Abstract
DNA replication is strictly regulated through a sequence of steps that involve many macromolecular protein complexes. One of them is the replicative helicase, which is required for initiation and elongation phases. A MCM helicase found as a prophage in the genome of Bacillus cereus is fused with a primase domain constituting an integrative arrangement of two essential activities for replication. We have isolated this helicase–primase complex (BcMCM) showing that it can bind DNA and displays not only helicase and primase but also DNA polymerase activity. Using single-particle electron microscopy and 3D reconstruction, we obtained structures of BcMCM using ATPγS or ADP in the absence and presence of DNA. The complex depicts the typical hexameric ring shape. The dissection of the unwinding mechanism using site-directed mutagenesis in the Walker A, Walker B, arginine finger and the helicase channels, suggests that the BcMCM complex unwinds DNA following the extrusion model similarly to the E1 helicase from papillomavirus.
INTRODUCTION
The functional information contained in the DNA must be conserved and transmitted to daughter cells. This is an essential process in all organisms indispensable for genome maintenance during cell division and proliferation. Evolution has developed a wide number of specific factors that handle the efficiency and fidelity of the DNA replication process. The replicative DNA polymerases require a single-stranded DNA (ssDNA) template to initiate synthesis; however, many lack double-stranded DNA (dsDNA) unwinding activity. To remediate this situation ssDNA is usually delivered by a replicative helicase, an enzyme capable to unwind a duplex DNA by a process dependent on nucleoside triphosphate hydrolysis. The replicative helicases are typically hexamers with a characteristic single-ring or double-ring structure (1–6). In archaea and eukaryotes the replicative helicases belong to the mini-chromosome maintenance (MCM) family of proteins (1,4,7,8). Most of the sequenced archaeal genomes have revealed the presence of only one MCM gene, in contrast with their eukaryotic counterparts, where the MCM complex is built-up by six related MCM proteins (MCM2-7) (9). The MCM2-7 complex plays an essential role during the initiation of DNA replication and the progression of the replisome. Dysregulation of these processes is linked to genomic instability and a variety of carcinomas (10–12). However, the mechanism by which helicases unwind dsDNA during initiation and elongation is not yet clear.
The crystal structures of the N-terminal domains of several archaeal MCM proteins have been solved, and hexameric models were built based in their crystallographic symmetry (13,14). Moreover, the corresponding full-length proteins have been crystallized in their monomeric form (15,16). Hence, all the hexameric models rely in arrangements generated from the N-terminal domain oligomers. Several studies using 3D-electron microscopy (3D-EM) have generated low resolution structures of the archaeal (5,17–21) and eukaryotic MCM complexes (6,22), providing snapshots of the helicases in different conformational states and shedding light in the functional mechanism of the complex.
Recently, a gene encoding a MCM homologue was identified in the genome of Bacillus cereus (BcMCM) (23). Interestingly, this gene is encoded within an integrated prophage in the bacterial genome. The N-terminal fragment of the protein (Figure 1A) contains a region that is homologous to the catalytic subunit of the archaeal–eukaryotic DNA primase (24,25). The N-terminal domain also shows homology with the primase–polymerase domain of the replication protein ORF904 of plasmid pRN1 from Sulfolobus islandicus (Supplementary Figure S1), which is also fused to a helicase domain (26). The BcMCM C-terminal section (Figure 1A) is homologous to the MCM AAA+ helicases, with typical Walker A and Walker B motifs in the ATP-binding site. This modular architecture is similar to that of the T7 phage primase–helicase protein where the primase and helicase domains are homologous to bacterial DnaG and DnaB (27), and to the replication protein ORF904, which combines an archaeal–eukaryotic primase–polymerase domain with a SF3 (superfamily 3) helicase domain (26) (Figure 1A). To our knowledge this is the first description of an archaeal–eukaryotic primase–polymerase and MCM helicase domains merged in the same polypeptide. Noteworthy, the BcMCM helicase fragment displays a high sequence similarity with the eukaryotic MCM2-7 proteins. The sequence identity is 22–24% among the MCM2-7 subunits and 20–24% similarity for BcMCM and the different MCM2-7 human subunits (Supplementary Figure S2).
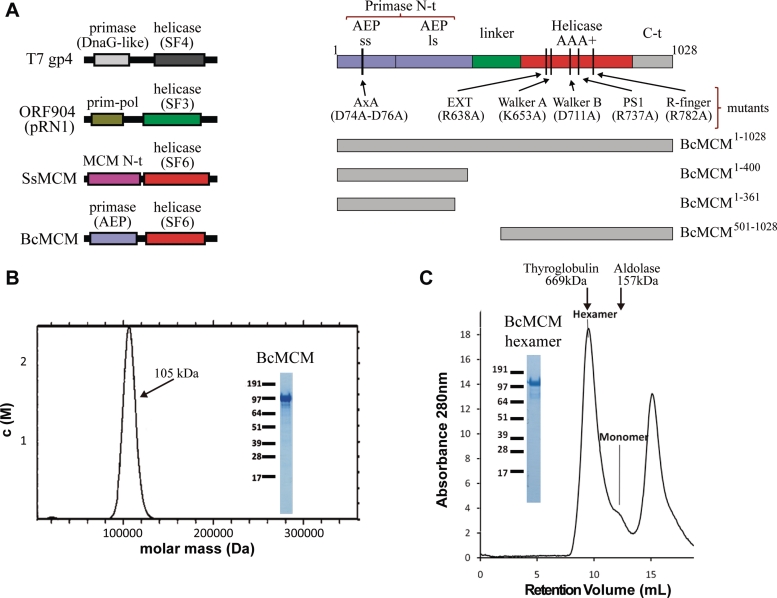
Figure 1.
BcMCM is purified as a monomer but can hexamerize. (A) Graphical representations of the domain architecture present in several multimodular helicases (left panel). The families of associated primases and helicases are indicated. BcMCM presents an exclusive combination of an archaeo–eukaryotic primase (AEP), including the small (ss) and large (ls) subunits, and a MCM-like helicase (Super Family 6). Scheme of the domain arrangement of BcMCM (right panel). A representation of the different constructions and mutants used along this article is depicted in the right lower panel. (B) Analytical ultracentrifugation experiments shows that the recombinant protein BcMCM expressed in E. coli is a monomer. Only one species is detected in the calculated molar mass distribution, c(M), which is consistent with the molecular mass of a monomer (see ‘Materials and Methods’ section). (C) Size-exclusion chromatography illustrates conversion of BcMCM monomers into hexamers. Upon addition of ATPγS and ssDNA (dT)40 and incubation with the purified monomer fractions the BcMCM protein elutes mainly as a hexamer from Superdex 200 column. The hexamer fractions were collected and analysed using SDS–PAGE (inset). The peak at the end of the chromatogram corresponds to the excess of ssDNA used in the assay. Elution positions of the molecular weight standards Thyroglobulin and Aldolase are indicated in the graph.
We have isolated, reconstituted and characterized a functional BcMCM hexamer, showing for the first time that BcMCM contains in vitro primase and polymerase activities, and revealing this enzyme as a unit gathering all the essential functions for replication. We analysed by 3D-EM the hexameric structure of the BcMCM complex to reveal the low-resolution structure of the helicase and its complex with DNA. The conformational changes of full-length BcMCM complex upon its binding to ADP, ATPγS with or without DNA were also examined by a combination of 3D-EM and biochemical experiments. The high similarity of the BcMCM with the eukaryotic MCM2-7 complex makes this helicase a simpler but representative candidate for biochemical and structural studies. Our findings could contribute to the understanding of the mechanistic details of the more complicated eukaryotic counterparts implicated in the process of DNA replication.
MATERIALS AND METHODS
Expression and purification of BcMCM full length; Walker A (K653A), Walker B (D711A), arginine finger (R782A), presensor I β-hairpin (R737A), external β-hairpin (R638A), AxA mutant (D74A–D76A) and BcMCM1–400 proteins
Full length, ATPase mutants, β-hairpins mutants, AxA mutant and BcMCM1–400 proteins were expressed in Escherichia coli BL21 Star (DE3; Invitrogen) strain using auto-inducing media ZYP-5052. Proteins were purified by affinity chromatography (GST-Trap HP column and HisTrap HP, GE Healthcare) and a size exclusion chromatography (Superdex 200 or Superdex 75, GE Healthcare in case of the BcMCM1–400 protein).
Expression and purification of BcMCM1–361 and BcMCM501–1028
BcMCM1–361 and BcMCM501–1028 were expressed in E. coli Rosetta strain (Invitrogen), using auto-inducing media ZYP-5052. Protein were purified by Histrap HP (GE Healthcare), Heparin HP (GE Healthcare) column and Superdex 75 (GE Healthcare) column or Superose 6 (GE Healthcare) column in the case of the BcMCM501–1028.
Analytical ultracentrifugation
The sedimentation velocity experiments were conducted in an XL-A analytical ultracentrifuge (Beckman-Coulter Inc.) at 42 000 rpm and 25°C, using an A50Ti rotor and a 1.2-mm charcoal-filled Epon double-sector centerpiece. Absorbance was measured at 280 nm. The protein concentration was 20 μM in 50 mM Tris–HCl, pH 7.5. Data were modelled as a superposition of Lamm equation solutions with SEDFIT (available at www.analyticalultracentrifugation.com/default.htm). The sedimentation coefficient distribution, c(s), was calculated at a P = 0.68 confidence level. The experimental sedimentation values were determined by integration of the main peak of c(s) and corrected to standard conditions to obtain the S20,w values with SEDNTERP. Calculation of the frictional coefficient ratio was performed with SEDFIT to yield the calculated molar mass distribution, c(M).
ATPase assay
ATP hydrolysis rates were measured by spectrophotometric coupled enzyme system composed of pyruvate kinase and lactate dehydrogenase to link the reaction of ATP hydrolysis to NADH oxidation (28). The reaction mixtures (20 µl) containing 1 µM of purified proteins were incubated at 30°C in a reaction buffer (10 mM Tris–HCl, pH 7.5, 10 mM MgCl2 and 0.1 mg/ml BSA) with 5 mM PEP, 2 mM NADH, 5 nM ssM13, 1 mM ATP, 75 µ/ml pyruvate kinase and 125 µ/ml of lactate dehydrogenase (SIGMA). Aliquots of 2 µl were withdrawn at time 0 and at 5 min intervals up to 30 min and the absorbance was measured at 340 nm by NanoDrop (ND-1000) spectrophotometer. ATPase rates were calculated as ATP molecules hydrolysed per minute per protein monomer (per minute).
Helicase assay
Oligonucleotides used for the preparation of the helicase substrates are listed in Supplementary Table SII. The 5′-ends of oligonucleotides were radiolabelled using T4 polynucleotide kinase (PNK) (Roche) and [γ32P]ATP (Perkin Elmer). The helicase reactions were performed at 30°C for 60 min in a buffer containing 20 mM Tris–HCl, pH 7.5, 100 µM EDTA, 100 mM ammonium glutamate, 5 mM ATP, 2 mM DTT, 0.1 mg/ml BSA, 10 mM Mg(OAc)2 10% glycerol, 0.5 nM DNA substrate and the amount of BcMCM protein or its mutants indicated in the figures. The reactions were stopped by addition of 6× stop solution (10 mM Tris–HCl, pH 7.6, 0.2% SDS, 60% glycerol, 100 mM EDTA, 0.03% bromophenol blue and 0.03% xylene cyanol). The products were separated by electrophoresis through 12% (w/v) non-denaturing polyacrylamide gel, and visualized using a Typhoon TRIO phosphorimager (GE Healthcare).
Primase assays
As a DNA template to assay primase activity, we first used M13 ssDNA (Amersham Bosciences-GE). The reaction mixture (10 μl) contained 50 mM Tris–HCl, pH 7.5, 75 mM NaCl, 1 mM MnCl2 or 5 mM MgCl2, 1 mM DTT, 2% glycerol, 0.05 mg/ml BSA, 200 ng M13 ssDNA, 1 µM wild-type BcMCM, and either ribonucleotides (100 µM ATP, GTP and UTP, and 16 nM [α-32P]CTP), or deoxynucleotides (100 µM dATP, dGTP and dTTP, and 16 nM of [α-32P]dCTP).
In an alternative primase assay, we used the 60-mer oligonucleotide GTCC: 5′ T36 CCTG T20 3′, that contains a putative priming initiation site. The reaction mixture (10 μl) contained 50 mM Tris–HCl, pH 7.5, 75 mM NaCl, 1 mM MnCl2 or 5 mM MgCl2, 1 mM DTT, 2% glycerol, 0.05 mg/ml BSA, 1 µM GTCC oligonucleotide, 10 µM of either GTP or dGTP, 16 nM [α-32P]ATP or 16 nM [α-32P]dATP, in the presence of 200 nM of either wild-type BcMCM, single mutant K653A or C-terminal deletion mutant BcMCM1–400. In both assays, after 60 min of incubation at 30°C, the reactions were stopped by addition of formamide loading buffer (10 mM EDTA, 95% v/v formamide, 0.3% w/v xylen-cyanol). Reactions were loaded in 8 M urea-containing 20% polyacrylamide sequencing gels. After electrophoresis, short polynucleotides (primers) were detected by autoradiography.
DNA polymerase assay
To prepare a conventional substrate to assay DNA polymerization, a 15-mer oligonucleotide (5′-GATCACAGTGAGTAC-3′) was 5′-labelled with T4 polynucleotide kinase and [γ-32P]ATP, as described by the manufacturer, and used as primer for DNA polymerization assays. The labelled primer was hybridized with the template 28-mer oligonucleotide (5-′AGAAGTGTATCTTGTACTCACTGTGATC-3′) by incubation for 10 min at 80°C, and cooled down to RT in the presence of 0.3 M NaCl. The resulting hybrid has a 3′-protrusion of 13 templating nucleotides. The reaction mixture (10 μl) contained 50 mM Tris–HCl, pH 7.5, 75 mM NaCl, 1 mM MnCl2 or 5 mM MgCl2, 1 mM DTT, 2% glycerol, 0.05 mg/ml BSA, 5 nM template/[γ-32P]-labelled primer DNA, 10 µM dNTPs, and either 100 nM wild-type BcMCM, single mutant K653A or C-terminal deletion mutant BcMCM1–400. After 60 min of incubation at 30°C, reactions were stopped by addition of formamide loading buffer (10 mM EDTA, 95% v/v formamide, 0.03% w/v bromophenol blue, 0.3% w/v xylen-cyanol). Reactions were loaded in 8 M urea-containing 20% polyacrylamide sequencing gels. After electrophoresis, the unextended and extended DNA primers were detected by autoradiography.
Electrophoretic mobility shift assays
Different 32P-labelled DNA structures were used in EMSA assays, dsDNA and ssDNA, the latter being (dT)40 or a 60-mer oligonucleotide. Several dsDNA were obtained by hybridization of that 32P-labelled 60-mer oligonucleotide with another oligonucleotides (shown in Supplementary Table SII). Protein–DNA-binding reactions were done by incubating 1 µM recombinant BcMCM with 1 nM of each probe in buffer EMSA [20 mM Tris–HCl, pH 7.5, 10 mM Mg(OAc)2, 10% glycerol, 1 mg/ml BSA, 100 mM NaCl, 1 mM DTT] at 25°C for 30 min. After incubation the mixtures were resolved in 5% polyacrylamide–TBE non-denaturing gel electrophoresis at 4°C. The gels were dried and binding was analysed by phosphorimager analysis.
BcMCM hexamerization
For hexamerization of BcMCM wild-type and its mutants, 9 µM of each protein, were incubated overnight at 4°C in oligomerization buffer (20 mM Tris–HCl, pH 7.5, 100 mM NaCl, 10 mM Mg(OAc)2, 100 mM ammonium glutamate, 10% glycerol, 1 mM DTT) with 5 mM ATPγS or 10 mM ADP, 18 µM ssDNA (dT)40. The oligomerization reactions were applied to a Superdex 200 10/300 (GE Healthcare) column pre-equilibrated with 20 mM Tris–HCl, pH 7.5, 100 mM NaCl, 100 mM ammonium glutamate, 10 mM Mg(OAc)2, 1 mM TCEP and/or diluted to 0.14 mg/ml, applied to glow discharged EM grid, negatively stained with 2% uranyl acetate (UAc) (w/v) and observed in a transmission electron microscope.
Generation of BcMCM–ssDNA complexes
DNA template used to obtain the BcMCM–ssDNA complexes consists of 5′ biotinylated primer (Biotin-EcoRI-5′-TCCCCCGAATTCCCCCCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT-3′) hybridized to the primer EcoRI- compl- 5′-GGGGGGAATTCGGGGG-3′. The EcoRI target sequence is highlighted in bold. Upon primers hybridization a 5′ biotinylated DNA was generated. The DNA is composed of a dsDNA fragment (16 bp) with an EcoRI restriction site followed by a (dT)50 section. DNA-bound BcMCM complexes were obtained upon incubation of BcMCM monomers in oligomerization buffer with the hybridized primers for 16 h at 4°C. The assembled complex was bound to streptavidin-coated magnetic beads (Dynabeads M280 streptavidin, Invitrogen) for 30 min at 4°C. After that, the magnetic beads were extensively washed in the oligomerization buffer containing 0.02% NP-40 (Calbiochem) and eluted after EcoRI digestion (for 30 min at 30°C). The eluted sample was tested on SDS–PAGE stained with Coomassie Blue and applied onto EM grid.
Electron microscopy and image analysis
For negative staining a few microlitres of oligomerized BcMCM–ADP, BcMCM–ATPγS, BcMCM–ATPγS–ssDNA, BcMCM–ADP–ssDNA and GST–BcMCM–ATPγS–ssDNA were diluted to an approximate concentration of 0.14 mg/ml. Samples were applied to glow-discharged carbon-coated grids and negatively stained with 2% UrAc. The sample was observed in a Tecnai G2 Spirit electron microscope (FEI, Netherlands) operated at 120 kV. In case of BcMCM–ATPγS and BcMCM–ATPγS–ssDNA, images were recorded on Kodak SO163 film using a JEOL 1230 operated at an accelerating voltage of 100 kV at a nominal magnification of 30 000. To increase the angular sampling, images were obtained at 0°, 20° and 30° tilt. Micrographs were digitized using a MINOLTA Dimage Scan Multi Pro scanner at 2400 dpi to get 3.5 Å/pixel at the specimen level. For BcMCM–ADP, BcMCM–ADP–ssDNA and GST–BcMCM–ATPγS–ssDNA images were recorded using TemCam-F416 4k × 4k pixel camera (TVIPS GmbH, Gauting, Germany) at calibrated magnification of 28 680 with 3.75 Å/pix at the specimen level. Several thousands of individual particles for each experiment (4853 for BcMCM–ADP, 3951 for BcMCM–ATPγS, 5748 for BcMCM–ATPγS–ssDNA and 6779 singles particles for GST–BcMCM–ATPγS–ssDNA complexes) were extracted using the ‘boxer’ program implemented in EMAN (29). These were masked, bandpass filtered, centered, normalized and subjected to angular refinement. The raw data was analysed using reference-free alignment and classification methods implemented in EMAN (29) and XMIPP (30). An initial 3D template for refinement was built using ‘startscym’ program implemented in EMAN (29) and the 6-fold symmetry of the molecule. BcMCM–ADP–ssDNA complex was obtained by incubating, in hexamerization buffer [20 mM Tris–HCl, pH 7.5, 100 mM NaCl, 10 mM Mg(OAc)2, 100 mM ammonium glutamate, 10% glycerol, 1 mM DTT], BcMCM monomers, ssDNA (1:4 molar ratio) and 10 mM ADP. The ‘multirefine’ from EMAN (29) allowed the BcMCM–ADP–ssDNA particle images to be classified simultaneously into two groups, the BcMCM–ADP–ssDNA and BcMCM–ADP set of images. For that BcMCM–ATPγS–ssDNA and BcMCM–ADP volumes filtered to 60 Å were used as a initial ‘multirefine models’. A total of 73.4% of the initial set of images correlated better with BcMCM–ATPγS–ssDNA structure. This new data set (8224 images) was processed independently. A reference-free initial model with 6-fold symmetry was generated using ‘startcsym’ program from EMAN. The data were further processed with no symmetry imposition following the same procedure as for the rest of the structures obtained here. As a control for the multirefine process we used BcMCM–ADP set of the particles and the same starting models used for classifying BcMCM–ADP–ssDNA images. We repeated the multirefine procedure with the same parameters as used to separate BcMCM–ADP–ssDNA data set. In this case 99.9% of the images were sorted into the group corresponding to the BcMCM–ADP. The resolution of the maps was estimated to be 36, 36, 33, 32 and 35 Å for BcMCM–ADP, BcMCM–ATPγS, BcMCM–ATPγS–ssDNA, BcMCM–ADP–ssDNA and GST–BcMCM–ATPγS–ssDNA, respectively, by Fourier Shell Correlation, using the criteria of a correlation coefficient of 0.5.
RESULTS
Reconstitution of BcMCM hexamer in vitro is nucleotide dependent
MCM helicases are AAA+ ATPases that function as hexamers. However, the recombinant BcMCM is isolated as a monomer (31). The protein is expressed as a N-terminal fusion with His6-GST tag. This protein tag is known to form dimers and may hamper oligomerization during expression. Nevertheless, after tag removal the isolated BcMCM still behaves as a monomer (Figure 1B, Supplementary Figure S3A and B). A similar behaviour could be observed when the protein was expressed with other tags such as His-tag or Strep-tag (data not shown). We developed an in vitro procedure to promote the assembly of the monomer into the hexameric complex (See ‘Materials and Methods’ section, Figure 1C and Supplementary Figure S3C). When the monomeric BcMCM was incubated at 4°C overnight in the presence of ATPγS or ADP, a shift was observed in the gel filtration chromatography profile indicating the presence of a high molecular weight oligomer. This oligomer was further stabilized in the presence of DNA (Figure 1C).
The dependence of nucleotide association for BcMCM functional assembly formation was dissected by site-directed mutagenesis in amino acid residues located in the Walker A and Walker B motifs. Mutations in these regions of the AAA+ domain are mechanistically related to different events occurring during nucleotide binding and hydrolysis. Thus, mutation of the conserved lysine of the Walker A abrogates nucleotide binding, whereas the mutation of the conserved aspartic in the Walker B affects nucleotide hydrolysis but not binding (32). To analyse the BcMCM helicase mechanism we generated the corresponding mutants, K653A and D711A, in conserved amino acids of the BcMCM Walker A and B motifs. In addition, we also mutated the arginine finger of BcMCM, generating a third mutant (R782A) whose nucleotide hydrolysis is affected. The effect of these mutations in the oligomeric behaviour of the BcMCM was analysed using the purified proteins (Supplementary Figure S3C). Whereas the R782A behaved similar to the wild-type, the D711A and the R737A (see PS1 section below) retained the capacity to form hexamers, although the mutations affect the monomer–hexamer ratio in the absence of DNA. However, the K653A mutant remains as a monomer indicating that nucleotide binding induces BcMCM assembly.
BcMCM binds to ssDNA and dsDNA
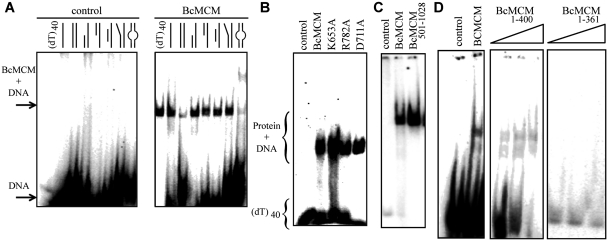
To assess the DNA-binding preferences of the BcMCM we performed electrophoretic mobility shift assays (EMSA) with different DNA probes resembling several replicative structures. A previous report showed that BcMCM binds ssDNA (31). However, BcMCM can also bind DNA probes that contain regions of single and double strand, such as template–primer molecules with 3′ and 5′ overhangs, and bubble-like structures (Figure 2A). It also displayed binding to dsDNA, although with lower affinity (Figure 2A). The ATPase domain mutants displayed similar binding ability to the DNA compared to the wild-type (Figure 2B). Hence, DNA binding by BcMCM is not dependent on ATP binding or hydrolysis as it has been shown for its archaeal homologues (1). Several deletion mutants were generated to identify the DNA-binding domain. The C-terminal region BcMCM501–1028, which contains the helicase activity, is sufficient to bind DNA (Figure 2C). Interestingly, the amino terminal BcMCM1–361, where the canonical primase domain is located, did not associate with DNA (Figure 2D) whereas the BcMCM1–400 construct binds DNA with low affinity, supporting previous results (31). Therefore these experiments suggest that the fragment comprising residues 361–400 contributes to DNA binding by the primase domain.
Figure 2.
The BcMCM protein binds DNA. (A) Native EMSA assays using different DNA structures. Sketches of the different DNA probes used in the assay are indicated above each lane. The assay without BcMCM was used as negative control. The 40-nt lane corresponds to a 40-nt poly-dT, (dT)40, a probe to discard binding to DNA secondary structures, and was used in subsequent assays. The sequences of the probes are described in Supplementary Table SII. Arrows and brackets indicate positions of the shifted protein–DNA complex and free DNA. All the assays were carried out using 1 µM of protein, unless otherwise indicated, and 1 nM of DNA. (B) Mutations in the ATPase site do not affect DNA binding. (C) The helicase domain BcMCM501–1028 is able to bind DNA. (D) The canonical primase domain BcMCM1–361 (1–3 µM) does not bind DNA. However, BcMCM1–400 (1–3 µM) shows weak binding compared to the wild-type protein.
The BcMCM protein and the isolated helicase domain hydrolyse ATP and unwind DNA
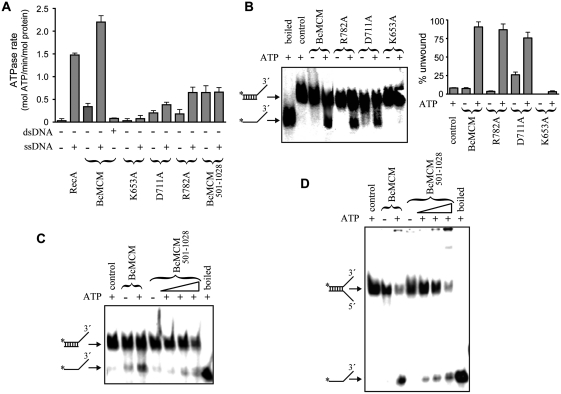
We monitored the ATPase activity associated with the purified BcMCM protein and analysed the AAA+ domain mutants and the primase truncated protein (BcMCM501–1028) for the coupling of the nucleotide hydrolysis to DNA unwinding activity. BcMCM presents ATPase activity that is stimulated by ssDNA but not by dsDNA (Figure 3A). The ATPase activity of the single-mutant proteins in either Walker B (D711A) or the arginine finger (R782A) was significantly lower but could still be stimulated by ssDNA. Remarkably, the ATPase activity of the helicase domain BcMCM501–1028 was significantly affected and was not stimulated by ssDNA, indicating an influence of the N-terminal domain. As expected, no ATPase activity was observed with the Walker A (K653A) mutant (Figure 3A). Next we compared the DNA helicase activity in the wild-type and mutant BcMCM proteins (Figure 3B). The polarity of the enzyme was 3′–5′ (31) and despite the loss in ATPase activity, the Walker B (D711A) and arginine finger (R782A) mutants displayed DNA helicase activity in agreement with the stimulation of nucleotide hydrolysis induced by ssDNA. This finding suggests that although hydrolysis is disturbed by the mutations, proper oligomerization still occurs (Supplementary Figure S3C), facilitating unwinding of the DNA duplexes. In contrast the Walker A (K653A) mutant, which does not bind the nucleotide and maintains a monomeric state, did not show DNA helicase activity in vitro (Figure 3B). In the isolated helicase domain BcMCM501–1028 nucleotide hydrolysis was not stimulated by ssDNA (Figure 3A), and consequently the helicase activity, measured using two different substrates, was significantly lower than in the full-length protein, suggesting again a coupling between the N-terminal and the helicase domains for proper BcMCM function (Figure 3C and D).
Figure 3.
BcMCM hydrolyses ATP and unwinds DNA. (A) For comparison the ATP hydrolysis activity of BcMCM and the different mutants was measured and represented as ATPase rates. The presence of ssDNA stimulates BcMCM ATPase activity. However, no effect was observed in the presence of dsDNA. The mutation in the Walker A region abolishes ATP hydrolysis, whereas the mutation in Walker B or the arginine finger reduces but not abolishes ATPase activity of BcMCM. No ssDNA stimulation was observed in the case of the BcMCM501–1028. An amount of 1 µM of protein was used in each reaction. The assay was performed by triplicate and the error bars indicate SEM. (B) BcMCM unwinds DNA with 3′→5′ polarity. The experiment compares the ATP dependent ability of BcMCM protein and the different mutants to separate the strands (% unwound DNA) of a specific dsDNA. Walker B or arginine finger mutants displayed a reduced ATP hydrolysis, although still show helicase activity. However, the Walker A mutant completely abrogated DNA unwinding. The assays were carried out using 0.5 nM of dsDNA and 0.5 µM of protein. (C and D) The helicase domain BcMCM501–1028 also presents helicase activity using fork or 3′-overhang DNA substrates. The assay for BcMCM501–1028 was performed using a gradient from 0.5 to 2 µM of protein.
BcMCM shows an intrinsic primase activity, preferentially using dNTPs as substrates
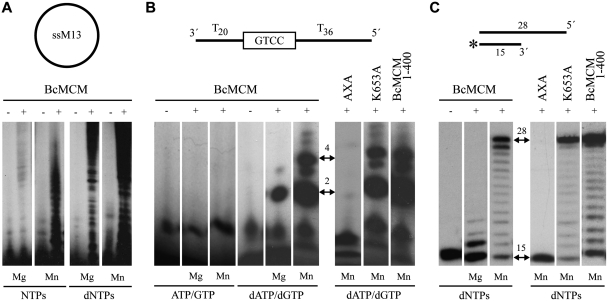
Based on amino acid sequence similarity analysis, the N-terminal domain of BcMCM has been proposed to contain a primase activity (Supplementary Figure S1). The primase activity was first assayed using a single-stranded circular DNA template (M13 ssDNA), and the hexameric form of BcMCM. In the presence of the four NTPs and magnesium, several bands reflecting primase activity were observed upon addition of BcMCM (Figure 4A). Interestingly, the same assay in the presence of manganese demonstrated a higher BcMCM primase activity using ribonucleotides as substrates. Furthermore primers were also synthesized using deoxynucleotides, both in the presence of magnesium and manganese activating ions, being the last one the preferred cation.
Figure 4.
BcMCM has intrinsic primase and polymerase activity. (A) Circular single-stranded M13 was used as template for BcMCM primase activity. BcMCM primase uses dNTPs more efficiently than NTPs and the reaction is favoured in the presence of manganese as cofactor. (B) A 60-mer ssDNA oligonucleotide (see scheme) was used as template with a putative priming site at the boxed sequence. The wild-type BcMCM displayed an intrinsic primase that preferentially uses dNTPs as substrates and manganese as metal activator. The primase activity is eliminated by the double mutation AxA in the catalytic site. In the monomeric Walker A mutant (K653A) and in the truncated version BcMCM1–400 the primase activity was similar. Arrows indicate the main formation of a dinucleotide and a 4-mer products. (C) Using a conventional template/primer substrate (see scheme), BcMCM displayed an intrinsic DNA polymerization activity, which was absent in the mutant AxA. The polymerase activity is preferentially activated by manganese ions. The DNA synthesis of the hexameric BcMCM is similar to the monomeric Walker A mutant (K653A), thus DNA polymerase activity does not involve hexamerization. The DNA polymerase activity of BcMCM, as in the case of the primase activity, is normal in the truncated version BcMCM1–400. Therefore both the primase and polymerase activities do not require the helicase domain. The initial substrate and the complete product positions are depicted with two-headed arrows. The asterisk in the substrate sketch indicates the labelled primer.
Furthermore, we evaluated the primase activity on a 60-mer oligonucleotide in which a potential primase recognition sequence (GTCC) is flanked by thymine residues (33). A tract of pyrimidines has been shown to be the preferred template context for initiation of the priming reaction by several viral, prokaryotic and cellular RNA primases (27,34,35). Again, when the conventional ribonucleotide substrates were used, the primase activity displayed by the BcMCM hexamer was very low when activated either by magnesium or manganese ions (Figure 4B). However, as observed on M13 ssDNA, the primase activity was much higher in the presence of deoxynucleotides as substrates, and particularly in the presence of manganese ions. The primase products obtained (dA-dG as the initiating dinucleotide, and its further elongation with dG and dA) are compatible with the use of the sequence GTCC as a preferred initiation site. That unusual primase activity using dNTPs preferentially activated by manganese ions was shown to be intrinsic to the BcMCM polypeptide by using a double mutant (AxA). Two of the three catalytic aspartates identified by amino acid sequence similarities among primases were mutated to alanines (Supplementary Figure S1). As shown in Figure 4, the double mutant was inactive as a primase, supporting the identification of the metal ligands at the catalytic site and the intrinsic nature of the primase, attributable to the BcMCM polypeptide. A primase activity with the same characteristics was also detected by using the monomeric form of BcMCM (data not shown), or by using mutant K653A, unable to form the BcMCM hexamer (Figure 4A). The putative location of the intrinsic primase at the N-terminal domain of the protein was confirmed by analysing the truncation derivative of BcMCM containing the first 400 residues that fully retained the primase activity.
BcMCM shows an intrinsic DNA-dependent DNA polymerase activity
In general, RNA primases make short RNA primers to be used by replicative DNA polymerases, to prime either the leading strand at an origin sequence, or the lagging strand at multiple sites to start Okazaki fragment synthesis. In bacteria, such a conventional primase activity corresponds to DnaG, the prokaryotic RNA primase. The length of the primers made in vitro by these RNA primases is variable. Consequently, in addition to the initiating product (dinucleotide) strictly corresponding to the primase activity, these enzymes behave also as RNA polymerases able to polymerize NTPs. In the case of BcMCM, the primase activity is unconventional, since it shows a preference for dNTPs to start synthesis. So, we tested whether in addition to the primase activity, this enzyme would be able to behave as a DNA-dependent DNA polymerase, using a short DNA hybrid with primer–template structure (see scheme in Figure 4C). Strikingly, BcMCM was able to extend the DNA primer by polymerizing dNTPs, both in the presence of either magnesium or manganese. Similarly to the primase assay, manganese ions (at an optimal concentration of 1 mM) led to higher activity, allowing the copying of the full-length template sequence. The catalytic AxA mutant demonstrated that this DNA polymerase activity was intrinsic to BcMCM, and not due to a possible contaminant polymerase. Moreover, the primase activity and the DNA-dependent DNA polymerase activity do not require the hexameric configuration (mutant K653A), and both reside at the N-terminal domain (BcMCM1–400, Figure 4C).
Architecture of the BcMCM. The BcMCM–ADP complex
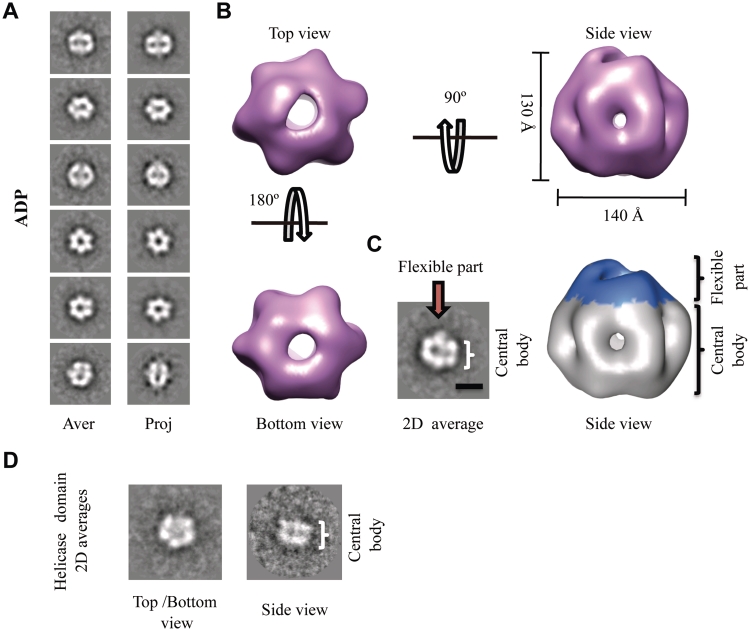
Electron microscopy (EM) studies of different MCM family members show that these proteins form oligomeric complexes, including hexamers, double hexamers, heptamers or filaments (1,6,17–19,36). Although the MCMs are loaded onto DNA as double hexamers in archaea and eukaryotes (1,6,17–19,36), the hexamer is supposed to represent the active form of the MCM helicase (37). Modelling studies show that the N-terminal domain forms a ring at one side of the hexamer while the AAA+ ATPase domain is located on the opposite side of the complex. BcMCM is isolated as a monomer; however, the addition of nucleotide induces hexamerization of the protein in vitro (Figures 1C, 5 and Supplementary Figure S3C). To elucidate the architecture of the BcMCM oligomer, we carried out single-particle negative stain EM analysis and 3D reconstruction. All structural analyses were performed on freshly oligomerized BcMCM complexes. First, we analysed samples of the BcMCM complex after incubation with ADP. A detailed examination of the EM field revealed a homogeneous area of particles. A reference-free classification was performed using the 4853 images collected of the BcMCM–ADP complex. To increase the signal to noise ratio the images corresponding to similar views of the complex were averaged. The averages revealed a donut-shaped 6-fold symmetry particle, which on the side view displayed two parallel layers of protein density separated by a sparse density section (Figure 5A, left panel). These data resembled the 2D averages from the single ring Methanothermobacter thermoautotrophicus (17,19) and Saccharomyces cerevisiae (6) MCM hexamers. A 3D reconstruction of the BcMCM–ADP hexamer was calculated starting with the initial model built using 2D averages of side and top–bottom views, and assuming a 6-fold rotational symmetry. The volume was refined with no symmetry imposition. The final reconstruction contains 2799 particles with a nominal resolution of 36 Å (calculated using a 0.5 cut-off of the Fourier Shell Correlation, as assessed by the EMAN Eotest program). 2D reprojections of the 3D model were in good agreement with the 2D class averages supporting the validity of the map (Figure 5A). The BcMCM–ADP complex forms a hexameric ring showing a roughly cylindrical shape with a maximum diameter of 140 Å and a height of ∼130 Å (Figure 5B). The overall architecture is consistent with six BcMCM monomers arranged around a 6-fold axis with a large central channel running through the whole molecule and six smaller side channels located at the monomer–monomer interface. The main channel has a diameter of 30 Å, large enough to accommodate dsDNA, whereas the side channels are displayed like slits wide enough to allow ssDNA but not dsDNA through. Despite its apparent 6-fold symmetry there is a clear asymmetry between the top and bottom sides of the molecule. The bottom view displays a more regular hexameric arrangement whereas the top view, the side view and the 2D averaged side view show different densities protruding from the complex (Figure 5B and C). The more compact hexameric part of the protein resembles in size and shape the MCM single ring hexamer. On the other hand the extra density displays features absent in previous structural studies of MCMs, indicating that it could correspond to the primase–polymerase domains of the BcMCM hexamer. Similarly to the primase domains of the T7 primase–helicase (38), this domain of the complex is most likely flexible and consequently partially visible. In addition 2D averages of the helicase fragment (BcMCM501–1028) resemble the 2D averages of the assembled BcMCM, but the extra density protruding from the hexamer is missing (Figure 5D).
Figure 5.
3D reconstruction of the BcMCM–ADP hexamer. (A) Gallery of selected reference-free 2D averages (left panel) compared to the corresponding reprojections of the final structure (right panel). (B) Surface representation of the 3D reconstruction of BcMCM–ADP hexamer filtered to 36 Å shown in different orientations. The protein monomers assemble into a single hexameric ring around a large central cavity displaying differences between the top and the bottom views. The overall dimensions of the complex are depicted in the figure. (C) The BcMCM–ADP hexamer (right panel) consists of a central body indicated (coloured in grey) and a flexible apical part (coloured in blue). In the 2D average side view (left panel) the central body and the flexible part are indicated with a bracket and an arrow respectively. A 100-Å bar is provided as a reference. (D) A typical reference-free 2D class averages of the BcMCM501–1028 helicase domain resembles the 2D averages of the BcMCM complex.
Structures of the BcMCM–ATPγS, BcMCM–ATPγS–ssDNA and BcMCM–ADP–ssDNA complexes
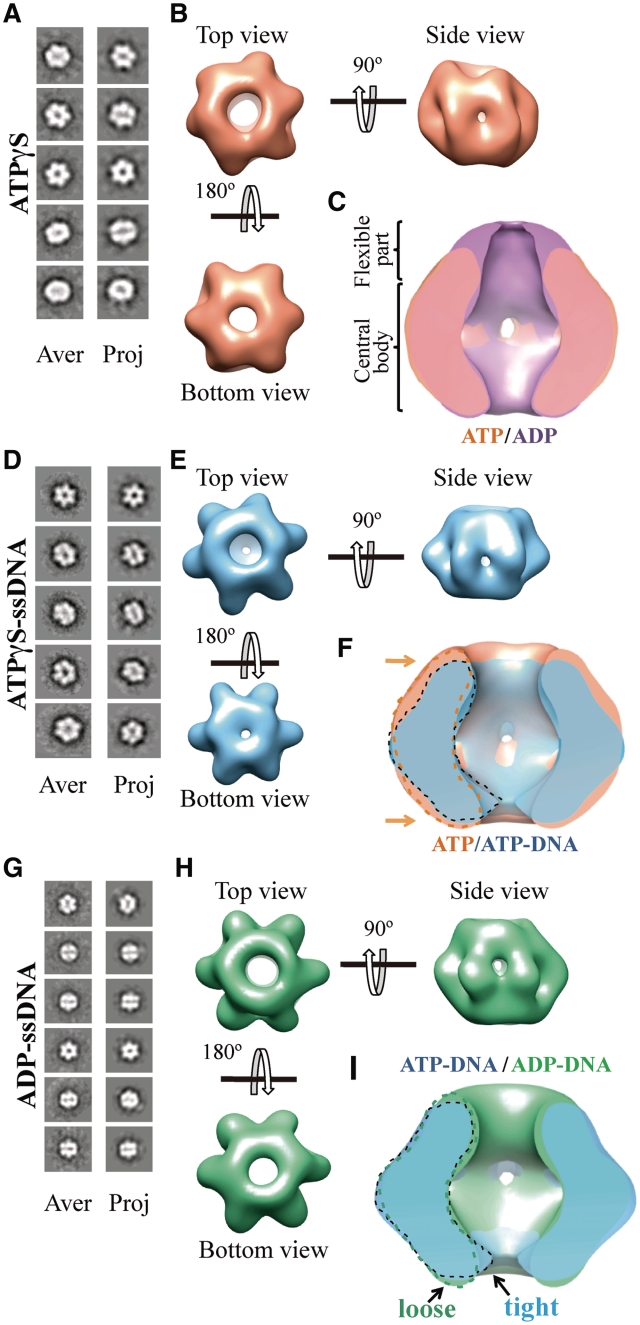
The BcMCM–ADP complex represents only one of the stages of the nucleotide cycle to unwind DNA. To cover as much as possible the conformational landscape of the complex during unwinding we have also obtained a BcMCM–ATPγS structure, representing the ATP-bound state as well as the BcMCM structures with ATPγS and ADP in complex with ssDNA.
We performed the BcMCM oligomerization experiment in the presence of ATPγS and examined this sample by single-particle EM and 3D reconstruction using the same procedure as above. The refined BcMCM–ATPγS 3D model revealed a cylinder-shaped hexamer (Figure 6A and B), which displays a flat part of the molecule on the bottom side and a wrinkled surface on the top side. The side views of the model are very similar to the ADP-bound structure (Figure 6B). However, the flexible primase–polymerase domain is less visible in the ATPγS compared to the ADP-bound structure (Figure 6C). Therefore at this resolution no significant conformational differences were observed in the central core of the BcMCM hexamer between the ATPγS and the ADP-bound structures.
Figure 6.
3D reconstruction of BcMCM–ATPγS, BcMCM–ATPγS-DNA and BcMCM–ADP complexes. (A) BcMCM–ATPγS reference-free class averages (left panel) and corresponding reprojections from the final structure (right panel). (B) Several views of different surface representations of a 3D reconstruction of BcMCM–ATPγS hexamer filtered to 36 Å. (C) Superposition of the symmetrized cut-open side views of BcMCM–ADP model in magenta and the symmetrized BcMCM–ATPγS in orange. ATP hydrolysis does not introduce large conformational changes in the BcMCM central body region. (D) BcMCM–ATPγS–ssDNA reference-free class averages (left panel) and corresponding reprojections from the structure (right panel). (E) Several views of different surface representations of BcMCM–ATPγS–ssDNA hexamer filtered to 33 Å. (F) Superposition of symmetrized cut-open side views of BcMCM–ATPγS model in orange and the symmetrized BcMCM–ATPγS–ssDNA in blue. One monomer of a BcMCM–ATPγS–ssDNA is highlighted with a dashed black line while one monomer of the BcMCM–ATPγS is highlighted with a dashed orange line. Conformational changes upon DNA binding (indicated by an arrow) permit a closer interaction of the DNA with the BcMCM hexamer. (G) BcMCM–ADP–ssDNA reference-free class averages (left panel) and corresponding reprojections from the structure (right panel). (H) Different views of a surface representation of a 3D reconstruction of BcMCM–ADP–ssDNA hexamer filtered to 33 Å. (I) Superposition of symmetrized cut-open side views of BcMCM–ATPγS–ssDNA model in blue and the symmetrized BcMCM–ADP–ssDNA in green. One monomer of a BcMCM–ATPγS–ssDNA is highlighted with a black line while one monomer of the BcMCM–ADP–ssDNA is indicated with a dashed green line.
To examine whether the interaction of ssDNA with BcMCM and the nucleotides could induce any conformational alterations of the BcMCM hexamer, we generated and examined the 3D structures of BcMCM–ATPγS–ssDNA and the BcMCM–ADP–ssDNA complexes. The monomeric BcMCM was incubated in the hexamerization buffer containing ATPγS and a 66-nt DNA probe containing a 50-nt 3′-overhang. The dsDNA section included an EcoRI restriction site coupled to a biotin molecule (see ‘Materials and Methods’ section and Supplementary Figure S4). The mixture was incubated with streptavidin-coated magnetic beads. The BcMCM–ATPγS–ssDNA complex was eluted from the beads by treating the washed beads with EcoRI that cleaves the DNA at the site near the biotin–streptavidin linkage. The freshly eluted fractions were directly applied on carbon-coated glow discharged EM grids and negatively stained for further structural analysis. BcMCM–ATPγS–ssDNA complexes visualized on a negative stained grid turned out to be single hexamers (Figure 6D, left panel).
We collected several thousands of negatively stained BcMCM–ATPγS–ssDNA specimens that were subjected to angular refinement. Reference free 2D averages show features similar to non-DNA-bound BcMCM hexamer. The top–bottom views displayed the characteristically six-petal flower shape, but each monomer looks more defined with respect to the hexamer in the absence of DNA (Figure 6D, left panel). Side views did not show a significant conformational difference when compared to the complex without DNA. We also generated a 3D structure of the BcMCM–ATPγS–ssDNA complex revealing conformational changes when compared to the cylindrical shape of the BcMCM–ATPγS hexamer in the absence of DNA (Figure 6B and C). Once the DNA is bound the bending of each monomer towards the central channel is coupled with a lateral movement of each moiety towards the central cavity (Figure 6F). As a result of these conformational changes the bottom accesses to the channel is remarkably reduced, allowing all BcMCM subunits to be in closer contact with the DNA. After these conformational changes the lateral channels are still wide enough to allow ssDNA through. In the BcMCM–ATPγS–DNA structure the flexible primase–polymerase domains of the complex are not clearly detected in this structure.
Hence to visualize and validate the assignment of the N-terminal domain within the BcMCM–ATPγS–ssDNA hexamer, we purified full length BcMCM protein avoiding the cleavage of the N-terminal GST-tag. The 27 kDa GST-tag was used as a marker for the primase domain. The purified GST–BcMCM monomer was fully functional (Supplementary Figure S5). The GST–BcMCM was incubated in hexamerization buffer with biotinylated DNA as previously described and after incubation with streptavidin-coated magnetic beads and digestion with EcoRI, the sample was used for negative-stain EM studies. The GST–BcMCM–ATPγS–ssDNA structure resembles the BcMCM–ATPγS structure with the bottom view of the protein presenting a clear 6-fold symmetry typical for MCMs helicases. The top view of the protein shows a disruption of the 6-fold symmetry, resulting in a distortion of the GST–BcMCM–ATPγS–ssDNA complex barrel shape, breaking the molecule between the N-terminal GST-tagged flexible side and the C-terminal helicase compact side (Supplementary Figure S6). This result indicates that the primase–polymerase domain of the BcMCM hexamer is a highly elastic region of the complex and therefore the GST tag cannot be clearly defined in the structure.
Finally, we analysed the molecular architecture of a BcMCM–ADP–ssDNA complex, providing a more complete view of the nucleotide induced conformational changes during DNA unwinding. The BcMCM–ADP–ssDNA complex was prepared following the same procedure as in the BcMCM–ATPγS–ssDNA complex. Although complex formation could be observed (Supplementary Figure S4), the number of particles obtained using this procedure was low, suggesting a decrease in the protein–DNA affinity. Therefore to generate a better quality structure the complex formation procedure was modified. The BcMCM monomer was incubated in hexamerization buffer containing ADP using a 4-fold molar excess of a 66-nt DNA probe with respect to the BcMCM hexamer. Freshly prepared sample was applied onto a carbon-coated glow discharged EM grid and negatively stained. More than 10 000 single particles images were collected for further analysis. To avoid a possible mixture of DNA-bound and DNA-free complexes in the data set the particles were classified in two groups using a supervised classification strategy and the multirefine command in EMAN (29). The BcMCM–ATPγS–ssDNA and the BcMCM–ADP structures were used as reference templates (for details see ‘Materials and Methods’ section). The final structure presented several conformational changes compared to the BcMCM–ADP complex in the absence of ssDNA (Figure 6G, H and Supplementary Figure S7I). Although the structure is similar to the BcMCM–ATPγS–ssDNA model, a clear difference can be observed between the BcMCM–ATPγS–ssDNA and the BcMCM–ADP–ssDNA structures in the diameter of the aperture located at the bottom part of the central channel of the complex. This entrance to the central channel is significantly narrower in the BcMCM–ATPγS–ssDNA complex, suggesting that it opens-up upon nucleotide hydrolysis (Figure 6E, H and I).
The BcMCM helicase domain forms hexamers in solution, but the BcMCM primase–polymerase domain does not oligomerize
Size-exclusion chromatography and negative-stain EM studies were used to test the oligomerization state of the purified BcMCM helicase domain (BcMCM501–1028). Again, the isolated fragment was incubated in oligomerization buffer in the same conditions established for the assembly of the full-length protein. Subsequently, the sample was applied on a glow-discharged EM grid and negatively stained with uranyl acetate. The single particles observed in the EM field appeared predominately ring-shaped, and were presumably top or tilted views of single rings. The 2D averages of the BcMCM501–1028 helicase domain show that it oligomerizes in a hexameric arrangement (Figure 5D), in agreement with the biochemical data (Figure 5D and Supplementary Figure S8). However, when the purified primase–polymerase domains (BcMCM1–361 and BcMCM1–400) were incubated in oligomerization buffer neither gel filtration (Supplementary Figure S8) nor the negative-stain EM studies detected hexameric oligomers of the BcMCM N-terminal domain, indicating that the 40-kDa primase–polymerase domain is not able to assemble as the full-length BcMCM protein and BcMCM501–1028 helicase domain.
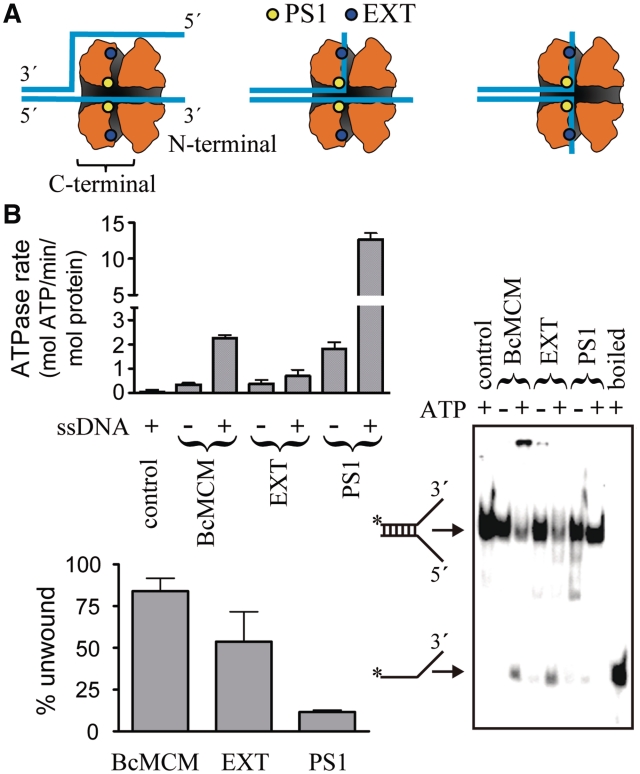
Mutations in the presensor 1 (PS1, R737A) and external hairpin (EXT, R638A) regions show different behaviour in the ATPase and helicase activities
New structural and site directed mutagenesis data in the archaeal system have provided a detailed background to examine the MCM mechanism. Recently three different variants have been proposed to explain the unwinding of DNA duplexes by the archaeal MCMs (Figure 7A) (16,39). Based on the monomer structure modelled onto the EM map, six side channels were observed in the interface between the subunits (16). Mutations in residues located in the side channel (EXT, R331A) and in the presensor 1 region (PS1, K430A) in Sulfolobus solfataricus MCM (SsMCM) decreased or abolished both the ATPase and the unwinding activities (39,40), suggesting that the displaced strand could be extruded, not only as previously proposed avoiding the entrance of the dsDNA through the central cavity (Figure 7A, left panel), but also using these side channels (Figure 7A, central and right panel). To investigate these hypotheses in BcMCM and to understand the unwinding mechanism, we performed similar mutations in the mesophilic BcMCM. In contrast with the archaeal MCMs, the corresponding mutations did not abolish the ATPase or helicase activities. In contrast with the archaeal counterpart the EXT mutant did not abolish the basal ATPase activity, and although the stimulation with ssDNA was diminished, it was not abrogated as in SsMCM. Moreover the PS1 mutant showed an ATPase activity higher than the wild-type, which was further stimulated by the presence of ssDNA (Figure 7B upper panel). In the helicase assay the EXT mutant conserved ∼60% of the wild-type activity whereas in the PS1 mutant the unwinding activity was diminished to ∼10% of BcMCM (Figure 7B lower and right panels), suggesting that in BcMCM while the EXT mutant conserves both activities although in a lower extent compared to the wild-type, the PS1 mutant decouples nucleotide hydrolysis from DNA unwinding.
Figure 7.
ATPase and helicase activity of the PS1 and the EXT mutants. (A) Schematic view of the location of the mutations in the BcMCM model and their implications in the different proposed extrusion mechanisms variants to unwind DNA (16). Each model threads the DNA strands in a different way through the helicase central and hypothetical lateral channels, which are depicted as a shaded inner surface. (B) ATPase (upper panel) and helicase (lower panel) activity of the PS1 and EXT BcMCM mutants. The histograms show the average of three experiments and the error bars indicate the SEM. A representative gel displaying the helicase experiment is depicted in the right side. Both assays were performed as in Figure 3.
DISCUSSION
Here, we describe a structure–function analysis of the multifunctional BcMCM, a protein coded as a prophage in the genome of B. cereus. This protein contains a N-terminal domain, which displays both primase and polymerase activities, and a C-terminal MCM helicase domain; constituting the first MCM protein that gathers in a single polypeptide the other key activities for genome replication. Although the sequence conservation between the archaeal and bacteriophage primase–polymerase domains is low, the BcMCM protein contains the crucial metal and nucleotide ligands forming the active site, characterized by the DXD motif, universally conserved in most families of nucleotidyl transferases. Concerning the specificity for DNA interactions, BcMCM showed a clear preference for the probes consisting exclusively of ssDNA or containing stretches of ssDNA (3′-, 5′-end and ‘bubble’) than for a probe consisting of only dsDNA (Figure 2A). Remarkably, a supershift was observed with the ‘bubble’ probe. This could be caused by the loading of more than one BcMCM hexamer on each ssDNA region. The enzyme presents ATP- dependent 3′–5′ helicase activity and similarly to other MCM helicases, BcMCM exhibits a basal ATPase activity that is stimulated by the addition of ssDNA (Figure 3A) in agreement with its binding preferences. However, the helicase domain, which binds ssDNA, did not show an increase in its ATPase activity in the presence of ssDNA; suggesting that the protein fragment containing the primase–polymerase domain and the linker facilitates DNA unwinding and consequently the hydrolysis of nucleotide (Figure 3).
In addition to its helicase domain BcMCM contains also a primase–polymerase activity able to start DNA synthesis (Figure 4). This primase–polymerase activity arising from the BcMCM N-terminal domain has the potential to initiate synthesis by using dNTPs as the favourite substrates. Similarly it has been shown that the ORF904 primase–polymerase domain coded in pRN1 plasmid also prefers dNTPs for polymerization, but requires both a ribonucleotide and a deoxynucleotide to synthesize the initiating dinucleotide (26).
Both the primase and polymerase activities show a preference for manganese in our assays. Manganese is an optional activating metal ion for polymerases that appears to be physiological in various processes, such as Non-Homologous End Joining (NHEJ). This cation has been considered mutagenic for a long time, however, renewed efforts in the study of polymerase function in vitro have led to demonstrate that some specialized polymerases, such as Polι, clearly prefer to utilize manganese even when magnesium is present in a large molar excess (41). In the case of Polµ, a DNA repair polymerase structurally and functionally related to terminal deoxynucleotidyltransferase (TdT) (42), the peak of polymerase activity with NHEJ substrates is reached at the physiological concentration of manganese ions, while the magnesium is inhibitory under the same conditions. Moreover, a physiological concentration of manganese does not have the negative effect on fidelity that can be observed at the high concentrations used previously (Maria Jose Martin & Luis Blanco, unpublished data).
A similar behaviour was observed in the case of Mycobacterium tuberculosis LigD, a bacterial enzyme involved in bacterial NHEJ, whose polymerization domain is related to AEPs (archeal/eukaryotic primases). LigD polymerization domain is involved in nucleotide additions associated to the repair of the DSBs, having manganese as the preferred metal activator (43). Therefore the activation of the primase and polymerase activities in the presence of manganese observed in BcMCM suggests that these synthetic activities may not be related to its conventional role in DNA replication, where the magnesium ion appears to be the metal of choice. It is tempting to speculate that the combination of these activities could be extremely useful to restart leading strand synthesis at stalled replication forks, as it has been recently proposed (44), providing bacteria with an advantageous mechanism to overcome this type of difficulties that arise during replication.
A comparison of the 3D EM structures of different MCMs shows that the homohexameric BcMCM and archaeal complexes display a regular hexamer in the C-terminal región while the eukaryotic MCM2-7 heterohexamer is less symmetrical in this area (Supplementary Figure S9). In adittion to the single hexameric forms, the archeal and eukaryotic MCM proteins can form double hexameric rings connected by their N-terminal domains (1,6,17–19,36). Despite the structural similarity of the EM structure of BcMCM to the other family members we have not detected a dodecameric structure in the presence or absence of DNA with the different nucleotides. Most likely the presence of the extra primase–polymerase domain located on the N-terminal of the protein instead of the N-terminal oligomerization domain avoids this feature observed in the archaeal and eukaryotic homologues.
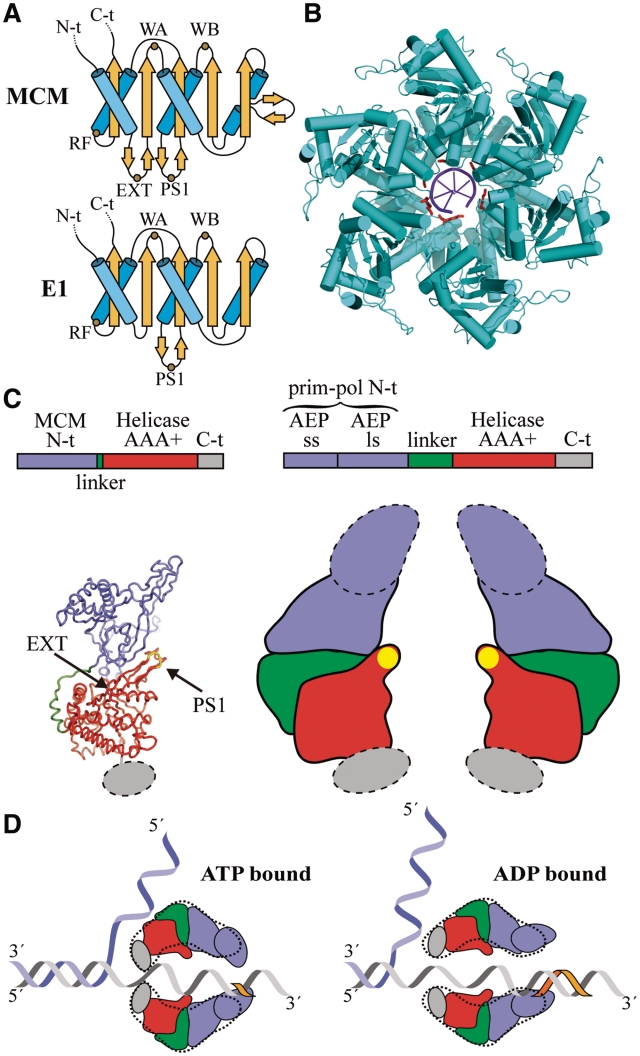
The BcMCM protein provides an excellent model to analyse the unwinding mechanism of the MCM family. Previous hypothesis on the eukaryotic replicative helicase function, based on steric exclusion (45,46) or rotary pumps (47,48) were focused on the MCM2-7 complex. So far no high-resolution structure of a MCM protein assembled in the hexameric form is available, neither in the apo state nor in complex with DNA to put the mutational data into a structural context. However, the E1 helicase from papilloma virus, a member of the SF3 helicase family whose structure has been solved in complex with ssDNA (49), shares similarities with the SF6 family, where the MCM proteins belong. These proteins are members of the AAA+ superfamily and certain structural features are conserved among them (Figure 8A). Superimposition of the E1 monomer in one of the subunits of the SsMCM hexamer model generated from the crystal N-terminal domain structure is in good agreement in half of the ATPase domain Cα atoms (4.01 Å r.m.s.d. for 162 out of 295 residues in the helicase domain). The overlay shows the structural conservation of the Walker A, Walker B motifs building the ATP-binding site and other structural elements. One of the absent elements is the hairpin that joins two β-strands and contains the residues from R323 to R329. The EXT R331A mutation in SsMCM is located at the end of one of these conserved β-strands in the neighbourhood of the ATP-binding site, and consequently its mutation could disturb the ATPase activity (Figures 8A and 7B, upper panel). In contrast, the β-hairpin, which contains the PS1 mutant K430A in SsMCM and R737A in BcMCM, is conserved in E1 (Figure 8A). This β-hairpin is crucial for DNA translocase activity in SF3 helicases, in particular the E1 K506 residue in its tip, which corresponds to K430 in SsMCM and R737 in BcMCM (Figure 8B). Moreover, the mutation of this residue (R737A) in BcMCM indicates that it could be involved in hexamer stabilization through its interaction with DNA. All these results may explain the decoupling effect observed between the ATPase and helicase activities in the BcMCM PS1 mutant. Moreover this residue is conserved in all the eukaryotic MCM proteins (Supplementary Figure S2), suggesting that the β-hairpin is structurally and functionally maintained during evolution.
Figure 8.
Unwinding mechanism of BcMCM. (A) Topology diagrams of the AAA+ domain of the MCM and E1 helicases. The sketch shows the conservation of the Walker A (WA), Walker B (WB), arginine finger (RF) and PS1 motifs between both helicases. The topology models were drawn based on the SsMCM (residues 304–484) and E1 (residues 408–545) structures. (B) Structure of the E1 helicase in complex with ssDNA. The lysine residues in the PS1 motifs contacting the DNA are coloured in red. (C) Model of the domain organization in BcMCM (right panel) based on the SsMCM crystal structure (left panel) (16) and the 3D-EM structures (Figures 5, 6 and Supplementary Figures S7 and S9). AEP denotes de archaeo–eukaryotic primase domain containing the small (ss) and large (ls) subunits. The PS1 β-hairpin is depicted with a yellow oval in the sketch. The dotted lines indicate the flexible protein regions. (D) Longitudinal sections of a hypothetical model for BcMCM helicase activity based on the steric exclusion model and the active role of the PS1 β-hairpin. A dotted line derived from the EM structures encircles the domain sketches. The BcMCM hexamer would embrace the DNA using the C-terminal regions in the ATP-bound state closing the helicase side aperture. The hydrolysis of the nucleotide would trigger the reorganization of the C-terminal, widening the helicase side and facilitating the translocation of the DNA by the PS1 β-hairpin through the central channel. Concomitantly with the helicase activity the flexible primase–polymerase domain would start primer synthesis (orange fragment). Therefore a unique enzyme might unwind dsDNA, and at the same time use one of the DNA strands as a template for DNA synthesis without the need for generating RNA as an intermediate product.
Based on the superposition with E1 we can model the position of the ATP molecules in the hexameric SsMCM assembly. The observation of the nucleotide sites reveals not only that the EXT R331A is in the vicinity of the nucleotide site, but also that the side channels proposed to be one of the possible DNA extrusion sites (Figure 7A, central and right panels) lie very close to the ATP-binding sites, suggesting that the movement of a displaced DNA strand through this narrow passage could interfere with the hydrolysis of the ATP.
The EM structures of the BcMCM provide snapshots of the conformational changes during different stages of the nucleotide hydrolysis cycle to unwind DNA. First we demonstrate that to form the hexameric assembly BcMCM must bind a nucleotide. This property is similar in other helicases of phage or viral origin (50,51). The presence of the nucleotide induces the oligomerization of the 700-kDa complex that was detected straightforward after negative stain by EM. A comparison between the ATPγS and the ADP structures in the absence of DNA reveals minor differences in the apertures of the hexameric cylinder, the diameter of the central cavity and the subunit interface cavities (Figure 5 and Figure 6A–C). The main difference is the lack of density in the flexible side corresponding to the primase–polymerase domain in the ATPγS-bound structure that suggests an increase in the flexibility in the primase–polymerase domain induced by the non-hydrolysable nucleotide analogue.
However, the conformational changes are larger in the central body when the nucleotide-bound complex is associated with DNA (Figure 6D–I). The bottom views of the DNA-bound BcMCM complexes show that the diameter of the helicase domain aperture undergoes a major rearrangement, narrowing the width of the DNA entrance in the ATP loaded complex. This region involves the C-terminal domain of the protein that has been described as a disordered fragment in the SsMCM (16), but could be rearranged in the presence of DNA in other archaeal MCM (Figure 8B) (19). In contrast when the assembly is loaded with ADP the diameter of the DNA entrance of the helicase moiety is wider. Moreover, the cavities located at the monomer–monomer interface do not display detectable rearrangements. Therefore the presence of the DNA and its interaction with the protein in the ATP loaded state seems to promote the conformational change.
The combination of these structural snapshots suggests that once that the helicase is associated with the DNA and loaded with ATP, the protein embraces the DNA tightly to avoid the sliding of the nucleic acid (Figure 8C). The hydrolysis of the nucleotide would force the DNA through the central channel using at least the PS1 β-hairpin as a mechanical flap to pull the strand in the central cavity, as proposed in the E1 helicase (Figure 8A), (49), towards the primase–polymerase domain region. After ATP hydrolysis the helicase in the ADP-bound state would open the aperture to embrace a new section of the DNA once the ADP nucleotide is exchanged by ATP in the ATPase domain (Figure 8C).
Our structure–function study is a first step to understand the mechanisms of BcMCM during DNA replication. This protein bears helicase, primase and polymerase activities in a single polypeptide, constituting a replicative ‘Swiss army knife’ that could be used in special situations for the bacteria such as reinitiating of leading strand synthesis at stalled replication forks. Therefore the presence of all these three activities in a single protein could have represented an evolving advantage for primitive organisms with small genomes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables SI–IV, Supplementary Figures S1–S9.
FUNDING
Ministerio de Ciencia e Innovación (BFU2008-01344, BFU2011-23815 to G.M., FIS PI080291 to J.B., BFU2010-21467, CSD2007-00015 to J.M. and BFU2009-10085 to L.B.); Basque Government predoctoral fellowships (to J.S.B. and A.I.). Funding for open access charge: Spanish Ministry of Science and Innovation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Jesús Prieto for performing the analytical ultracentrifugation experiment and Prof. Oscar Llorca for the use of the electron microscope and helpful comments, and Prof. J.M. Valpuesta, Dr S. Ramón-Maiques and Dr D. Lietha for discussion.
REFERENCES
- 1.Chong JP, Hayashi MK, Simon MN, Xu RM, Stillman B. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl Acad. Sci. USA. 2000;97:1530–1535. doi: 10.1073/pnas.030539597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shechter DF, Ying CY, Gautier J. The intrinsic DNA helicase activity of Methanobacterium thermoautotrophicum delta H minichromosome maintenance protein. J. Biol. Chem. 2000;275:15049–15059. doi: 10.1074/jbc.M000398200. [DOI] [PubMed] [Google Scholar]
- 3.Niedenzu T, Roleke D, Bains G, Scherzinger E, Saenger W. Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010. J. Mol. Biol. 2001;306:479–487. doi: 10.1006/jmbi.2000.4398. [DOI] [PubMed] [Google Scholar]
- 4.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol. Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 5.Gomez-Llorente Y, Fletcher RJ, Chen XS, Carazo JM, San Martin C. Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J. Biol. Chem. 2005;280:40909–40915. doi: 10.1074/jbc.M509760200. [DOI] [PubMed] [Google Scholar]
- 6.Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelman Z, Lee JK, Hurwitz J. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DeltaH contains DNA helicase activity. Proc. Natl Acad. Sci. USA. 1999;96:14783–14788. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpentieri F, De Felice M, De Falco M, Rossi M, Pisani FM. Physical and functional interaction between the mini-chromosome maintenance-like DNA helicase and the single-stranded DNA binding protein from the crenarchaeon Sulfolobus solfataricus. J. Biol. Chem. 2002;277:12118–12127. doi: 10.1074/jbc.M200091200. [DOI] [PubMed] [Google Scholar]
- 9.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol. Biol. Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Going JJ, Keith WN, Neilson L, Stoeber K, Stuart RC, Williams GH. Aberrant expression of minichromosome maintenance proteins 2 and 5, and Ki-67 in dysplastic squamous oesophageal epithelium and Barrett’s mucosa. Gut. 2002;50:373–377. doi: 10.1136/gut.50.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shetty A, Loddo M, Fanshawe T, Prevost AT, Sainsbury R, Williams GH, Stoeber K. DNA replication licensing and cell cycle kinetics of normal and neoplastic breast. Br. J. Cancer. 2005;93:1295–1300. doi: 10.1038/sj.bjc.6602829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honeycutt KA, Chen Z, Koster MI, Miers M, Nuchtern J, Hicks J, Roop DR, Shohet JM. Deregulated minichromosomal maintenance protein MCM7 contributes to oncogene driven tumorigenesis. Oncogene. 2006;25:4027–4032. doi: 10.1038/sj.onc.1209435. [DOI] [PubMed] [Google Scholar]
- 13.Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat. Struct. Biol. 2003;10:160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, Pucci B, Rossi M, Pisani FM, Ladenstein R. Structural analysis of the Sulfolobus solfataricus MCM protein N-terminal domain. Nucleic Acids Res. 2008;36:3235–3243. doi: 10.1093/nar/gkn183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bae B, Chen YH, Costa A, Onesti S, Brunzelle JS, Lin Y, Cann IK, Nair SK. Insights into the architecture of the replicative helicase from the structure of an archaeal MCM homolog. Structure. 2009;17:211–222. doi: 10.1016/j.str.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadia M, Klein MG, Chen XS. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc. Natl Acad. Sci. USA. 2008;105:20191–20196. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pape T, Meka H, Chen S, Vicentini G, van Heel M, Onesti S. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4:1079–1083. doi: 10.1038/sj.embor.7400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YJ, Yu X, Kasiviswanathan R, Shin J-H, Kelman Z, Egelman EH. Structural polymorphism of Methanothermobacter thermautotrophicus MCM. J. Mol. Biol. 2005;346:389–394. doi: 10.1016/j.jmb.2004.11.076. [DOI] [PubMed] [Google Scholar]
- 19.Costa A, Pape T, van Heel M, Brick P, Patwardhan A, Onesti S. Structural basis of the Methanothermobacter thermautotrophicus MCM helicase activity. Nucleic Acids Res. 2006;34:5829–5838. doi: 10.1093/nar/gkl708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa A, van Duinen G, Medagli B, Chong J, Sakakibara N, Kelman Z, Nair SK, Patwardhan A, Onesti S. Cryo-electron microscopy reveals a novel DNA-binding site on the MCM helicase. EMBO J. 2008;27:2250–2258. doi: 10.1038/emboj.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenkinson ER, Costa A, Leech AP, Patwardhan A, Onesti S, Chong JP. Mutations in subdomain B of the minichromosome maintenance (MCM) helicase affect DNA binding and modulate conformational transitions. J. Biol. Chem. 2009;284:5654–5661. doi: 10.1074/jbc.M806973200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGeoch AT, Bell SD. Eukaryotic/archaeal primase and MCM proteins encoded in a bacteriophage genome. Cell. 2005;120:167–168. doi: 10.1016/j.cell.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 24.Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGeoch AT, Bell SD. Eukaryotic/archaeal primase and MCM proteins encoded in a bacteriophage genome. Cell. 2005;120:167–168. doi: 10.1016/j.cell.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 26.Lipps G, Weinzierl AO, Scheven,von G, Buchen C, Cramer P. Structure of a bifunctional DNA primase-polymerase. Nat. Struct. Mol. Biol. 2004;11:157–162. doi: 10.1038/nsmb723. [DOI] [PubMed] [Google Scholar]
- 27.Frick DN, Richardson CC. DNA primases. Annu. Rev. Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 28.Panuska JR, Goldthwait DA. A DNA-dependent ATPase from T4-infected Escherichia coli. Purification and properties of a 63,000-dalton enzyme and its conversion to a 22,000-dalton form. J. Biol. Chem. 1980;255:5208–5214. [PubMed] [Google Scholar]
- 29.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 30.Sorzano CO, Marabini R, Velazquez-Muriel J, Bilbao-Castro JR, Scheres SH, Carazo JM, Pascual-Montano A. XMIPP: a new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Samuels M, Gulati G, Shin J-H, Opara R, Mcsweeney E, Sekedat M, Long S, Kelman Z, Jeruzalmi D. A biochemically active MCM-like helicase in Bacillus cereus. Nucleic Acids Res. 2009;37:4441–4452. doi: 10.1093/nar/gkp376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell. Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 33.Cavanaugh NA, Kuchta RD. Initiation of new DNA strands by the herpes simplex virus-1 primase-helicase complex and either herpes DNA polymerase or human DNA polymerase alpha. J. Biol. Chem. 2009;284:1523–1532. doi: 10.1074/jbc.M805476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmes AM, Cheriathundam E, Bollum FJ, Chang LM. Initiation of DNA synthesis by the calf thymus DNA polymerase-primase complex. J. Biol. Chem. 1985;260:10840–10846. [PubMed] [Google Scholar]
- 35.Parker WB, Cheng YC. Inhibition of DNA primase by nucleoside triphosphates and their arabinofuranosyl analogs. Mol. Pharmacol. 1987;31:146–151. [PubMed] [Google Scholar]
- 36.Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl Acad. Sci. USA. 2009;106 doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol. Biol. Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toth EA, Li Y, Sawaya MR, Cheng Y, Ellenberger T. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol. Cell. 2003;12:1113–1123. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- 39.Brewster AS, Slaymaker IM, Afif SA, Chen XS. Mutational analysis of an archaeal minichromosome maintenance protein exterior hairpin reveals critical residues for helicase activity and DNA binding. BMC Mol. Biol. 11:62. doi: 10.1186/1471-2199-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat. Struct. Mol. Biol. 2005;12:756–762. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 41.Frank EG, Woodgate R. Increased catalytic activity and altered fidelity of human DNA polymerase iota in the presence of manganese. J. Biol. Chem. 2007;282:24689–24696. doi: 10.1074/jbc.M702159200. [DOI] [PubMed] [Google Scholar]
- 42.Domínguez O, Ruiz JF, Laín de Lera T, García-Díaz M, González MA, Kirchhoff T, Martínez-A C, Bernad A, Blanco L. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19:1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pitcher RS, Brissett NC, Picher AJ, Andrade P, Juarez R, Thompson D, Fox GC, Blanco L, Doherty AJ. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J. Mol. Biol. 2007;366:391–405. doi: 10.1016/j.jmb.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 44.Gabbai CB, Marians KJ. Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair. 2010;9:202–209. doi: 10.1016/j.dnarep.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JK, Hurwitz J. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl Acad. Sci. USA. 2001;98:54–59. doi: 10.1073/pnas.98.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaplan DL, Davey MJ, O’Donnell M. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J. Biol. Chem. 2003;278:49171–49182. doi: 10.1074/jbc.M308074200. [DOI] [PubMed] [Google Scholar]
- 47.Laskey RA, Madine MA. A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. EMBO Rep. 2003;4:26–30. doi: 10.1038/sj.embor.embor706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Méndez J, Stillman B. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays. 2003;25:1158–1167. doi: 10.1002/bies.10370. [DOI] [PubMed] [Google Scholar]
- 49.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 50.Hingorani MM, Patel SS. Cooperative interactions of nucleotide ligands are linked to oligomerization and DNA binding in bacteriophage T7 gene 4 helicases. Biochemistry. 1996;35:2218–2228. doi: 10.1021/bi9521497. [DOI] [PubMed] [Google Scholar]
- 51.Dean FB, Borowiec JA, Eki T, Hurwitz J. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J. Biol. Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.