Abstract
We have investigated the DNA-binding nature as well as the function of a putative Alba (Acetylation lowers binding affinity) family protein (PfAlba3) from Plasmodium falciparum. PfAlba3 possesses DNA-binding property like Alba family proteins. PfAlba3 binds to DNA sequence non-specifically at the minor groove and acetylation lowers its DNA-binding affinity. The protein is ubiquitously expressed in all the erythrocytic stages of P. falciparum and it exists predominantly in the acetylated form. PfAlba3 inhibits transcription in vitro by binding to DNA. Plasmodium falciparum Sir2 (PfSir2A), a nuclear localized deacetylase interacts with PfAlba3 and deacetylates the lysine residue of N-terminal peptide of PfAlba3 specific for DNA binding. PfAlba3 is localized with PfSir2A in the periphery of the nucleus. Fluorescence in situ hybridization studies revealed the presence of PfAlba3 in the telomeric and subtelomeric regions. ChIP and ChIP ReChIP analyses further confirmed that PfAlba3 binds to the telomeric and subtelomeric regions as well as to var gene promoter.
INTRODUCTION
Human malaria parasite Plasmodium falciparum (P. falciparum) causes one of the most life-threatening diseases, accounting for nearly 1.5–2.7 million deaths annually (1). Unfortunately, the management of malaria is becoming a difficult task due to the development of resistance against currently used drugs (2,3). Identification and characterization of novel protein from P. falciparum are accepted as a fundamental approach to understand parasite biology. Genome sequencing and transcriptome analysis of P. falciparum provides an excellent opportunity to identify important protein from the parasite (4). Comprehensive genome analysis of P. falciparum reveals that it encodes each of core histone together with histone variants H2A.Z, H3.3, centromere-specific H3 (CenH3), H4 and H2Bv, expressed differentially during different intra-erythrocytic stages (5,6). Besides histones, Plasmodium genome constitutes other expanded family of DNA-binding proteins involved in regulation of gene expression (7–13). Plasmodium falciparum genome encodes novel DNA–RNA-binding proteins show homology with Alba super-family conserved domain (Archeal chromatin protein family Alba), well characterized in hyperthermophilic Sulfolobus spp. (14–19). Genome search reveals the presence of three homologs of DNA–RNA-binding protein belonging to Alba family termed as Alba 1(Pf08_0074), Alba 2 (MAL13P1.233) and Alba 3 (Pf10_0063), which may play an important role in parasite biology (http://www.genedb.org). Alba super family proteins are originated as RNA-binding proteins that unify the Alba proteins with RNase P subunit (20). The Alba protein (previously named as Sso10b) identified as a major architectural DNA-binding protein (15), which organizes and regulates the genome of both euryarchea (having histone) as well as crenarchea (having no histone) (21,22). Alba is controlled by acetylation and deacetylation, where acetylation at specific N-terminal lysine residue lowers its binding affinity toward double-strand (ds) DNA (14,19). In Sulfolobus solfatataricus, Alba was found to be reversibly acetylated at lys 16 by a homolog of Pat (protein acetyltransferase) and deacetylated by Sir2 protein (a sirtuin family deacetylase) (14,17,23). Alba proteins also contain RGG repeats at C-terminal and the affinity of these repeats for RNA is modulated through the methylation of arginine in many RGG-box proteins (20,24). Apart from this phosphoproteome analysis also revealed the presence of phosphorylated form of Alba protein (25). Alba binds dsDNA cooperatively without significant compaction (26) in a sequence-independent manner and distributed uniformly on the chromosomes (14,18). Crystal structure of Alba protein reveals homodimer (dimeric) nature (common α/β fold), sharing the N-terminal domain of DNase I and the C-terminal domain of translation initiation factor (14,27). In S. solfatataricus, two copies of Alba protein (Alba 1 and Alba 2) are reported, which exist exclusively as heterodimer (28). Plasmodium falciparum genome also encodes a homolog of yeast Sir2 well characterized in the apicomplexians (13,29,30). In P. falciparum, Sir2 (PfSir2A) is nuclear localized (telomeric and subtelomeric) and involved in silencing of different var gene subsets (11,31,32). Plasmodium falciparum var gene family encodes the variant surface antigen PfEMP1 (P. falciparum erythrocyte membrane protein 1) (33,34) and associated with various endothelial receptors (i.e. ICAM-1, CD36 and CSA) (35). Studies on var gene expression revealed that histone modification pattern as well as interactions of different nuclear proteins (unique to either subtelomeric or internal chromosomal var gene promoters) is associated with var gene silencing (36–38). PfHP1 (P. falciparum heterochromatin 1) and histone variant H2A.Z were also shown to be linked with mutually exclusive expression of var genes (39,40). PfSir2A plays a key role in heterochromatin-mediated silencing and comprehensive repression and switching expression of var genes (31,41). Sir2 from P. falciparum and Saccharomyces cerevisiae is associated with silent chromatin and binds to regions of the chromosomes that are transcriptionally suppressed comprising the telomeric and subtelomeric regions (42,43). The crucial role of PfSir2A and histone acetylation in var gene regulation was also established by gene disruption studies (32,41). Recent studies documented the presence of an additional plasmodial histone deacetylase, termed PfSir2B, involved exclusively in the expression of the ups B subtype (32). These findings validated the involvement of two paralogs of Sir2 (termed as PfSir2A and PfSir2B) that cooperate to regulate the expression of the var gene repertoire in P. falciparum. Furthermore, Sir2 paralogs also played an important role in the regulation of other antigenically variant genes (such as rif gene families) as well as maintained telomere repeat length (32). Apart from this, a broad range of histone acetyltransferse has also been identified in Plasmodium genome, only a few of them have been characterized till date (44). The wide presence of these acetylase and deacetylase in Plasmodium suggests the crucial role performed by them and their target proteins. PfAlba3 may be one of the possible targets of this global machinery for the regulation of cellular growth and development. Information regarding PfAlba3 is currently insufficient to elucidate its role in P. falciparum.
Here, we have characterized DNA-binding properties of PfAlba3 and shown the association of this protein with PfSir2A at telomeric and subtelomeric region of chromosomes. It has been confirmed that PfAlba3 binds dsDNA sequence non-specifically and acetylation of this protein lowers its DNA-binding affinity; a feature shared by Alba family proteins. PfAlba3 is expressed throughout all the asexual blood stages and binding of PfAlba3 to DNA inhibits transcription. We also provide evidence that protein exists as an acetylated form and the N-terminal peptide of PfAlba3 could be deacetylated by PfSir2A, specifically at lysine 22 of KKP motif. This protein interacts with PfSir2A and occupies mainly the periphery of the nucleus encompassing the telomeric and subtelomeric region including var gene promoter.
MATERIALS AND METHODS
Cloning and expression
RT–PCR amplified cDNA fragment corresponding to the PfAlba3 (Pf10_0063, 324 bp) and PfSir2A (Pf13_0152, 822 bp) was cloned into the pET-28a (PfAlba3) or pET28a and pGEX-5X-3 (PfSir2A) vector to express the recombinant protein. Identity of resultant recombinant clone was confirmed by DNA sequencing on an automated sequencer (3730 Analyzer-17121-005, Applied Bio-system) at gene sequencing facility, University of Delhi, South campus. The sequence was compared with the existing sequences from the Malaria Genome Consortium (www.plasmodb.org) using BLASTN program. For recombinant protein expression, pET-28a-PfAlba3 clone was transformed into the Escherichia coli BL21 (Rosetta) strain and protein synthesis was induced with 1 mM IPTG when OD600 reached at 0.4–0.6. Culture was further grown for 6 h under same conditions. Cells were harvested by centrifugation, resuspended in 15 ml of lysis buffer (50 mM Tris–HCl, pH 8.0, 300 mM NaCl, 10 mM imidazole and 10% glycerol) containing 1 mg/ml lysozyme and protease inhibitor cocktail. Cells were incubated for 30 min on ice and lysed by sonication with pulse-rest cycle (10 cycles; 10 s pulse at 25 W with 1 min interval after each pulse). The protein was purified by Ni-NTA column affinity chromatography. Recombinant protein was concentrated by using Millipore filtration units. The protein was further purified by HPLC using protein-pak™ 125 A gel-filtration column (Waters, 7.8 × 300 mm) by running in 20 mM Tris–HCl buffer (pH 8.0) containing 100 mM NaCl at a flow rate of 0.5 ml/min. Purified protein was subjected to extensive dialysis (10 kDa cut off membrane, Pierce) against 10 mM Tris–HCl, pH-8.0, 20% glycerol and 50 mM NaCl and 2 mM DTT. For PfSir2A expression, PfSir2A-pET-28a and PfSir2A-pGEX-5X-3 clones were transformed in BL21 (DE3)/pLys E. coli and BL21 (DE3) cells, respectively. Recombinant protein synthesis was induced by the addition of 0.1 mM IPTG at OD600 = 0.25. Cells were grown overnight at room temperature. Recombinant His and GST fusion PfSir2A was purified using Ni-NTA and GST column chromatography respectively. Homogeneity of the recombinant protein was checked by SDS–PAGE and western blotting using anti-PfAlba3 and anti-PfSir2A antibodies. The concentration of the protein was determined as described (45).
Agarose gel-retardation assay
DNA-binding activity of PfAlba3 was analyzed by using agarose gel-retardation assay as described previously (27). In brief, supercoiled ϕX174 dsDNA (250 ng) was incubated with various concentrations of native (non-acetylated)/acetylated PfAlba3 in binding buffer (20 mM MES, pH 6.5, 100 mM potassium glutamate, 1 mM MgCl2 and 0.1 mg/ml bovine serum albumin) in a total volume of 10 µl. After 15 min at 20°C, one-sixth volume of loading buffer (6 × loading dye 0.25% bromophenol blue, 0.25% xylene cyanol FF, 35% ficoll type 400) was added and samples were electrophoresed (in 0.7% agarose, 1 × TBE) at 10 V/cm for 90 min. After electrophoresis, gels were stained in ethidium bromide and visualized under UV light.
Electrophoretic mobility shift assay
Sequences of oligonucleotides and their corresponding complements used for electrophoretic mobility shift assay (EMSA) were as follows: 30-bp-5′CATGGATGAAGAAAGATAGAGAACCAATAG 3′ and 5′ CTATTGGTTCTCTATCTTTCTTCATCCATG 3′. The 20-bp-5′AGAAAGATAGGGATGGGATG 3′ and 5′CATCCCATCCCTATCTTTCT 3′, respectively (Sigma Aldrich). Synthetic oligonucleotides were annealed to create ds and 5′-end-labeled with [γ-32P] ATP using T4 polynucleotide kinase, according to standard protocols. Unincorporated [γ-32P] ATP was removed by ethanol precipitation at −20°C (overnight) followed by washing with 70% ethanol. Reactions were performed in a volume of 10 µl containing 32P-labeled ds oligonucleotide probes (50 nM) incubated with recombinant PfAlba3 in binding buffer (10 mM HEPES buffer, pH 7.6, 50 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 0.1 mM dithiothreitol, 1 mg/ml BSA and 0.05% Triton X-100) for 30 min on ice. Complexes were separated by native gel electrophoresis (7%) running under 10 V/cm at 4°C with 0.5 × TBE (Tris/borate/EDTA; 1 × TBE = 45 mM Tris–borate and 1 mM EDTA) running buffer. Supershift assay was performed by adding anti-PfAlba3 polyclonal antibody in the reaction. For competition assay, unlabeled competitor oligonucleotides (same oligonucleotides in 1- to 100-fold molar excess) were added at the start of the incubation. The gel was dried and exposed to an X-ray film overnight at −80°C before film development. The intensity of free and bound DNA was quantified by using phosphoimager and auto-radiograph analysis (Image J software).
Dissociation constant of protein–DNA complexes was determined using linear regression according to the Scatchard equation as described previously (46). In short, 50 nM of ds oligos (20 bp) were incubated with increasing concentrations of PfAlba3 for 30 min on ice and the products were resolved in 7% native PAGE. Data were analyzed using Scatchard equation r/[A] = −(1/ Kdapp) r + n/Kdapp, where the ratio of protein bound oligos to total oligos [{PfAlba3-oligo}/ oligoT] denoted as r, [A] referred to PfAlba3 concentration and Kdapp is apparent dissociation constant, while n is the number of binding sites for protein. Specificity of PfAlba3 binding to major or minor groove of dsDNA was studied by using groove specific dye as a competitor as described earlier (47). In brief, 32P-labeled ds oligos (30 bp) were pre-incubated with increasing concentrations of actinomycin D (Gibco Biosciences) and methyl green (Sigma) for 30 min at 20°C in a reaction volume of 20 µl in binding buffer. Following incubation, PfAlba3 (500 nM) was added to the reaction mixture, and allowed to an additional incubation on ice for 30 min. The DNA–protein complex was analyzed on a 7% native polyacrylamide gel in 0.5 × TBE at 150 V at 4°C for 2–3 h.
RT–PCR and western blotting for stage-specific expression
Stage-specific expression of PfAlba3 and PfSir2A was checked by using RT–PCR and western blotting. Parasites from different stages were obtained after culturing ring-synchronized P. falciparum. Synchronization of culture was done as described earlier (48–50). To check the stage-specific expression, equivalent amount of RNA (1 µg) from different stages was used as template to amplify PfAlba3, PfSir2A and seryl tRNA synthetase. Seryl tRNA synthetase expresses equally in each stage of the P. falciparum used as a positive control (51). Primer sequences used for seryl tRNA synthetase were 5′-GAGGAATTTTACGTGTTCATCAA-3′ (forward) and 5′-GATTACTTGTAGGAAAGAATCCTTC-3′ (reverse). RT–PCR products were analyzed through electrophoresis on 1% agarose gel in TAE buffer at 10 V/cm. For immunodetection, parasites (10% parasitemia) from ring, trophozoite and schizont stages were prepared by lysis of infected RBC with 0.05% saponin. Released parasite was centrifuged and washed thrice with PBS, and suspended in 1× PBS. The isolated parasite was lysed in PBS containing protease inhibitor cocktail (Sigma) by mild sonication (5 s pulse, bath type sonicator) at 4°C and the whole lysate was then resolved in 12% SDS–PAGE and electroblotted onto the nitrocellulose membrane. After incubating the blots with rabbit polyclonal anti-PfAlba3 and anti-PfSir2A antibodies (1:5000 dilutions) or rabbit pre immune sera at room temperature for 2 h with constant shaking, it was washed five times with PBS containing 0.05% (v/v) Tween-20. Membrane was further incubated with HRP conjugated anti-rabbit/anti-mouse antibody for 1 h with constant shaking, blot was washed and developed using a chemiluminescence detection system. Anti-tubulin antibody was used as control.
Immunoprecipitation
PfAlba3 and PfSir2A from the parasite lysate were immunoprecipitated by anti-PfAlba3 and anti-PfSir2A antibody, respectively. In brief, parasite (∼10% parasitemia) was released by 0.05% saponin treatment, washed with PBS and subsequently lysed in ice chilled RIPA buffer (0.1% SDS, 0.5% Triton X-100, 0.5% sodium deoxycholate, 1 mM PMSF and protease inhibitor cocktail) by incubation on ice for 45 min. The lysate was centrifuged at 14 500g for 10 min and the supernatant was pre-cleared by the addition of Protein A sepharose beads. The clear supernatant was incubated with primary antibody (rabbit anti-PfAlba3/mouse anti-PfSir2A antibodies) for 6 h at 4°C with simultaneous mixing on a rotating wheel and centrifuged at 14 500g for 20 min. The supernatant was further incubated for 12–14 h with Protein A sepharose at 4°C and beads were pelleted by centrifugation at 13 000 g for 5 min at 4°C. Beads were then washed with ice chilled RIPA buffer followed by a PBS wash. Immunoprecipitated protein was obtained by treating the beads with SDS Lamelli buffer and samples were electrophoresed on a 15% SDS–PAGE.
Immunoflourescence studies
For immunodetection, parasite pellets containing ring, trophozoite and schizont stages were washed with 1× PBS and then boiled with 1× sodium dodecylsulfate (SDS) gel loading buffer and loaded on 10% SDS–polyacrylamide gel. Western blot analysis was done using affinity purified anti-PfSir2A and anti-PfAlba3 antibodies (1:5000) using the standard protocol. For localization studies, synchronized parasites were cultured according to standard procedures. Immunofluorescence (IF) analysis was performed in different asexual stages of the parasite. Synchronized cultures of P. falciparum were washed in phosphate-buffered saline (PBS), and fixed in 4% paraformaldehyde and 0.0075% glutaraldehyde in PBS for 20 min. Cells were subsequently washed with PBS and permeabilized with 0.1% Triton X-100 in PBS for 10 min. Slides were then blocked with 3% bovine serum albumin (BSA) in PBS for 1 h at room temperature and rinsed with PBS. Parasites were then incubated with the primary antibodies (rabbit anti-PfAlba3 or mouse anti-PfSir2A antibodies) diluted in 3% BSA overnight at 4°C. After that cells were washed three times (10 min for each wash) with PBS and incubated with secondary antibody (Alexa Fluor 633 and Alexa Fluor 488 tagged goat anti-rabbit IgG or goat anti-mouse IgG, Invitrogen) diluted (1:500) in blocking solution for 1 h at room temperature. After final washes, slides were mounted in prolong gold antifade reagent (Invitrogen) with 4-6-diamidino-2-phenylindole (DAPI). Confocal images were obtained with a Laser Confocal Microscope, Olympus FluoView™ FV1000 at 100× magnification with oil immersion objective.
Nuclear and cytoplasmic extract preparation
Nuclear and cytoplasmic fractions were prepared essentially as described earlier (52). In brief, Parasites from mixed stage culture were isolated from infected red blood cells by saponin lysis and washed twice in PBS, resuspend in ice-chilled lysis buffer (20 mM HEPES, pH 7.8, 10 mM KCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.65% NP-40) and incubated for 10 min on ice. Nuclei were pelleted by centrifugation (2500g) for 5 min, followed by separation of the supernatant (containing cytoplasmic proteins) and stored at −80°C. The pellet washed thrice in lysis buffer and suspended in extraction buffer (20 mM HEPES, pH 7.8, 800 mM KCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 3 μM pepstatin A and 10 μM leupeptin). The extract was subjected to vigorous shaking at 4°C for 30 min and cleared by centrifugation at 13 000g for 30 min. The supernatant (containing nuclear proteins) was diluted with one volume of dilution buffer (20 mM HEPES, pH 7.8, 1 mM EDTA, 1 mM DTT, 30% glycerol) and stored at −80°C.
Deacetylation assays
Deacetylation assays were performed using synthetic peptide substrate following standard protocol in presence of 2.5–10 µg of recombinant PfSir2A. Reaction mixtures were incubated at 37°C for 6 h. All assays were performed in triplicate. PfSir2A deacetylase activity was monitored in 50 µl of buffer containing 50 mM Tris–HCl, pH 8.0, 4 mM MgCl2, 0.2 mM dithiothreitol (DTT), variable concentrations of nicotinamide adenine dinucleotide (NAD+, 0–500 µM), 2.5–10 µg of the purified recombinant PfSir2A and 10 µg of the N-terminal peptide of PfAlba3 (at lysine 22 and 23 at KKP sequence). Reaction was initiated by the addition of enzyme and incubated for 4–6 h at 37°C. To check the effect of NAD+ on the PfSir2A-mediated lysine deacetylase activity, assays were performed with or without NAD+. After the incubation, the products were precipitated at −20°C overnight by adding 50 µl of distilled water and 25 µl of 100% trichloroacetic acid (TCA) solution. For HPLC, the precipitates were reconstituted in 5% CH3CN and 0.1% trifluoroacetic acid (TFA) and run through a gradient concentration of 0.05% TFA to 0.043% TFA plus 80% CH3CN on Waters 1525 Binary HPLC pump system with Deltapak C4 column (300 Å, 3.9 × 300 mm, Waters, USA) and analyzed by Empower 2 software (version 6.10.01.00). The chromatograms were collected by dual–wavelength diode array with chromatograms analyzed at wavelength of 210/215 nm. Reactions were quantified by integrating area of peaks corresponding to deacetylated peptides (identities of peptides confirmed by mass spectrometry). MALDI-mass analysis of deacetylation reaction products were performed using Kratos Kampact MALDI III machine (Shimadzu, Duisburg, Germany).
Immunofluorescence combined with fluorescence in situ hybridization
IF combined with fluorescence in situ hybridization (IF–FISH) was performed as described previously (31,41) with a slight modification. Briefly, parasites were air dried on glass slides for 30 min at room temperature followed by fixation in 4% paraformaldehyde/PBS for 10 min. The fixed parasites were permeabilized with 0.1% Triton X-100 and then incubated with the primary (anti-PfAlba3) and secondary (Alexa flour 488 conjugated) antibodies, followed by another time fixation using 4% paraformaldehyde/PBS for 10 min to preserve antibody complexes. To visualize telomeric cluster, Rep20 probe was used. Hybridization was performed overnight with 0.5–1 ng/ml of probe in 50% formamide, 10% dextran sulfate (Quantum), 2× SSPE and 250 mg/ml herring sperm DNA.
ChIP assay
The chromatin immunoprecipitation assay was performed as described earlier (31) with minor modifications. In brief, parasites were isolated from infected erythrocytes by saponin lysis and incubated with formaldehyde in a final concentration of 1% for 10 min at 37°C. Cells were collected after three washes with PBS and parasites were resuspended in chilled immunoprecipitation buffer (25 mM Tris–HCl, pH 8.0, 300 mM NaCl, 1 mM EDTA, 0.25% NP-40 and protease inhibitor cocktail) and kept on ice for 30–60 min for lysis. Parasites were subsequently lysed by homogenizer and the nuclei were recovered by centrifugation. This is followed by sonication at 40% maximum amplitude (10 s on, 60 s off) for 12 pulses to obtain DNA fragments in the range of 200–1000 bp. The samples were centrifuged for 10 min at 15 000 g at 4°C. For input DNA samples, an aliquot of lysate used in the immunoprecipitation was processed along with the rest of the samples.
Prior to immunoprecipitation, chromatin was diluted 10-fold in ChIP dilution buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl (pH 8.0) and 167 mM NaCl] and pre-cleared using rabbit IgG bound to protein A Sepharose for 2 h at 4°C. Pre-cleared chromatin was used for immunoprecipitation reactions with a polyclonal antibody against PfAlba3 and PfSir2A protein and non-immune serum was used as control. The reactions were incubated on a rotor overnight at 4°C. Chromatin complexes were precipitated with 80 μl of salmon sperm DNA–protein agarose slurry for 2 h at 4°C, washed once in low salt immune complex wash buffer, once in high salt immune complex wash buffer, once in LiCl immune complex wash buffer and finally two times in 10 mM Tris–HCl, pH 8.0, 1 mM EDTA. Precipitated complexes were removed from the beads by incubating for 30 min in 250 μl of 1% SDS with 100 mM NaHCO3. The protein–DNA cross-links were reversed by heating samples at 65°C overnight in 250 μl TE with 400 μg Pronase (Roche). DNA was purified by phenol chloroform extraction. DNA from input and ChIP samples were resuspended in 100 and 20 µl TE, respectively. For PCR amplification, 0.5 µl of input DNA and 2 µl of the ChIP sample were used as template. Primers were used for the telomeric, TARE1, TARE2, TARE2-3, (11) TARE6 (53) and promoter region of LSA (Liver stage antigen), CSP (circumsporozoite protein) and GBP 130 (31) as previously published. The primers for 5′-UTR region of var genes (ups A, ups B, ups C and ups E) were used as described previously (31,54). The PCR products were analyzed on 1% agarose gel. All buffers used in assay contained protease inhibitor cocktail.
ChIP ReChIP assay
ChIP ReChIP experiment was performed to further confirm the association between PfSir2A and PfAlba3 as well as the status of PfAlba3. In brief, PfAlba3 or PfSir2A antibodies were used for the first round of immunoprecipitation and the order was reversed for the second round of immunoprecipitation. While, to check the acetylation status of promoter bound PfAlba3, anti-acetyl lysine antibody used for first round and anti-PfAlba3 antibody was used for second round.
In order to analyze the mutual association between PfAlba3 and PfSir2A, we divided soluble chromatin fractions (which were derived from cross-linked parasites) into two aliquots. We immunoprecipitated the first chromatin aliquot with antibody to PfAlba3, washed it, released the bound immune–DNA complexes using 25 mM dithiothreitol solution for 30 min at 37°C and resuspended them in one volume of immunoprecipitation dilution buffer for ReChIP assay with antibody to PfSir2A. In reciprocal way, we treated the second chromatin aliquot identically except that immune precipitated it with PfSir2A antibody before ReChIP with antibody to PfAlba3. To test antibody specificity, we used either a no antibody control or an unrelated antibody.
RESULTS
Identification and molecular modeling of putative DNA–RNA-binding protein
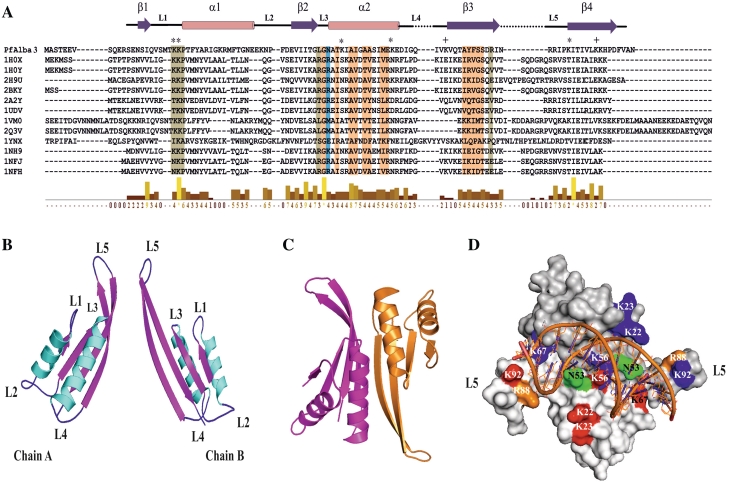
Plasmodium falciparum genome analysis reveals that it encodes a putative DNA–RNA-binding protein, shows sequence homology and structural similarity with Alba family proteins (Figure 1A and Supplementary Figure S1). Sequence alignment of PfAlba3 with other known Alba sequences (with known 3D structure) (19,27,55–57) revealed the presence of conserved residues involved in DNA binding and dimer interface (Figure 1A). Multiple sequence alignments indicated the wide presence of Alba including Plasmodium, Leishmania, Trypanosoma, Archaea and Arabidopsis (Supplementary Figure S1). Like Archeal Alba, PfAlba3 also possesses conserved sequence motif K(X)KP in the loop between helix1 and strand 1, which comprises critical lysine involved in DNA binding (Figure 1A and B) (20). In order to visualize structural details of PfAlba3, we build the 3D model of PfAlba3 using an Alba protein from Archaeoglobus fulgidus (PDB code: 1NFH) as reference structure (19). PfAlba3 monomer model structure shows reasonably good structural similarities with the known 3D structures of Alba family (Supplementary Table S1). Alba family protein is mainly dimeric in nature. Therefore, we modeled dimeric structure of PfAlba3 to understand the structural complexity and DNA-binding mode. Monomer models of PfAlba3 were further used in protein–protein docking to generate the probable dimerization model. Even though predicting the dimerization mode through protein–protein docking is a difficult task, we could generate a decent dimer model of PfAlba3 (Figure 1B and C). This model shows strong structural similarity (overall root mean square deviation of 0.9 A°) with the 1NFH dimer generated using X-ray crystallography (Supplementary Table S1). Modeled structure of PfAlba3 belongs to mix αβ structure with four β strands (β1–β4) and two α helices (α1−α2) (Figure 1B) as observed in other Alba family proteins (19,20,27,55). Like other Alba proteins, PfAlba3 also has extended β hairpin arm between strands 3 and 4 (Figure 1B and C), which is suggested to interact with minor groove of DNA (14). Dimer structure of PfAlba3 showed an asymmetric unit and allied by 2-fold symmetry. The two β-hairpin of both monomers are separated from each other by ∼54 Å (Figure 1C) and can accommodate ∼15 bp of B-form DNA. Helix α2 and strands β3–β4 of both monomers is engaged in monomer association and concomitant dimer formation (Figure 1C). Docking studies provide an apparent image of PfAlba3–DNA interaction and probable mode of DNA binding. It suggests that PfAlba3 dimer binds with DNA in side by side fashion and covers an area greater than one complete turn of DNA. Loop L1 of both monomers is located in the central part of the dimer, well placed to accommodate DNA duplex. Loops L1, L3 and L5 play an important role in DNA binding, whereas surface exposed lysines present in KKP motif in the loop L1 (K22 and K23) are found in very close proximity with the DNA double helix (Figure 1D). Lysine (K) 56 and lysine (K) 67 of helix α2 are also located beneath the docked DNA, hence may be involved in DNA binding. Apart from this, proline (P24) from both chains and asparagine (N) 53, arginine (R) 88 and lysine (K) 92 are also found to be engaged in PfAlba3–DNA interaction (Figure 1D).
Figure 1.
Molecular modeling and docking studies of PfAlba3. (A) PfAlba3 sequence is aligned with other known Alba fold containing proteins whose 3D structures are known. DNA-binding residues (colored in pale green) and dimer interface residues (colored in light orange) are mapped onto the alignment. Residues that involved in both DNA binding and dimer interface are marked in blue. Secondary structure of PfAlba3 derived from the model displayed onto the alignment. Surface exposed lysines involved in DNA binding are marked by ‘*’, whereas those not involved in DNA binding represented by ‘+’. The level of sequence homology is represented by using the bar chart. Key to the identities of the Alba sequences aligned with PfAlba3 is represented as color-coded matrix (See the Supplementary Figure S1B). (B–D) Dimerization and DNA-binding studies of PfAlba3. (B) Cartoon representation of 3D structures of chains A and B of PfAlba3 as derived from template structure 1NFH. (C) Cartoon representation of the dimerization model of PfAlba3 chains A and B. (D) Plausible mode of DNA-PfAlba3 (dimer) interaction. Surface exposed lysines (involved in DNA binding) from chains A and B are marked in red and blue, respectively, while other residues involve in DNA binding (from chains A and B) are marked (N53, K56, K67, R88 and K92) as indicated in figure.
Purification and characterization of PfAlba3
PfAlba3 was overexpressed as soluble His-tag fusion protein of expected size ∼13.0 kDa and readily purified to homogeneity by the combination of Ni-NTA affinity and HPLC gel filtration chromatography (Figure 2A). Specificity and homogeneity of PfAlba3 were further confirmed by western blotting using anti-His and anti-PfAlba3 antibodies (Supplementary Figure S2A). Secondary structure analysis of PfAlba3 by circular dichroism (CD) spectropolarimetry suggests that the α and β content of the recombinant PfAlba3 are ∼26% and ∼32%, respectively, and remaining part is assumed to be randomly coiled and β turn structures. These results are in close agreement with the predicted secondary structure as calculated from HHpred and 3D model (PfAlba3) predictions for helix contents (Supplementary Table S2).
Figure 2.
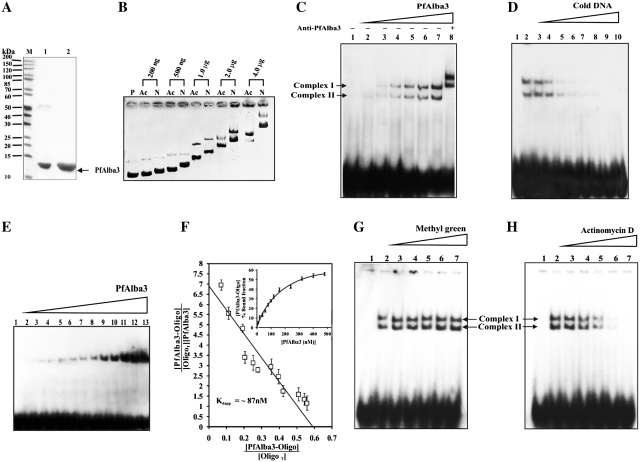
Characterization of DNA-binding properties of PfAlba3. (A) Recombinant PfAlba3 was purified by Ni-NTA affinity chromatography and subsequently by HPLC. Lane M; Protein molecular weight marker, lane 1; Ni-NTA purified fraction and lane 2; HPLC purified protein. (B) DNA-binding property of PfAlba3 was characterized by agarose gel retardation assay. Samples were run pair wise (acetylated and non-acetylated protein at the same concentration) to evaluate the effect of lysine modification (acetylation) on DNA-binding affinity of PfAlba3. Lane P; ϕX174 dsDNA alone, while subsequent lanes contain ϕX174 dsDNA plus increasing concentrations of native or mild acetylated PfAlba3, respectively. Ac; mild acetylated PfAlba3 and N; native PfAlba3. (C) EMSA and supershift assay. Lane 1; probe alone; lanes 2–7; probe incubated with increasing concentrations of PfAlba3 (20, 40, 80 120, 240 and 300 nM) and lane 8, probe plus PfAlba3 (300 nM) and anti-PfAlba3 antibody. (D) Competition assay. Lane 1; probe alone, lane 2; probe plus PfAlba3 (300 nM), lanes 3–10; probe plus PfAlba3 (300 nM) along with an excess cold competitor (1, 5, 10, 20, 40, 60, 80 and 100 fold) DNA. (E) Determination of dissociation constant using 20-bp oligonucleotide. Lane 1; probe, lanes 2–13; probe along with 10, 20, 40, 60, 80, 120, 180, 240, 320, 400 and 480 nM of PfAlba3. (F) PfAlba3 concentrations were plotted against corresponding percent-bound fractions (inset). Dissociation constant (Kdapp) was calculated from the plot using Scatchard equation. Data for each signal (free and bound DNA) was performed in triplicate and reported error bar represent the standard deviation. (G and H) PfAlba3 interacts with minor groove of dsDNA. Complementary oligonucleotide was annealed, radiolabeled with γ P32 ATP and subsequently incubated with increasing concentration of (G) methyl green (major groove-binding dye) lanes 3–7; 0.01, 0.1, 5, 50 and 500 µM and (H) Actinomycin D (minor groove-binding dye) Lanes 3–7; 0.01, 0.1, 5, 50 and 500 µM. The reactions were further incubated with 500 nM PfAlba3 and analyzed on 7% native PAGE. Lanes 1 and 2 (both in Figure G and H) represented only labeled DNA (control) and labeled DNA incubated with PfAlba3 (500 nM), respectively.
DNA-binding property of PfAlba3: modification of lysines interferes with DNA binding
The DNA-binding property of PfAlba3 was investigated by following agarose gel retardation assay. The mobility of super coiled DNA (using ϕX174 DNA) incubated with PfAlba3 was shifted sharply at PfAlba3–DNA mass ratio of 0.8. The shifting was gradually increased with the increase of PfAlba3–DNA mass ratio, showing maximum shift at PfAlba3–DNA mass ratio of 16 (Figure 2B). PfAlba3 has sequence homology with Alba domain and docking studies suggest the role of surface exposed lysine residues in protein–DNA interaction (Figure 1). Therefore, to verify the role of lysine in DNA binding, we modified lysines of PfAlba3 by acetylation and also by trinitrobenzene sulfonate (TNBS, a lysine modifier). Acetylation was confirmed by western blotting using anti-acetyl lysine antibody, which selectively recognized acetylated PfAlba3 but not native PfAlba3 (non-acetylated) (Supplementary Figure S2B). We have acetylated PfAlba3 in a controlled fashion to prepare mild and high acetylated forms to follow DNA binding. Acetylation of PfAlba3 lowers its DNA-binding affinity in comparison to the native (non-acetylated) PfAlba3 as indicated by the reduced shifting at mild acetylation at varying concentrations of the protein (Figure 2B). However, the highly acetylated PfAlba3 lost its DNA-binding activity completely (Supplementary Figure S2C and D). A similar result was observed with TNBS-modified PfAlba3, where no shifting was detected (Supplementary Figure S2C). In summary, our data of protein modification reveal that non-acetylated PfAlba3 binds well with DNA; mild acetylation lowers its binding whereas no binding was observed at high acetylation. Modification of PfAlba3 did not significantly alter the structural integrity of PfAlba3 as evident from CD and fluoremetric analysis (Supplementary Figure S3 and Supplementary Table S2).
PfAlba3 binds DNA sequence non-specifically
DNA-binding nature of PfAlba3 was also followed by EMSA using 5′-end-labeled ds oligos. The smallest binding-site length of PfAlba3 was determined by using ds oligonucleotide probes of 20 and 30 bp. When the DNA (using 30-bp ds oligos) incubated with PfAlba3 and resolved in native gel showed two distinct bands of proteins: DNA complexes (complex I and II) (Figure 2C). These bands became intense with the increasing concentration of PfAlba3 (Figure 2C, lanes 2–7). When anti-PfAlba3 antibody was added in the reaction, it super shifted both proteins: DNA complexes (Figure 2C, lane 8). Competition experiments were also carried out in order to reconfirm the binding affinity of PfAlba3 using 5′-end labeled and non-radiolabeled (cold DNA) ds oligos (30 bp) (Figure 2D). At 20-fold excess of cold competitor oligos the complex I (upper shifted band) is almost vanished but not complex II (Figure 2D, lane 6). However, 60-fold excess or onwards of cold competitor probe completely prevented the formation of both the protein: DNA complexes (Figure 2D, lanes 8–10). On the other hand, when 20 bp ds oligos were used only one shift equivalent to protein: DNA complex was observed (Figure 2E). These analyses suggest the possible binding site length is ∼15 bp of substrate per dimer of PfAlba3. The affinity of the PfAlba3 for DNA was determined by calculation of Kdapp (apparent dissociation constant) of the single complex obtained with the 20-bp probe applying linear regression by using Scatchard equation. The nucleotide binding of PfAlba3 revealed Kdapp value in nano molar range (∼87 nM) (Figure 2F).
We further check whether PfAlba3 has any major or minor groove specificity for binding. Groove specific dyes are used that specifically interact with either the major or minor groove of DNA and compete with the protein binding. Pre-incubation of DNA with increasing concentrations of methyl green (major groove binder) did not affect the binding activity of PfAlba3 (Figure 2G, lanes 3–7), even at a very higher concentration (500 µM) (Figure 2G, lane 7). In contrast, actinomycin D (minor groove binder) initiated partial inhibition of DNA binding at 5 µM (Figure 2H, lane 5) with considerable inhibition at 50 µM (Figure 2H, lane 6) and complete inhibition at 500 µM (Figure 2H, lane 7), indicating that PfAlba3 binds to minor groove of DNA. However, the concentration of actinomycin D required for inhibition was very high.
PfAlba3 prevents transcription after DNA binding
Existing studies of Alba proteins suggest that this family proteins are not involved in DNA folding and chromatin packaging (14,26). Since, acetylation has an impact on DNA-binding activity; it is assumed that Alba may have some role in the regulation of gene expression and transcriptional modulation. In order to verify this hypothesis, we performed in vitro transcription assay in the presence of native (non-acetylated) and mild acetylated PfAlba3. Data indicate that PfAlba3 prevents transcription concentration dependently (Supplementary Figure S4A). PfAlba3 at a lower concentration (100 ng) partially prevented transcription of tRNAlysine, resulting in additional bands corresponding to aborted short transcripts. However, at higher concentrations (0.5–4 µg), PfAlba3 showed gradual inhibition of transcription resulting in the formation of numerous shorter transcripts (Supplementary Figure S4A). In contrast, complete inhibition of transcription was observed at 6–10 µg of PfAlba3, generating only very small short transcripts (∼6–10 nt) instead of full-length transcripts. Transcription under control conditions (without PfAlba3 or heat inactivated PfAlba3), and in the presence of only buffer produced full-length transcript (tRNAlysine) without any aborted product. This suggests that transcription abortion was not due to buffer or any hindrance to polymerase. Interestingly, mild acetylated PfAlba3, even at a very high concentration (10 µg) was unable to inhibit transcription (Supplementary Figure S4B). These findings indicate that PfAlba3 by binding dsDNA blocks transcription and acetylated PfAlba3 is unable to prevent transcription due to reduced DNA-binding affinity.
PfAlba3 expression and localization
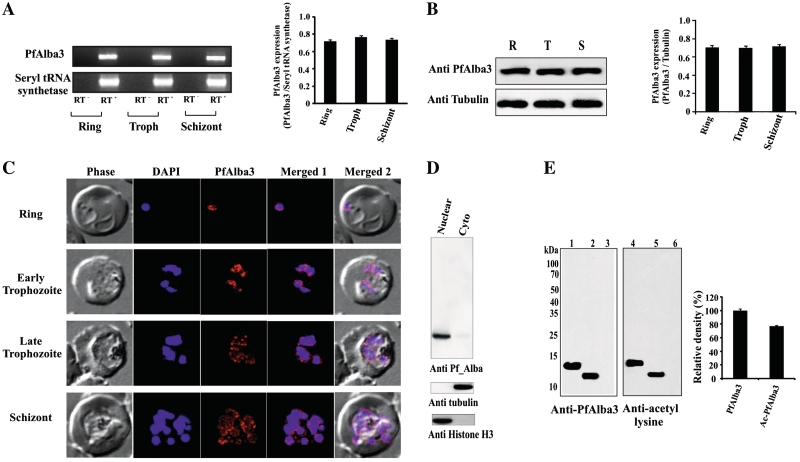
Stage-specific expression of PfAlba3 was checked by RT–PCR and western blotting using anti-PfAlba3 antibody. Amplification of a fragment of 324 bp, equal to the size of PfAlba3, was found in RT–PCR using RNA from ring, trophozoite and schizont stages (Figure 3A). Seryl tRNA synthetase was used as control, which is expressed uniformly throughout all the stages. The quantitative analysis of the expression (RT–PCR data) indicated almost equal level of expression of PfAlba3 throughout all the asexual stages (Figure 3A, side panel). This finding was further supported by western blot analysis of parasite lysate prepared from different stages. Western blot analysis using anti-PfAlba3 antibody showed the presence of a single band of ∼12 kDa, matched with the calculated molecular weight of parasite expressed PfAlba3 (Figure 3B), uniformly at all the blood stages of malaria parasite (Figure 3B, side panel). We further investigated the subcellular localization of PfAlba3 during the asexual developmental cycle by confocal microscopy (Figure 3C) and western blotting using the cytoplasmic and nuclear fraction isolated from mixed stages culture (Figure 3D). IF analysis showed that PfAlba3 (alexa fluor 633, red) signal merged well with the DAPI signal indicating its nuclear localization and reconfirmed its constitutive expression (Figure 3C). PfAlba3 appeared as making distinct subnuclear spot and concentrated mainly toward the periphery of the nucleus during ring stages, whereas during trophozoite and schizont stages PfAlba3 seems distributed throughout the nucleus with more visible punctate staining pattern (Figure 3C). Immunoblot analysis of parasite nuclear extracts with anti-PfAlba3 antibody detected a band (∼12 kDa), suggesting the major nuclear compartmentalization (Figure 3D). However, a very weak signal was also detected in the cytoplasmic fraction in western blot (Figure 3D).The purity of isolated subcellular fractions (cytoplasmic and nuclear) was confirmed by western blot analysis using anti-histone H3 (nuclear) and anti-tubulin (cytoplasmic) antibody (Figure 3D). Presence of PfAlba3 in parasite was further ensured by immunoprecipitation from the whole parasite lysate using anti-PfAlba3 antibody. Antibodies raised against recombinant PfAlba3 specifically pulled out a ∼12 kDa protein that was specifically recognized by rabbit anti-PfAlba3 antibody (data not shown). To check the existence of acetylated form of PfAlba3 in parasite, the whole PfAlba3 was first immunoprecipitated by anti-PfAlba3 antibody and then divided into two equal parts for western blotting using anti-PfAlba3/anti-acetyl lysine antibodies (Figure 3E). Results document the presence of a detectable band in both the fractions (Figure 3E, lanes 2 and 4) proving that PfAlba3 also exists in an acetylated form in P. falciparum. However, no signal was detected when rabbit pre-immune serum was used for immunoprecipitation (Figure 3E, lanes 3 and 6). Recombinant PfAlba3 and acetylated PfAlba3 were used as positive control (Figure 3E, lanes 1 and 4). Quantitative analysis of the immunoprecipitation experiments indicates that PfAlba3 exists predominantely in an acetylated form (∼80% of the total PfAlba3 population) in P. falciparum (Figure 3E, side panel).
Figure 3.
Stage-specific expression and subcellular localization of PfAlba3. (A and B) Stage-specific expression of PfAlba3 transcript was confirmed from total RNA isolated from sorbitol synchronized P. falciparum culture at ring (R), trophozoit (T) and schizont (S) stages. RT− control represents reaction without reverse transcriptase. Side panel shows densitometric analysis of PfAlba3 expression at different stages of P. falciparum. Band intensities were quantified by spot densitometry using AlphaImager 3400 imaging software (Alpha Innotech Corporation) and the intensity of PfAlba3 transcript was normalized to that of seryl tRNA synthetase. (B) Western blot analysis of PfAlba3 expression in P. falciparum intra-erythrocytic stages. The sorbitol synchronized parasite lysate was resolved in 15% SDS–PAGE and subjected to western blotting using anti-PfAlba3 antibody. To confirm the equal amount of protein in parasite lysate, blot was reprobed with anti-tubulin antibody. Side panel shows densitometric analysis and intensity of PfAlba3 was normalized against tubulin. (C) Subcellular localization of PfAlba3 was confirmed by IF analysis of PfAlba3 (Red, Alexa flour 633) throughout the parasite asexual blood stages (ring, early and late trophozoite, schizont stages). (D) Nuclear localization of PfAlba3 was further confirmed by western blotting using cytoplasmic and nuclear fraction from asynchronous parasite culture. To cross check the possibilities of contamination of cytoplasm and nuclear fraction with each other, we also performed the experiment using cytoplasmic (anti-tubulin antibody) and nuclear (anti-Histone H3) marker proteins. (E) PfAlba3 also exists in the acetylated form. To confirm acetylation status of PfAlba3 in parasite, immunoprecipitated batch of protein was equally divided in two fractions and subjected to western blotting using rabbit anti-PfAlba3 antibody for first and mouse anti-acetyl lysine antibody for second fraction. Rabbit pre-immune serum was used as a control in all the experiments. Left panel showing lane 1; purified recombinant PfAlba3 (control), lane 2; immunoprecipitation with rabbit anti-PfAlba3 antibody, lane 3; immunoprecipitation with rabbit pre-immune serum. Right panel represents, Lane 4; acetylated PfAlba3 (control), lane 5; immunoprecipitation with rabbit anti-PfAlba3 antibody; lane 6; immunoprecipitation with rabbit pre-immune serum. Side panel shows the densitometric analysis of acetylated and non-acetylated PfAlba3.
PfSir2A interacts with PfAlba3 and deacetylates N-terminal peptide specific for DNA binding
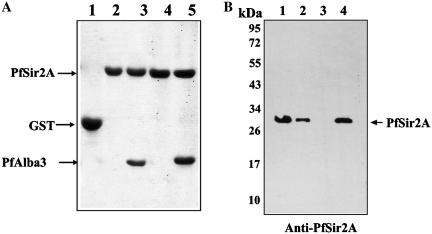
To explore the function of PfAlba3, identification of critical proteins involved for acetylation–deacetylation in P. falciparum is essential. Previous studies reported that Alba binding is regulated by Sir2 through deacetylation, specifically at N-terminal lysine residue (14,17). Interestingly, P. falciparum genome also encodes a homolog of Sir2, a NAD+-dependent deacetylase (13).Therefore, we have cloned and overexpressed GST and His fusion tagged PfSir2A to verify the interaction of PfSir2A with PfAlba3 (as a non-histone substrate). GST pull-down assays were conducted by using purified GST-PfSir2A and His-tagged PfAlba3 (Figure 4A). The data indicate that PfSir2A interacts well with PfAlba3 as evident from the presence of PfAlba3 in pulled out material (Figure 4A, lanes 3 and 5). Whereas, no interaction was found when PfMIF (P. falciparum macrophage migration inhibitory factor, His-tag fusion protein, used as control) was used (Figure 4A, lanes 2 and 4). However, GST alone did not interact with PfAlba3 suggesting its specificity toward PfSir2A not to GST (Figure 4A, lane 1). These results were further supported by far western analyses (Supplementary Figure S5B).
Figure 4.
PfAlba3 interacts with PfSir2A. (A) GST pull-down assay. Lane 1; GST alone, Lanes 2 and 4; 25 and 50 µl of pulled out material of PfSir2A (GST tag) incubated with PfMIF (His tagged), Lanes 3 and 5; 25 and 50 µl pulled out material of PfSir2A (GST tagged) incubated with PfAlba3 (His tag). (B) His tag pull-down assay and immunoprecipitaion. PfSir2A was pulled down from the whole parasite lysate (mixed stage) using His tag PfAlba3 as well as immunoprecipitated using anti-PfSir2A antibody. The samples were then subjected to SDS–PAGE analysis, followed by western blotting using anti-PfSir2A antibody. Lane 1; Immunoprecipitated PfSir2A using anti-PfSir2A antibody, lane 2; whole parasite lysate, lane 3; pull down material using His tag PfMIF, lane 4; pull down material using His tag PfAlba3. Purified His tag PfMIF used as a control in all the experiments.
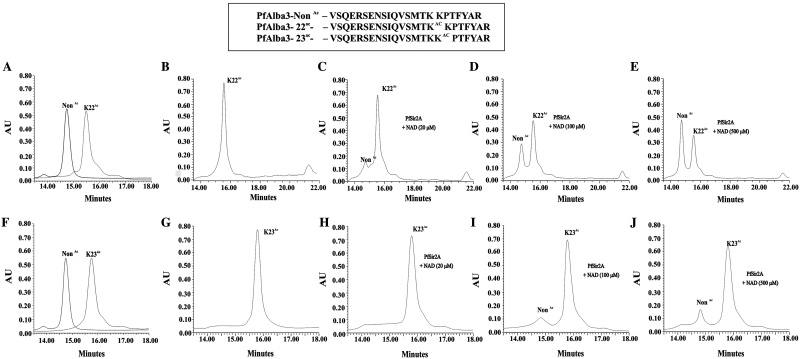
To further investigate the interaction between PfAlba3 and PfSir2A, we raised polyclonal antibodies against the recombinant PfSir2A and performed His-tag pull-down assay using His-tagged PfAlba3 as bait and incubated it with parasite lysate to pull out an interacting partner using Ni-NTA. Pulled down material was analyzed by western blot using anti-PfSir2A antibody, which indicated the presence of a band ∼30 kDa (Figure 4B, lane 4), specific to parasite encoded PfSir2A.To detect the specificity of antibody, we immunoprecipitated PfSir2A from the parasite lysate as well (Figure 4B, lane 1). Immunoblot analysis of parasite extracts with anti-PfSir2A antibody also detected a 30-kDa protein at the same position (Figure 4B, lane 2). Whereas, no band was detected in His fusion PfMIF pulled out material (Figure 4B, lane 3). Docking studies suggested the role of critical N-terminal lysines (in KKP motif) in DNA binding, which may be regulated by acetylation and deacetylation similar to Archea (14,17). To test the significance of N-terminal lysine in DNA binding by PfAlba3, we used 23 residue synthetic peptide [residues 7–29] corresponding to N-terminal of PfAlba3, acetylated at lysine (K) 22/lysine (K) 23 and followed deacetylation using PfSir2A (Figure 5). In the absence of NAD+ and PfSir2A, these peptides showed one prominent peak corresponding to the non-acetylated or acetylated peptides (K22 or K23), respectively (Figure 5A and F). When acetylated peptides (K22/ K23) were incubated with only PfSir2A, no additional peak was observed (Figure 5B and G). However, when acetylated (K22/K23) peptides were incubated with PfSir2A in the presence of NAD+ additional peak was observed, which became more prominent with increasing concentrations of NAD+ (Figure 5C–E and H–J). This additional peak was collected and mass was analyzed, which corresponds to deacetylated species of the peptide (non-acetylated ∼2687 Da) (data not shown). However, deacetylation was selectively favored at K22, but not at K23 as evident from comparative HPLC chromatograms of deacetylation reaction with K22Ac and K23Ac peptides (Figure 5E and J). About 60% of the K22Ac peptide (Figure 5E) was deacetylated, whereas <20% of K23Ac peptide was deacetylated under the same condition (Figure 5J). Fractions of deacetylation reaction using K22Ac/K23Ac peptides were collected and processed for mass analysis to further confirm the deacetylation (Supplementary Figure S6A–C). Mass data show the presence of mixed population containing two main peaks corresponding to acetylated and deacetylated forms (Supplementary Figure S6B–C).
Figure 5.
In vitro deacetylation of the N-terminal peptide (specific to PfAlba3) by recombinant PfSir2A. HPLC chromatograms showing absorbance at 210 nm using (A) non-acetylated and K22Ac peptide only, (B) K22Ac peptide incubated with PfSir2A, without NAD+, (C) K22Ac peptide with PfSir2A, NAD+ (20 µM), (D) K22Ac peptide with PfSir2A, NAD+ (100 µM), (E) K22Ac peptide with PfSir2A, NAD+ (500 µM), (F) non-acetylated and K23Ac peptide only, (G) K23Ac peptide incubated with PfSir2A, without NAD+, (H) K23Ac peptide with PfSir2A, NAD+ (20 µM), (I) K23Ac peptide with PfSir2A, NAD+ (100 µM), (J) K23Ac peptide with PfSir2A, NAD+ (500 µM), respectively. Box representing the peptide sequence used in deacetylation reaction.
Co-localization and ChIP studies
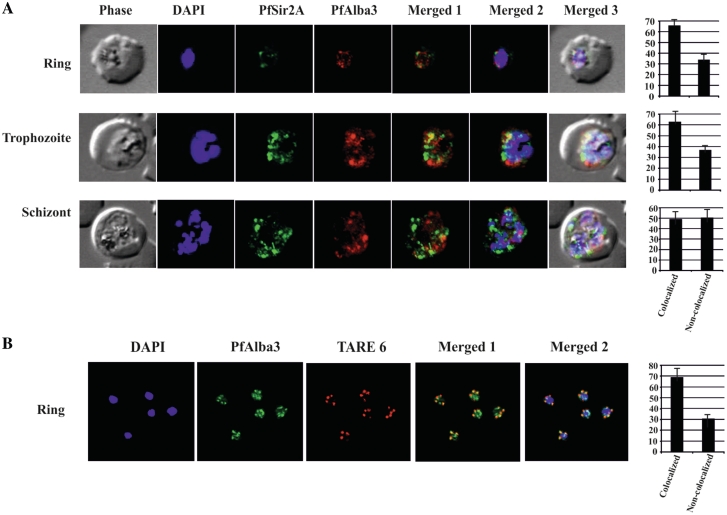
Since the recombinant PfSir2A can deacetylate synthetic peptide specific to N-terminus of PfAlba3 as substrates, we checked whether PfSir2A and PfAlba3 are positioned in the same genomic location or not. To test this, we performed dual IF studies using rabbit anti-PfAlba3 and mouse anti-PfSir2A antibodies. The data indicated the expression of both these proteins at different intra-erythrocytic stages; however, low level of expression of PfSir2A at ring stage was detected in comparison to trophozoite and schizont stages (Figure 6A). A similar result was also observed in western immunoblot with parasite lysate using anti-PfSir2A antibodies (Supplementary Figure S5A). IF studies show that PfAlba3 (red) and PfSir2A (green) fluorescence signals merge at several places (Figure 6A) foremost at the peripheral region of the nucleus. However, co-localization was partial and not significant during schizont stage as evident from the overlap and non-overlap scores (Figure 6A, side panel). PfSir2A and PfAlba3 signals occupied mainly toward the periphery of the nucleus and making discrete foci. PfSir2A is not considered as a specific marker for telomeric clusters and also reported to stain the nucleolus as well (31). Hence, we carried out IF–FISH to analyze the association of PfAlba3 at the telomeric region (Figure 6B). Visualization of the telomeric clusters during the blood-stage cycle of malaria parasite was performed by IF–FISH using TARE6 sequence as a probe. The co-localization of PfAlba3 with telomeric cluster at the nuclear periphery was also observed suggesting the telomeric association of PfAlba3 (Figure 6B).
Figure 6.
Co-localization studies. (A) Plasmodium falciparum infected red blood cells were treated with DAPI (blue), mouse anti-PfSir2A antibody (secondary antibody Alexa Fluor 488, green) and rabbit anti-PfAlba3 antibody (secondary antibody Alexa Fluor 633, red), respectively. Overlay panels show the merged images in different combinations. Side panel shows the score values of co-localized and non-co-localized fluorescence signals at ring, trophozoite and schizont stages, (B) FISH analysis combined with IF labeling of PfAlba3 to visualize telomeric region of chromosomes. Immunolocalization (IF) of PfAlba3 was performed at ring stage parasites using an Alexa 488-conjugated secondary antibody (green). FISH analysis was performed simultaneously. The nuclei were stained with DAPI (blue) and hybridized with TARE6 (Red) probe to visualize the chromosome ends on ring stage. The extent of co-localization in the selected image was measured (side panel) using the Pearson correlation coefficient and expressed as a percentage using Image Pro Plus software. For co-localization studies (IFA and IFA–FISH), approximately 100 nuclei were counted and represented as mean ± SD. The images presented are a representation of one of the experiments.
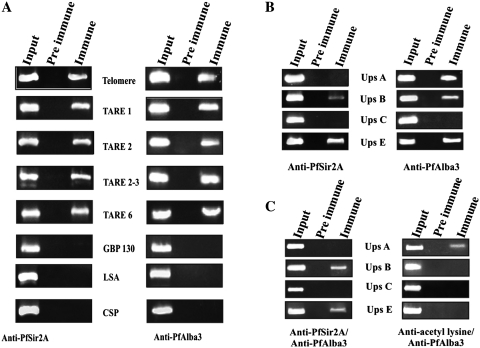
Previous studies showed a key role of P. falciparum Sir2 in heterochromatin-mediated silencing and comprehensive repression and switching expression of var genes and telomeric maintenance (31,41). To get a preliminary idea of the possible role of PfAlba3 in PfSir2A-mediated regulation of gene expression, we moved for ChIP and ChIP ReChIP assays using highly synchronized ring stage parasite (Figure 7). ChIP analyses show that PfSir2A binds to telomeric region, including telomere, telomeric repeats and rep 20 (TARE 6) but not at promoter regions of GBP130 (blood stage expressed), LSA and CSP (not expressed during blood stage) (Figure 7A, left panel), as also reported earlier (11,31,39). Input DNA (nuclear DNA prepared immediately after sonication) was used as a control. Results of ChIP assay using anti-PfAlba3 also followed a similar pattern (Figure 7A, right panel), which again suggested the telomeric and subtelomeric location of PfAlba3. The regulation of var genes expression is controlled by distinct promoters (symbolize as ups A–C and E) located in the subtelomeric region (A, B and E) and center of the chromosomes (ups C) (34,58). Role of PfSir2A is well established in the regulation of var gene expression; therefore, we performed ChIP assays against var gene promoter using anti-PfSir2A and anti-PfAlba3 antibodies (Figure 7B) to check whether PfAlba3 has any association with these promoters. We found that anti-PfSir2A antibodies immunoprecipitated ups B and ups E var gene promoter, but not ups A and ups C (Figure 7B, left panel). However, ChIP analysis using anti-PfAlba3 showed a slightly different picture and immunoprecipitated ups A, B and E, but how this can be attributed is not clear (Figure 7B, right panel). To further make the picture clear, we performed ChIP ReChIP assay using a combination of anti-PfSir2A/anti-PfAlba3 as well as anti-acetyl lysine/anti-PfAlba3 antibodies. The ChIP ReChIP studies suggest that ups E and ups B promoters are enriched with both PfAlba3 and PfSir2A but not ups C and A promoters as evident from successive immunoprecipitation (Figure 7C, left panel) using anti-PfAlba3/anti-PfSir2A antibodies. Interestingly, ChIP ReChIP using anti-acetyl lysine/anti-PfAlba3 antibodies immunoprecipitated ups A only suggesting that PfAlba3 is in an acetylated form at ups A promoter but not at ups B and E promoters (Figure 7C, right panel), hence not picked up in ChIP Re ChIP.
Figure 7.
ChIP and ChIP ReChIP analysis (A) ChIP analysis performed from ring stage parasites using antibodies against PfSir2A (left panel) and PfAlba3 (right panel). Immunoprecipitated DNA was analyzed by PCR amplification specific to telomeric and subtelomeric sequences (TARE1, TARE2, TARE2-3 and TARE6), GBP130, LSA and CSP 5′-UTR region. Input DNA material was used as a positive control, whereas material immunoprecipitated by the pre-immune sera or IgG used as control. PfAlba3 and PfSir2A are assembled at the telomeric and subtelomeric regions of chromatin, as shown by ChIP analysis. (B) PfAlba3 is also associated to the 5′-UTR of var genes. ChIP analysis of ring stage parasites was performed using antibodies against PfSir2A (left panel) and PfAlba3 (right panel). (C) PfAlba3 and PfSir2A are assembled at same promoter region and the association between PfSir2A and PfAlba3 causes its deacetylation, as shown by ChIP ReChIP analysis. Soluble chromatin was prepared from isolated parasites and divided into two chromatin aliquots, which were immunoprecipitated with antibodies to either anti-PfSir2A/PfAlba3 antibody or anti-acetyl lysine/PfAlba3 antibody, respectively. Controls for the ChIP ReChIP fractions include the input chromatin and pre-immune serum. Input corresponds to DNA prepared from fragmented chromatin prior to immunoprecipitation. Each experiment was performed in triplicate and the data presented were a representation of one out of three experiments showing similar result.
DISCUSSION
In the present study, we have identified and characterized an Alba family protein (PfAlba3) from P. falciparum. We have presented evidence that PfAlba3 is expressed in all the erythrocytic developmental stages of P. falciparum. PfSir2A interacts with PfAlba3 and deacetylates N-terminal peptide of PfAlba3 specific for DNA binding. Our results also documented the association between PfAlba3 and PfSir2A at telomeric, subtelomeric regions and var gene promoter.
Alba protein originates from a group of ancient nucleic acid-binding protein and predominantly present in Archaea suggested the evolutionary divergence of this family of proteins among other groups, including Plasmodium. PfAlba3 retains all the structural properties shared by other proteins belonging to Alba family. Structural studies with Alba protein show that each dimer of Alba has two extended β-hairpins flanking a central body which is suggested to interact with DNA (27). This extended anti parallel β-hairpin structure plays a central role in DNA interaction by several DNA-binding proteins (59–61). Molecular modeling and docking studies of PfAlba3 also revealed a similar kind of structure and mode of interaction with DNA. PfAlba3 binds well with DNA without any sequence specificity and acetylation lowers the binding affinity of PfAlba3, a characteristic feature of proteins belonging to Alba family. PfAlba3 contains large numbers of basic amino acid (10 lysines and 6 arginines), out of which few are surface exposed and accessible to modification. Previous studies explored the presence of N-terminal lysines in KKP motif (in loop L1, well conserved among Alba family proteins) and their critical role in DNA binding (14,17, 20). Docking of dsDNA with PfAlba3 suggested a similar mode of DNA binding and critical role of N-terminal lysines (K22 and K23) as well as other residues (K56, K67, K92, R88, N53, etc.) in stabilizing the interaction. However, due to lack of proper strategy of in vivo modification of protein at specific lysines, we did in vitro modification of PfAlba3 to analyze the effect of modification on DNA binding. The reduction or loss of binding after chemical modification of lysine validated the essential role of lysine in DNA binding. Reaction with acetic anhydride is expected to acetylate all surface lysine (total 7) side chains and not just the specific residues whose enzymatically catalyzed acetylation–deacetylation is expected to control DNA-binding affinity in vivo. It is true that in vivo acetylation is a highly specific process and acetylation under in vitro condition produced heterogeneous population. Therefore, mutagenesis studies along with identification of corresponding acetyltransferase are essential for proper identification of specific lysine whose acetylation controls PfAlba3 interaction with DNA.
PfAlba3 is uniformly expressed at all the intra-erythrocytic stages of P. falciparum. This suggests that PfAlba3 may play an essential role in all developmental stages of the intra-erythrocytic life cycle of P. falciparum. PfAlba3 also exists in the acetylated form in P. falciparum indicating that the interaction and affinity of PfAlba3 with DNA are possibly regulated by its acetylation/deacetylation mode like other Alba family proteins (14,17). Since we have quantified acetylated PfAlba3 using parasite from mixed stages, future studies are necessary to see whether the acetylation status throughout all the individual asexual stages remains constant or not. PfAlba3 significantly inhibited transcription under in vitro condition. However, acetylated PfAlba3 did not inhibit transcription even at a very high concentration. This finding suggests that PfAlba3 binds DNA with low affinity under acetylated status as compared with non-acetylated PfAlba3 and therefore, unable to inhibit transcription. Sequence non-specific DNA binding and its control by acetylation are common ways for the regulation of gene expression (62). These findings suggest that PfAlba3 might have some role in gene expression. Recently, Mair and coworkers (63) have identified Alba family proteins in maternal mRNP complex and suggested the involvement of these proteins in zygote development as well as post-transcriptional regulation of gene expression in Plasmodium. PfSir2A (NAD+ -dependent deacetylase) interacts well with PfAlba3 as evident from protein–protein interaction studies and deacetylates synthetic N-terminal peptides (acetylated specifically at lysine K22Ac/K23Ac) preferably at lysine 22. These studies along with earlier reports suggest the importance of N-terminal lysine residues in K(X) KP motif in DNA binding and acetylation /deacetylation regulation. However, in our studies, we have analyzed the role of N-terminal lysines, but we cannot exclude the possibilities of involvement of other lysines in DNA binding.
Data accumulated in recent years suggested an active role of PfSir2A in controlling var genes expression (64). PfSir2A is associated with the telomeric rep20 sequences and therefore, spreads gene repression to ∼50-kb internal regions of chromosome comprising var genes (11,31,41). PfSir2A has been reported to be involved in regulating subtelomeric var genes (ups A and ups E) whereas centrally located var genes (ups C) are not under direct control of PfSir2A as also observed from PfSir2A knockout studies (31,32,41). Confocal microscopic and FISH studies documented that both PfAlba3 and PfSir2A are localized in the nucleus and occupies mainly at the telomeric and subtelomeric region. These data suggest a possible in vivo association and functional regulation of the PfAlba3 by PfSir2A through its deacetylation. Since, PfAlba3 lacks any organelle specific signal sequence but its existence in acetylated/deacetylated forms advocates the possibilities of its subcellular localization regulated by acetylation and deacetylation. PfAlba3 and PfSir2A co-localization is not significant during schizontic stages and during other stages it may be inevitably overlapped due to peripheral localization of the nucleus. Recent studies by Mani et al. (25) suggested that Alba proteins from Trypanosoma brucei are cytoplasmic and involved in translation regulation. However, in our microscopic studies, we observed that during asexual blood stages PfAlba3 is mainly nuclear in localization. But, in the western blot analysis, we also observed a faint band in the cytoplasmic fraction. This faint band is definitely not due to any contamination of nuclear fraction as confirmed by western blotting using nuclear and cytoplasm specific antibodies. These studies postulated the nucleo-cytoplasmic localization of PfAlba3, regulated probably by acetylation/deacetylation mechanism. Therefore, it may be possible that PfAlba3 may be dual in localization based on need and cellular stage, biological stress as well as regulation mediated by corresponding acetylase and deacetylase counterpart.
ChIP data revealed the presence of PfSir2A at the telomeric, subtelomeric region and at var gene promoters. The data also documented the presence of PfSir2A during ring stage at ups B and E var promoter region but not at ups C and E. The presence of PfSir2A at promoter regions attributes silencing of downstream genes and its absence suggests active transcription. However, the transcription profiles of var genes vary within individuals or in a population. Differential expression of var genes are associated with several factors, including disease severity (65), exposure to biological stress (66) and disease phenotype (54,65,67). ChIP assay using PfAlba3 antibody shows its presence at telomere, TARE repeat sequence but not at GBP, LSA and CSP promoter. Interestingly, PfAlba3 immunoprecipitated ups A, B and E promoter but not ups C suggesting the absence of PfAlba3 at centrally located var gene (ups C) as well as far away gene promoters (∼100 kb). These results suggested the correlation between PfSir2A and PfAlba3. Since, PfSir2A binding spreads out to the subtelomeric repeat Rep20 and is not associated with the genes located too far (>100 kb) from telomere end. To elucidate the association between PfSir2A and PfAlba3, we further performed ChIP ReChIP assay. Results validate the association of PfAlba3 and PfSir2A at ups B and E promoters but not at ups A and ups C promoters. However, PfAlba3 is present at these promoters (ups A, B and E) but differs in acetylation status. At ups A it is in acetylated status but not at ups B and E, where it is in deacetylated status and this deacetylation is probably mediated by PfSir2A.
Indeed, var genes silencing involves heterochromatin formation mediated by PfSir2A and involves reversible modification in the histone tail. In our studies, we found the presence of PfAlba3 at telomeric and subtelomeric regions shared with PfSir2A and presence of PfSir2A employed deacetylation of PfAlba3 as evident from ChIP and ChIP ReChIP assays. However, a marginal co-localization in IF studies was evident but co-immunoprecipitation and chromatin immunoprecipitation experiments showed the physical association between PfAlba3 and PfSir2A. Again, PfSir2A could deacetylate peptides specific for N-terminal DNA-binding region of PfAlba3. Therefore, it is highly possible that PfAlba3 may be involved in telomere position effect and epigenetic contribution to the regulation of gene expression mediated by PfSir2A. Future studies using antigenic homogeneous parasitic population combined with knockout and microarray analysis should be needed for better understanding of the precise role of PfAlba3. In conclusion, our studies reveal that PfAlba3, an Alba family protein serves as a non-histone substrate for PfSir2A and may cooperate for var gene expression in P. falciparum.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–2, Supplementary Figures 1–6, Supplementary Methods and Supplementary References [68–86].
FUNDING
Council of Scientific and Industrial Research (CSIR), New Delhi. CSIR fellowship to M.G. Funding for open access charge: Council of Scientific and Industrial Research (CSIR), New Delhi.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr S.K. Puri (Division of Parasitology, Central Drug Research Institute, Lucknow) for providing P. falciparum culture. The authors also thank Prof. Rahul Banerjee (SINP), Dr Alok Dutta and Dr Hemanta K. Majumdar (IICB) for critical reading of the manuscript.
REFERENCES
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam A, Goyal M, Iqbal MS, Pal C, Dey S, Bindu S, Maity P, Bandyopadhyay U. Novel antimalarial drug targets: hope for new antimalarial drugs. Expert Rev Clin. Pharmacol. 2009;2:469–489. doi: 10.1586/ecp.09.28. [DOI] [PubMed] [Google Scholar]
- 3.White NJ. Antimalarial drug resistance. J. Clin. Invest. 2004;113:1084–1092. doi: 10.1172/JCI21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 5.Miao J, Fan Q, Cui L, Li J. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene. 2006;369:53–65. doi: 10.1016/j.gene.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Trelle MB, Salcedo-Amaya AM, Cohen AM, Stunnenberg HG, Jensen ON. Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum. J. Proteome. Res. 2009;8:3439–3450. doi: 10.1021/pr9000898. [DOI] [PubMed] [Google Scholar]
- 7.Cui L, Fan Q, Li J. The malaria parasite Plasmodium falciparum encodes members of the Puf RNA-binding protein family with conserved RNA binding activity. Nucleic Acids Res. 2002;30:4607–4617. doi: 10.1093/nar/gkf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Silva EK, Gehrke AR, Olszewski K, Leon I, Chahal JS, Bulyk ML, Llinas M. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc. Natl Acad. Sci. USA. 2008;105:8393–8398. doi: 10.1073/pnas.0801993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gissot M, Briquet S, Refour P, Boschet C, Vaquero C. PfMyb1, a Plasmodium falciparum transcription factor, is required for intra-erythrocytic growth and controls key genes for cell cycle regulation. J. Mol. Biol. 2005;346:29–42. doi: 10.1016/j.jmb.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 10.Kumar K, Singal A, Rizvi MM, Chauhan VS. High mobility group box (HMGB) proteins of Plasmodium falciparum: DNA binding proteins with pro-inflammatory activity. Parasitol. Int. 2008;57:150–157. doi: 10.1016/j.parint.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Mancio-Silva L, Rojas-Meza AP, Vargas M, Scherf A, Hernandez-Rivas R. Differential association of Orc1 and Sir2 proteins to telomeric domains in Plasmodium falciparum. J. Cell. Sci. 2008;121:2046–2053. doi: 10.1242/jcs.026427. [DOI] [PubMed] [Google Scholar]
- 12.McAndrew MB, Read M, Sims PF, Hyde JE. Characterisation of the gene encoding an unusually divergent TATA-binding protein (TBP) from the extremely A+T-rich human malaria parasite Plasmodium falciparum. Gene. 1993;124:165–171. doi: 10.1016/0378-1119(93)90390-o. [DOI] [PubMed] [Google Scholar]
- 13.Merrick CJ, Duraisingh MT. Plasmodium falciparum Sir2: an unusual sirtuin with dual histone deacetylase and ADP-ribosyltransferase activity. Eukaryot. Cell. 2007;6:2081–2091. doi: 10.1128/EC.00114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002;296:148–151. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- 15.Forterre P, Confalonieri F, Knapp S. Identification of the gene encoding archeal-specific DNA-binding proteins of the Sac10b family. Mol. Microbiol. 1999;32:669–670. doi: 10.1046/j.1365-2958.1999.01366.x. [DOI] [PubMed] [Google Scholar]
- 16.Guo R, Xue H, Huang L. Ssh10b, a conserved thermophilic archaeal protein, binds RNA in vivo. Mol. Microbiol. 2003;50:1605–1615. doi: 10.1046/j.1365-2958.2003.03793.x. [DOI] [PubMed] [Google Scholar]
- 17.Marsh VL, Peak-Chew SY, Bell SD. Sir2 and the acetyltransferase, Pat, regulate the archaeal chromatin protein, Alba. J. Biol. Chem. 2005;280:21122–21128. doi: 10.1074/jbc.M501280200. [DOI] [PubMed] [Google Scholar]
- 18.Xue H, Guo R, Wen Y, Liu D, Huang L. An abundant DNA binding protein from the hyperthermophilic archaeon Sulfolobus shibatae affects DNA supercoiling in a temperature-dependent fashion. J. Bacteriol. 2000;182:3929–3933. doi: 10.1128/jb.182.14.3929-3933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao K, Chai X, Marmorstein R. Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA binding. J. Biol. Chem. 2003;278:26071–26077. doi: 10.1074/jbc.M303666200. [DOI] [PubMed] [Google Scholar]
- 20.Aravind L, Iyer LM, Anantharaman V. The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 2003;4:R64. doi: 10.1186/gb-2003-4-10-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allers T, Mevarech M. Archaeal genetics - the third way. Nat. Rev. Genet. 2005;6:58–73. doi: 10.1038/nrg1504. [DOI] [PubMed] [Google Scholar]
- 22.Reeve JN. Archaeal chromatin and transcription. Mol. Microbiol. 2003;48:587–598. doi: 10.1046/j.1365-2958.2003.03439.x. [DOI] [PubMed] [Google Scholar]
- 23.Starai VJ, Escalante-Semerena JC. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 2004;340:1005–1012. doi: 10.1016/j.jmb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mani J, Guttinger A, Schimanski B, Heller M, Acosta-Serrano A, Pescher P, Spath G, Roditi I. Alba-domain proteins of Trypanosoma brucei are cytoplasmic rna-binding proteins that interact with the translation machinery. PLoS One. 2011;6:e22463. doi: 10.1371/journal.pone.0022463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lurz R, Grote M, Dijk J, Reinhardt R, Dobrinski B. Electron microscopic study of DNA complexes with proteins from the Archaebacterium Sulfolobus acidocaldarius. EMBO J. 1986;5:3715–3721. doi: 10.1002/j.1460-2075.1986.tb04705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wardleworth BN, Russell RJ, Bell SD, Taylor GL, White MF. Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J. 2002;21:4654–4662. doi: 10.1093/emboj/cdf465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jelinska C, Conroy MJ, Craven CJ, Hounslow AM, Bullough PA, Waltho JP, Taylor GL, White MF. Obligate heterodimerization of the archaeal Alba2 protein with Alba1 provides a mechanism for control of DNA packaging. Structure. 2005;13:963–971. doi: 10.1016/j.str.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 29.Kowieski TM, Lee S, Denu JM. Acetylation-dependent ADP-ribosylation by Trypanosoma brucei Sir2. J. Biol. Chem. 2008;283:5317–5326. doi: 10.1074/jbc.M707613200. [DOI] [PubMed] [Google Scholar]
- 30.Sereno D, Vergnes B, Mathieu-Daude F, Cordeiro da Silva A, Ouaissi A. Looking for putative functions of the Leishmania cytosolic SIR2 deacetylase. Parasitol Res. 2006;100:1–9. doi: 10.1007/s00436-006-0280-2. [DOI] [PubMed] [Google Scholar]
- 31.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 32.Tonkin CJ, Carret CK, Duraisingh MT, Voss TS, Ralph SA, Hommel M, Duffy MF, Silva LM, Scherf A, Ivens A, et al. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 2009;7:e84. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kyes SA, Kraemer SM, Smith JD. Antigenic variation in Plasmodium falciparum: gene organization and regulation of the var multigene family. Eukaryot. Cell. 2007;6:1511–1520. doi: 10.1128/EC.00173-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 36.Chookajorn T, Dzikowski R, Frank M, Li F, Jiwani AZ, Hartl DL, Deitsch KW. Epigenetic memory at malaria virulence genes. Proc. Natl Acad. Sci. USA. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez Rivas R, Scherf A. 5' flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol. 2007;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voss TS, Kaestli M, Vogel D, Bopp S, Beck HP. Identification of nuclear proteins that interact differentially with Plasmodium falciparum var gene promoters. Mol. Microbiol. 2003;48:1593–1607. doi: 10.1046/j.1365-2958.2003.03528.x. [DOI] [PubMed] [Google Scholar]
- 39.Perez-Toledo K, Rojas-Meza AP, Mancio-Silva L, Hernandez-Cuevas NA, Delgadillo DM, Vargas M, Martinez-Calvillo S, Scherf A, Hernandez-Rivas R. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 2009;37:2596–2606. doi: 10.1093/nar/gkp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petter M, Lee CC, Byrne TJ, Boysen KE, Volz J, Ralph SA, Cowman AF, Brown GV, Duffy MF. Expression of P. falciparum var Genes Involves Exchange of the Histone Variant H2A.Z at the Promoter. PLoS Pathog. 2011;7:e1001292. doi: 10.1371/journal.ppat.1001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 42.Deitsch KW. Malaria virulence genes controlling expression through chromatin modification. Cell. 2005;121:1–2. doi: 10.1016/j.cell.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 43.Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 44.Cui L, Miao J. Chromatin-mediated epigenetic regulation in the malaria parasite Plasmodium falciparum. Eukaryot. Cell. 2010;9:1138–1149. doi: 10.1128/EC.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 46.Miao F, Bouziane M, O'Connor TR. Interaction of the recombinant human methylpurine-DNA glycosylase (MPG protein) with oligodeoxyribonucleotides containing either hypoxanthine or abasic sites. Nucleic Acids Res. 1998;26:4034–4041. doi: 10.1093/nar/26.17.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Basu D, Khare G, Singh S, Tyagi A, Khosla S, Mande SC. A novel nucleoid-associated protein of Mycobacterium tuberculosis is a sequence homolog of GroEL. Nucleic Acids Res. 2009;37:4944–4954. doi: 10.1093/nar/gkp502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choubey V, Guha M, Maity P, Kumar S, Raghunandan R, Maulik PR, Mitra K, Halder UC, Bandyopadhyay U. Molecular characterization and localization of Plasmodium falciparum choline kinase. Biochim. Biophys. Acta. 2006;1760:1027–1038. doi: 10.1016/j.bbagen.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Choubey V, Maity P, Guha M, Kumar S, Srivastava K, Puri SK, Bandyopadhyay U. Inhibition of Plasmodium falciparum choline kinase by hexadecyltrimethylammonium bromide: a possible antimalarial mechanism. Antimicrob. Agents Chemother. 2007;51:696–706. doi: 10.1128/AAC.00919-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- 51.Ben Mamoun C, Gluzman IY, Hott C, MacMillan SK, Amarakone AS, Anderson DL, Carlton JM, Dame JB, Chakrabarti D, Martin RK, et al. Co-ordinated programme of gene expression during asexual intraerythrocytic development of the human malaria parasite Plasmodium falciparum revealed by microarray analysis. Mol. Microbiol. 2001;39:26–36. doi: 10.1046/j.1365-2958.2001.02222.x. [DOI] [PubMed] [Google Scholar]
- 52.Voss TS, Mini T, Jenoe P, Beck HP. Plasmodium falciparum possesses a cell cycle-regulated short type replication protein A large subunit encoded by an unusual transcript. J. Biol. Chem. 2002;277:17493–17501. doi: 10.1074/jbc.M200100200. [DOI] [PubMed] [Google Scholar]
- 53.O'Donnell RA, Freitas-Junior LH, Preiser PR, Williamson DH, Duraisingh M, McElwain TF, Scherf A, Cowman AF, Crabb BS. A genetic screen for improved plasmid segregation reveals a role for Rep20 in the interaction of Plasmodium falciparum chromosomes. EMBO J. 2002;21:1231–1239. doi: 10.1093/emboj/21.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaestli M, Cockburn IA, Cortes A, Baea K, Rowe JA, Beck HP. Virulence of malaria is associated with differential expression of Plasmodium falciparum var gene subgroups in a case-control study. J. Infect. Dis. 2006;193:1567–1574. doi: 10.1086/503776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chou CC, Lin TW, Chen CY, Wang AH. Crystal structure of the hyperthermophilic archaeal DNA-binding protein Sso10b2 at a resolution of 1.85 Angstroms. J. Bacteriol. 2003;185:4066–4073. doi: 10.1128/JB.185.14.4066-4073.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumarevel T, Sakamoto K, Gopinath SC, Shinkai A, Kumar PK, Yokoyama S. Crystal structure of an archaeal specific DNA-binding protein (Ape10b2) from Aeropyrum pernix K1. Proteins. 2008;71:1156–1162. doi: 10.1002/prot.21807. [DOI] [PubMed] [Google Scholar]
- 57.Wang G, Guo R, Bartlam M, Yang H, Xue H, Liu Y, Huang L, Rao Z. Crystal structure of a DNA binding protein from the hyperthermophilic euryarchaeon Methanococcus jannaschii. Protein Sci. 2003;12:2815–2822. doi: 10.1110/ps.03325103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lavstsen T, Salanti A, Jensen AT, Arnot DE, Theander TG. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malar. J. 2003;2:27. doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rice PA. Making DNA do a U-turn: IHF and related proteins. Curr. Opin. Struct. Biol. 1997;7:86–93. doi: 10.1016/s0959-440x(97)80011-5. [DOI] [PubMed] [Google Scholar]
- 60.Suck D, Lahm A, Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988;332:464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- 61.White SW, Appelt K, Wilson KS, Tanaka I. A protein structural motif that bends DNA. Proteins. 1989;5:281–288. doi: 10.1002/prot.340050405. [DOI] [PubMed] [Google Scholar]
- 62.Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mair GR, Lasonder E, Garver LS, Franke-Fayard BM, Carret CK, Wiegant JC, Dirks RW, Dimopoulos G, Janse CJ, Waters AP. Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 2010;6:e1000767. doi: 10.1371/journal.ppat.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ralph SA, Scherf A. The epigenetic control of antigenic variation in Plasmodium falciparum. Curr. Opin. Microbiol. 2005;8:434–440. doi: 10.1016/j.mib.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 65.Rottmann M, Lavstsen T, Mugasa JP, Kaestli M, Jensen AT, Muller D, Theander T, Beck HP. Differential expression of var gene groups is associated with morbidity caused by Plasmodium falciparum infection in Tanzanian children. Infect. Immun. 2006;74:3904–3911. doi: 10.1128/IAI.02073-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenberg E, Ben-Shmuel A, Shalev O, Sinay R, Cowman A, Pollack Y. Differential, positional-dependent transcriptional response of antigenic variation (var) genes to biological stress in Plasmodium falciparum. PLoS One. 2009;4:e6991. doi: 10.1371/journal.pone.0006991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jensen AT, Magistrado P, Sharp S, Joergensen L, Lavstsen T, Chiucchiuini A, Salanti A, Vestergaard LS, Lusingu JP, Hermsen R, et al. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J. Exp. Med. 2004;199:1179–1190. doi: 10.1084/jem.20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katoh K, Kuma K, Miyata T, Toh H. Improvement in the accuracy of multiple sequence alignment program MAFFT. Genome Inform. 2005;16:22–33. [PubMed] [Google Scholar]
- 69.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 71.Hildebrand A, Remmert M, Biegert A, Soding J. Fast and accurate automatic structure prediction with HHpred. Proteins. 2009;77(Suppl. 9):128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 72.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 73.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 74.Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 75.Tosatto SC, Bindewald E, Hesser J, Manner R. A divide and conquer approach to fast loop modeling. Protein Eng. 2002;15:279–286. doi: 10.1093/protein/15.4.279. [DOI] [PubMed] [Google Scholar]
- 76.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 78.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 79.Kumar S, Das SK, Dey S, Maity P, Guha M, Choubey V, Panda G, Bandyopadhyay U. Antiplasmodial activity of [(aryl)arylsulfanylmethyl]Pyridine. Antimicrob. Agents Chemother. 2008;52:705–715. doi: 10.1128/AAC.00898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar S, Guha M, Choubey V, Maity P, Srivastava K, Puri SK, Bandyopadhyay U. Bilirubin inhibits Plasmodium falciparum growth through the generation of reactive oxygen species. Free Radic. Biol. Med. 2008;44:602–613. doi: 10.1016/j.freeradbiomed.2007.10.057. [DOI] [PubMed] [Google Scholar]
- 81.Kyes S, Pinches R, Newbold C. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol. Biochem. Parasitol. 2000;105:311–315. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- 82.Alcalde M, Plou FJ, Andersen C, Martin MT, Pedersen S, Ballesteros A. Chemical modification of lysine side chains of cyclodextrin glycosyltransferase from Thermoanaerobacter causes a shift from cyclodextrin glycosyltransferase to alpha-amylase specificity. FEBS Lett. 1999;445:333–337. doi: 10.1016/s0014-5793(99)00134-9. [DOI] [PubMed] [Google Scholar]
- 83.Fraenkel-Conrat H. Methods for investigating the essential groups. Methods Enzymol. 1957;4:247–269. [Google Scholar]
- 84.Fojo AT, Reuben PM, Whitney PL, Awad WM., Jr Effect of glycerol on protein acetylation by acetic anhydride. Arch Biochem. Biophys. 1985;240:43–50. doi: 10.1016/0003-9861(85)90006-2. [DOI] [PubMed] [Google Scholar]
- 85.Lobley A, Whitmore L, Wallace BA. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 86.Alam A, Goyal M, Iqbal MS, Bindu S, Dey S, Pal C, Maity P, Mascarenhas NM, Ghoshal N, Bandyopadhyay U. Cysteine-3 and cysteine-4 are essential for the thioredoxin-like oxidoreductase and antioxidant activities of Plasmodium falciparum macrophage migration inhibitory factor. Free Radic. Biol. Med. 2011;50:1659–1668. doi: 10.1016/j.freeradbiomed.2011.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.