBM11 directly modulates splicing events by interacting with target sequences in the pre-mRNA. In support of this hypothesis, deletion of 78 bp located between the two alternative 5′ splice sites in exon 2 strongly reduced crosslink of RBM11 to the BCL-X RNA and suppressed its effect on splicing. Importantly, binding of RBM11 to BCL-X exon 2 RNA impaired recruitment of SRSF1 and competed with its ability to enhance BCL-XL splicing, suggesting that RBM11 might antagonize SR proteins like an hnRNP.
Our study has not addressed the physiological role of RBM11. However, its highly restricted pattern of expression allows some speculations. We found that RBM11 expression increases at developmental stages when major differentiation events occur in brain and testis (30–32). Furthermore, in the testis RBM11 is strongly expressed in post-mitotic spermatocytes and spermatids, and to a lower extent in mitotic spermatogonia. Thus, it is conceivable that RBM11 participates to changes in gene expression required to insure correct differentiation of neurons and germ cells. In this regard, it is noteworthy that its putative target BCL-X is an essential gene for the development of the nervous system, where its ablation leads to massive cell death in post-mitotic neurons but not in neural progenitor cells (42). It will be interesting to investigate whether changes in the expression levels of RBM11 affect development of the nervous system, as many neurodegenerative diseases seem to depend on aberrant AS and/or splicing factors expression (43). RBM11 is among the genes putatively linked to Down syndrome (21–23), and it will also be interesting to test whether its unbalanced expression in patients with chromosome 21 trisomy can lead to changes in the splicing pattern of genes that affect the pathology.
In conclusion, our study describes the characterization of a novel tissue-specific splicing factor that might be required for the proper developmental program of neurons and germ cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table S1, Supplementary Figures S1–S5.
FUNDING
Funding for open access charge: Agenzia Spaziale Italiana (ASI); Associazione Italiana Ricerca sul Cancro (AIRC); Telethon (GGP09154).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank Dr Benoit Chabot for the BCL-X minigenes and Dr Stephan Stamm for the SMN2 and CD44 minigenes, Dr Pamela Bielli for critical reading of the manuscript and production of reagents.
REFERENCES
- 1.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell. Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 4.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 5.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 6.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun S, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat. Struct. Mol. Biol. 2007;17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valacca C, Bonomi S, Buratti E, Pedrotti S, Baralle FE, Sette C, Ghigna C, Biamonti G. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J. Cell. Biol. 2010;191:87–99. doi: 10.1083/jcb.201001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ule J, Darnell RB. Functional and mechanistic insights from genome-wide studies of splicing regulation in the brain. Adv. Exp. Med. Biol. 2007;623:148–160. doi: 10.1007/978-0-387-77374-2_9. [DOI] [PubMed] [Google Scholar]
- 11.Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 12.Kim KK, Kim YC, Adelstein RS, Kawamoto S. Fox-3 and PSF interact to activate neural cell-specific alternative splicing. Nucleic Acids Res. 2011;39:3064–3078. doi: 10.1093/nar/gkq1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott DJ, Grellscheid SN. Alternative RNA splicing regulation in the testis. Reproduction. 2006;132:811–819. doi: 10.1530/REP-06-0147. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Bourgeois CF, Pang S, Kudla M, Dreumont N, Kister L, Sun YH, Stevenin J, Elliott DJ. The germ cell nuclear proteins hnRNP G-T and RBMY activate a testis-specific exon. PLoS Genet. 2009;5:e1000707. doi: 10.1371/journal.pgen.1000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dreumont N, Bourgeois CF, Lejeune F, Liu Y, Ehrmann IE, Elliott DJ, Stévenin J. Human RBMY regulates germline-specific splicing events by modulating the function of the serine/arginine-rich proteins 9G8 and Tra2-{beta} J. Cell Sci. 2010;123:40–50. doi: 10.1242/jcs.055889. [DOI] [PubMed] [Google Scholar]
- 16.Venables JP, Vernet C, Chew SL, Elliott DJ, Cowmeadow RB, Wu J, Cooke HJ, Artzt K, Eperon IC. T-STAR/ETOILE: a novel relative of SAM68 that interacts with an RNA-binding protein implicated in spermatogenesis. Hum. Mol. Genet. 1999;8:959–969. doi: 10.1093/hmg/8.6.959. [DOI] [PubMed] [Google Scholar]
- 17.Richard S, Torabi N, Franco GV, Tremblay GA, Chen T, Vogel G, Morel M, Cléroux P, Forget-Richard A, Komarova S, et al. Ablation of the Sam68 RNA binding protein protects mice from age-related bone loss. PLoS Genet. 2005;1:e74. doi: 10.1371/journal.pgen.0010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paronetto MP, Messina V, Bianchi E, Barchi M, Vogel G, Moretti C, Palombi F, Stefanini M, Geremia R, Richard S, et al. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J. Cell Biol. 2009;185:235–249. doi: 10.1083/jcb.200811138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paronetto MP, Zalfa F, Botti F, Geremia R, Bagni C, Sette C. The nuclear RNA-binding protein Sam68 translocates to the cytoplasm and associates with the polysomes in mouse spermatocytes. Mol. Biol. Cell. 2006;17:14–24. doi: 10.1091/mbc.E05-06-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paronetto MP, Messina V, Barchi M, Geremia R, Richard S, Sette C. Sam68 marks the transcriptionally active stages of spermatogenesis and modulates alternative splicing in male germ cells. Nucleic Acids Res. 2011;39:4961–4974. doi: 10.1093/nar/gkr085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardiner K, Slavov D, Bechtel L, Davisson M. Annotation of human chromosome 21 for relevance to Down syndrome: gene structure and expression analysis. Genomics. 2002;79:833–843. doi: 10.1006/geno.2002.6782. [DOI] [PubMed] [Google Scholar]
- 22.Brun ME, Ruault M, Ventura M, Roizès G, De Sario A. Juxtacentromeric region of human chromosome 21: a boundary between centromeric heterochromatin and euchromatic chromosome arms. Gene. 2003;312:41–50. doi: 10.1016/s0378-1119(03)00530-4. [DOI] [PubMed] [Google Scholar]
- 23.Hu YH, Warnatz HJ, Vanhecke D, Wagner F, Fiebitz A, Thamm S, Kahlem P, Lehrach H, Yaspo ML, Janitz M. Cell array-based intracellular localization screening reveals novel functional features of human chromosome 21 proteins. BMC Genomics. 2006;7:155. doi: 10.1186/1471-2164-7-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J. Cell. Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garneau D, Revil T, Fisette JF, Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 26.Revil T, Pelletier J, Toutant J, Cloutier A, Chabot B. Heterogeneous nuclear ribonucleoprotein K represses the production of pro-apoptotic Bcl-xS splice isoform. J. Biol. Chem. 2009;284:21458–21467. doi: 10.1074/jbc.M109.019711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denegri M, Chiodi I, Corioni M, Cobianchi F, Riva S, Biamonti G. Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol. Biol. Cell. 2001;12:3502–3514. doi: 10.1091/mbc.12.11.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busà R, Geremia R, Sette C. Genotoxic stress causes the accumulation of the splicing regulator Sam68 in nuclear foci of transcriptionally active chromatin. Nucleic Acids Res. 2010;38:3005–3018. doi: 10.1093/nar/gkq004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedrotti S, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Stamm S, Manley JL, Sette C. The splicing regulator Sam68 binds to a novel exonic splicing silencer and functions in SMN2 alternative splicing in spinal muscular atrophy. EMBO J. 2010;29:1235–1247. doi: 10.1038/emboj.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brumwell CL, Curran T. Developmental mouse gene expression maps. J. Physiol. 2006;575:343–346. doi: 10.1113/jphysiol.2006.112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Ann. Rev. Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- 32.Bellvé AR, Millette CF, Bhatnagar YM, O'Brien DA. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J. Histochem. Cytochem. 1977;25:480–494. doi: 10.1177/25.7.893996. [DOI] [PubMed] [Google Scholar]
- 33.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;72:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 34.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol. Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 37.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 38.Biamonti G, Caceres JF. Cellular stress and RNA splicing. Trends Biochem. Sci. 2009;34:146–153. doi: 10.1016/j.tibs.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Busà R, Sette C. An emerging role for nuclear RNA-mediated responses to genotoxic stress. RNA Biol. 2010;7:390–396. doi: 10.4161/rna.7.4.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de la Grange P, Gratadou L, Delord M, Dutertre M, Auboeuf D. Splicing factor and exon profiling across human tissues. Nucleic Acids Res. 2010;38:2825–2838. doi: 10.1093/nar/gkq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grosso AR, Gomes AQ, Barbosa-Morais NL, Caldeira S, Thorne NP, Grech G, von Lindern M, Carmo-Fonseca M. Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res. 2008;36:4823–4832. doi: 10.1093/nar/gkn463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roth KA, D'Sa C. Apoptosis and brain development. Ment. Retard. Dev. Disabil. Res. Rev. 2001;7:261–266. doi: 10.1002/mrdd.1036. [DOI] [PubMed] [Google Scholar]
- 43.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.