Abstract
The Leishmania tarentolae Parrot-TarII strain genome sequence was resolved to an average 16-fold mean coverage by next-generation DNA sequencing technologies. This is the first non-pathogenic to humans kinetoplastid protozoan genome to be described thus providing an opportunity for comparison with the completed genomes of pathogenic Leishmania species. A high synteny was observed between all sequenced Leishmania species. A limited number of chromosomal regions diverged between L. tarentolae and L. infantum, while remaining syntenic to L. major. Globally, >90% of the L. tarentolae gene content was shared with the other Leishmania species. We identified 95 predicted coding sequences unique to L. tarentolae and 250 genes that were absent from L. tarentolae. Interestingly, many of the latter genes were expressed in the intracellular amastigote stage of pathogenic species. In addition, genes coding for products involved in antioxidant defence or participating in vesicular-mediated protein transport were underrepresented in L. tarentolae. In contrast to other Leishmania genomes, two gene families were expanded in L. tarentolae, namely the zinc metallo-peptidase surface glycoprotein GP63 and the promastigote surface antigen PSA31C. Overall, L. tarentolae's gene content appears better adapted to the promastigote insect stage rather than the amastigote mammalian stage.
INTRODUCTION
Leishmania is an early-branching unicellular eukaryote that belongs to the Kinetoplastida order and the family Trypanosomatidae. Leishmania species are transmitted by the bite of female phlebotomine sand flies as extracellular flagellated metacyclic promastigotes and replicate in mammalian macrophages as intracellular aflagellated amastigotes. Leishmania infections represent a global health problem with 350 million people at risk, an annual incidence of 2 million and an overall prevalence estimated at 12 million people worldwide (1). At least 20 Leishmania species cause a large spectrum of clinical manifestations ranging from self-resolving skin lesions (L. major) to mucocutaneous manifestations (L. braziliensis) reaching to life-threatening visceral diseases (L. donovani/L. infantum).
Through phylogenetic analyses, Leishmania was divided in three distinct subgenera: the Leishmania, the Viannia and the Sauroleishmania (2). The classification of lizard Leishmania as a distinct genus was once debated (3,4) but the molecular evidence did not support this assumption. Leishmania (Sauroleishmania) tarentolae was first isolated from the lizard Tarentola mauritanica in 1921 (5) and is probably the most widely studied Leishmania (Sauroleishmania) species. In lizards, the parasites live predominantly as promastigotes in the lumen of the cloacae and intestine or in the bloodstream (6). Amastigotes, either free or inside monocytes, are rarely observed in lizards (4,6), although both free promastigotes and amastigotes from the blood were reported (7). The ability of L. tarentolae to develop into amastigote forms in the lizard is still debated but, as for several Leishmania species infecting lizards (6), L. tarentolae is able to enter human phagocytic cells and differentiate into amastigote-like forms. However, there is no clear evidence for their efficient replication within macrophages (8–10). Because of its rapid growth in defined media and lack of pathogenicity to humans, L. tarentolae has been used widely as a model organism for studies on gene amplification (11–13) or RNA editing (14). Furthermore, L. tarentolae has been used as a platform for the production of recombinant proteins (15) and as a potential vaccine candidate (9,10).
The genomes of two Leishmania (Leishmania) species, L. major and L. infantum, and one Leishmania (Viannia) species, L. braziliensis, have already been completed and annotated (16,17), and more are being sequenced (www.tritrypdb.org). The 32.8 Mb genome of L. major clone Friedlin spreads over 36 chromosomes and is presently the best annotated Leishmania genome (16). A 5-fold shotgun sequencing was provided for L. braziliensis clone M2904 and L. infantum clone JPCM5. The genomes of the various Leishmania species contain a similar number of genes, estimated at 8200 (Table 1). Despite the 20–100 million years of divergence within the Leishmania genus, a recent sequence comparison of the genomes of L. major, L. infantum and L. braziliensis has revealed a strong conservation of gene content and synteny across the genus (17). Comparative genomics of L. major, L. infantum and L. braziliensis have shown that approximately 200 genes were differentially distributed between the species and that only 78 genes were unique to one species (17). Notable differences were observed in L. braziliensis, which contains a putative RNA interference pathway and two types of transposons (TATES) and retroposons (SLACS) absent in the two other species.
Table 1.
Summary of sequenced Leishmania spp. genomes
| Leishmania tarentolae | Leishmania major (V5.2) | Leishmania infantum (V3a) | Leishmania braziliensis (V2) | |
|---|---|---|---|---|
| Strain | Parrott-TarII | Friedlin | JPCM5 | WHOM/BR/75/M2904 |
| Number of chromosomes | 36 | 36 | 36 | 35 |
| Genome size (bp) | 30 440 719 | 32 816 678 | 32 134 935 | 32 005 207 |
| Overall G+C content (%) | 57.2 | 59.7 | 59.3 | 57.8 |
| Coding G+C content (%) | 58.4 | 62.5 | 62.5 | 60.4 |
| Number of coding genes in database (time of study) | 8201 | 8304 | 8216 | 8133 |
| Number of OGs | 7449 | 7530 | 7506 | 7353 |
| Mean nucleotide identity with L. tarentolae (%) | – | 84.9 | 85.0 | 79.2 |
| Mean amino acid identity with L. tarentolae (%) | – | 81.9 | 82.4 | 74.8 |
| Reference | Current study | (16) | (17) | (17) |
Here, we sequenced and assembled de novo the L. tarentolae strain Parrot-TarII using next-generation high-throughput DNA sequencing technologies. The comparison of the L. tarentolae genome with the published genomes of pathogenic Leishmania species revealed a high degree of synteny. We identified 95 predicted coding sequences unique to L. tarentolae and 250 genes present in the pathogenic species but absent in L. tarentolae. Interestingly, several of the genes lacking from L. tarentolae are expressed preferentially in the intracellular amastigote stage of pathogenic species. This may explain in part why L. tarentolae is less well adapted to infect human macrophages and why these parasites are mostly reported as free organisms in the lizards.
MATERIALS AND METHODS
Sample preparation and sequencing
Promastigotes of L. tarentolae (strain Parrot-TarII) were grown up to the late log phase in SDM-79 (Schneider's Drosophila medium) supplemented with 5 µg/ml hemin and 5% heat-inactivated fetal calf serum (FCS) (Multicell, Wisent Inc.). High molecular weight genomic DNA was extracted from parasites after chemical lysis with SDS (1%) followed by two sequential phenol extractions, proteinase K and RNase A treatments. Chromosomal DNA was further purified on cesium chloride gradients to limit the extent of kinetoplastid DNA inclusion (18). Purified L. tarentolae genomic DNA was sequenced with next-generation sequencing technologies using paired (2.5 kb inserts) and unpaired GS-FLX or Titanium sequencing procedures (Roche 454). GS-FLX and Titanium sequencing were performed at the McGill University and Génome Québec Innovation Centre, Montréal, Canada. Genomic sequences using unpaired Solexa/Illumina sequencing were performed at the Netherlands Cancer Institute. Sequencing runs are described in Supplementary Table S1.
Assembly
Sequences produced on the GS-FLX and Titanium sequencers were assembled using Newbler 2.0 (Roche). The N50 scaffold length of the L. tarentolae assembly was 61 619 bp. Sequences produced by Solexa/Illumina were assembled with the Velvet version 0.7.55 software (19). Additional validation of the assembly was performed using our own de novo assembler Ray (20). The assemblies were merged using the minimus2 scaffolder found in the AMOS package version 2.0.8 (21). Chromosome-like scaffolds were created by comparing gene order in the de novo assembled scaffolds and contigs to the gene order in the version 5.2 of the reference L. major genome using BLAST and custom python scripts.
Annotation
Gene identification was performed by comparing the assembled L. tarentolae genome sequence to the putative genes of L. infantum, L. major and L. braziliensis using BLAST 2.2.21 (22). Sequences from the assembly were compared to the coding sequences (CDS) of the three other species and all L. tarentolae open reading frames (ORFs) larger than 100 nucleotides were translated and compared to the gene sequences of the three other species. Similar analysis was also performed on T. cruzi and T. brucei. Another set of genes was predicted with Augustus using evidence from protein homology to Trypanosomatidae and an ab initio model trained for L. tarentolae (23–25). A consensus set of genes was compiled by pooling the two annotation sets, which were filtered in order to identify the most probable genes in the L. tarentolae sequence. All putative genes were mined for domain structures and motifs using HMMER 2.0 and the pfam database (26,27). Further gene ontology (GO) analyses were performed using Blast2GO (28). Additional phylogenetic analyses were conducted in MEGA4 (29). Synteny maps were drawn using the R software (http://www.r-project.org/) based on chromosome comparisons with BLAST (blastn). The genome sequence and annotation of L. tarentolae Parrot-TarII is available on TriTrypDB (www.tritrypdb.org).
Orthologous group identification
Putative L. tarentolae genes were compared to genes from other sequenced Leishmania species using the OrthoMCL 2.0 web tool (30,31). Genes from L. major, L. infantum and L. braziliensis have already been clustered in groups of orthologs, which are publicly available in the OrthoMCL database (30). Each group of orthologous genes contains orthologs (related genes found in different species) and paralogs (related genes found within a species). Orthologous groups (OGs) may include whole gene families, subfamilies or single genes, depending on the extent and variability of the gene family. Genes qualified as absent from L. tarentolae were validated manually by searching the sequence of this gene in the original reads and by assembling the matching reads using CAP3 (32).
Validation of copy number variation
Reads were mapped onto the L. tarentolae scaffolds using the BWA software (33). The number of reads corresponding to each nucleotide position was calculated and the mean read coverage for each gene was estimated. For each OG, the read coverages of all genes were summed to estimate the total coverage for each OG. The total read coverage of OG were compared to the number of genes in the OG. Orthologous groups for which copy number variation were observed between L. tarentolae and the other species were manually inspected to validate that total coverage of OG confirmed the number of genes.
Southern blot hybridizations
Promastigotes of L. tarentolae strain Parrot-TarII, L. tarentolae strain S125, L. major strain Friedlin, L. infantum strain JPCM5 and L. braziliensis strain WHOM/BR/75/M2904 were grown at 25°C in SDM-79 medium supplemented with 10% heat-inactivated FCS and 5 µg/ml of hemin. Total DNA from each culture was prepared with DNAzole (Invitrogen), digested with XhoI restriction enzyme (New England Biolabs, Pickering, ON, Canada) and run-on standard agarose gels. Southern blot analyses with [α-32P]-dCTP labelled DNA probes were performed according to standard protocols (18). All probes were generated by polymerase chain reaction using primer sets listed in Supplementary Table S2. For each target, we generated a specific probe for each Leishmania species, which were co-hybridized on a blot of total digested genomic DNAs. Equal amount of DNA was layered for each strain and monitored by hybridization with the single copy PTR1 gene.
Comparative genomic hybridization
Leishmania whole-genome DNA microarrays used in CGH experiments, which included 8100 60-mer probes that were designed to hybridize all genes of L. major 5.2 and L. infantum 3.0a, were obtained from Agilent Technologies (Mississauga, ON, Canada). Microarray platform details and probe sequences were deposited in the GEO database under the accession GPL11330. Sample preparation, pre-hybridization and hybridization steps were performed as previously described (34). Normalization and data analysis were done in R with LIMMA 2.7.3. (35). Multiple testing correction was done using the false discovery rate (FDR) method and probes were considered significant when P < 0.05 and log-2 ratio > 2. The entire data set was deposited in GEO under the reference number series GSE27184.
H2O2 IC50 assay
Promastigotes of L. major LV39, L. infantum MHOMMA#67#ITMAP-263 and L. tarentolae S125 were grown at pH 7.0 and 25°C in SDM-79 medium supplemented with 10% fetal bovine serum, 5 μg/ml hemin and 5 μM biopterin (Sigma-Aldrich, St-Louis, MO). Leishmania promastigotes (5 × 106) were inoculated in 5 ml medium and H2O2 (Rougier Pharma, Mirabel, QC, Canada) was added at various concentration (100 µM, 250 µM, 500 µM, 1000 µM, 2500 µM and 5000 µM). OD600nm values were taken after 72 h. Each curve was performed in triplicate. IC50 were calculated for each curve and the mean and 95% confidence intervals were calculated.
RESULTS
Leishmania tarentolae genome sequencing
The genome of L. tarentolae strain Parrot-TarII was resolved using high-throughput sequencing technologies (Supplementary Table S1) to a 16-fold mean coverage and 23-fold peak coverage. The assembled L. tarentolae genome contains a total of 30 440 719 bases with 95.1% of the GS-FLX and Titanium reads found in 773 scaffolds and the remaining 4.9% distributed in 2499 contigs. Reads obtained by Illumina sequencing were also incorporated in the final sequence to assist in the assembly. After de novo assembly, sequences of specific chromosomes were built using contigs and scaffolds based on their homology to L. major. Directed assembly on L. major allowed the mapping of 29 862 062 bases (98.1%) leaving 578 657 bases (1.9%) in 1315 small contigs. A summary of the sequencing statistics of the L. tarentolae genome and the other published Leishmania spp. genomes are presented in Table 1. CHEF analysis of the L. tarentolae genome showed 24–28 chromosomal bands (36). Similar analysis of the L. infantum genome showed a comparable number of band, which were further resolved in 36 chromosomes using hybridization of genome fragments (37).
Leishmania tarentolae is syntenic with other Leishmania species
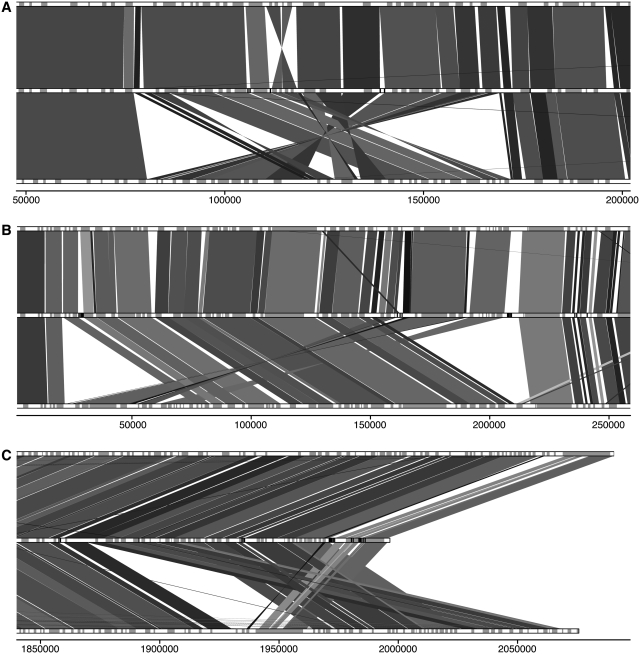
De novo assembled contigs and scaffolds were generally syntenic with both L. major and L. infantum. Most differences between L. tarentolae and the other species consisted in gene insertions or deletions distributed randomly or in tandem arrays throughout the genome. Full synteny maps comparing L. tarentolae to L. infantum and L. major are provided in Supplementary Figures S1–S36. Although L. tarentolae possesses similar percent identity with L. infantum and L. major (Table 1), its synteny is closer to the latter. For example on chromosome 28, a stretch of 90 kb is syntenic between L. tarentolae and L. major but the gene ordering is different in L. infantum (Figure 1A). De novo assembled scaffolds are, in this case, long enough to confirm that L. tarentolae is more syntenic to L. major. Similar differences were found at the proximal region of chromosome 7 (Figure 1B) and at the distal end of chromosome 35 (Figure 1C), both suggesting greater synteny to L. major than to L. infantum. Other loci where L. tarentolae de novo assembled scaffolds were syntenic to L. major but not to L. infantum can be found in Supplementary Figures S7, S9, S11–13, S32 and S35. Overall, the synteny of L. tarentolae is closer to the L. (Leishmania) species than to the L. (Viannia) subgenus. Indeed, several loci are in common between L. tarentolae and L. major, although they are absent from L. braziliensis. Breaks in synteny are also more frequent with L. braziliensis than L. major (data not shown).
Figure 1.
Synteny map of L. tarentolae (middle) compared to L. major (top) and L. infantum (bottom). Genes on chromosome tracks are grey. Leishmania tarentolae contig delimitation is indicated in black in the middle lane. Shade of synteny blocks is proportional to sequence identity, the darker the more similar are the sequences. The scale represents nucleotide position on the chromosome. (A) 5′ region of chromosome 28. (B) 5′ region of chromosome 7. (C) 3′-end of chromosome 35.
Leishmania tarentolae gene content
Genome annotation of L. tarentolae indicated a total of 8201 putative protein-coding genes, a number similar to the other sequenced Leishmania species (Table 1). Annotation was performed by comparing the L. tarentolae genome to other Leishmania and Trypanosoma species along with ab initio annotation using the Augustus software trained for Leishmania gene detection (23–25). Given that assembly of repeated gene clusters is more difficult, the count of genes found in L. tarentolae may be biased, especially for genes present in high copy number.
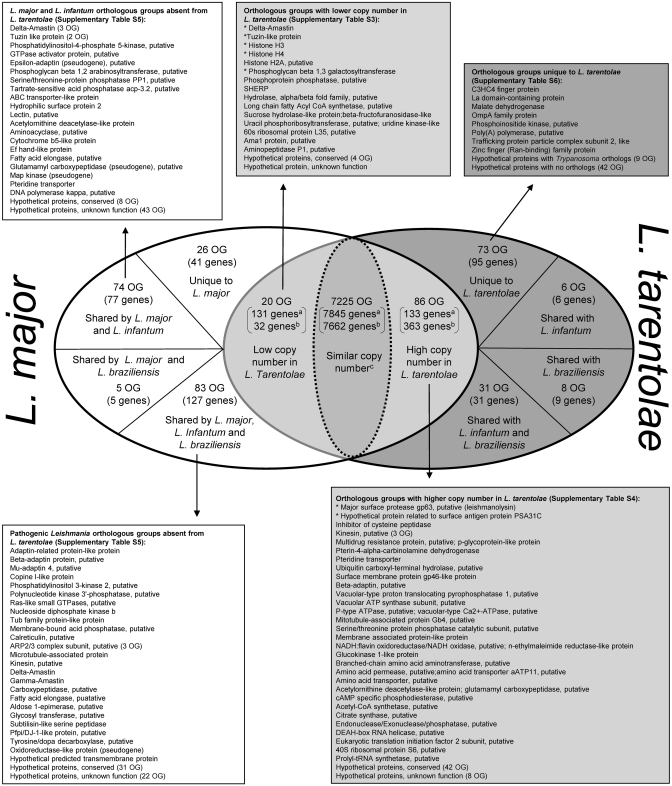
Using the OrthoMCL web tool, the set of L. tarentolae putative genes was compared to the OrthoMCL database (www.orthomcl.org) in order to assign each gene to a group of orthologs (30). This allowed us to readily compare the gene content of the four sequenced Leishmania species, determine which genes are unique to a given species and calculate which OG of genes vary in copy number between the different species. These results were further confirmed by interspecies comparative genomics hybridization (CGH) microarrays, read depth analysis and, in selected cases, by Southern blots analyses. The gene content of L. tarentolae is highly similar to the three pathogenic Leishmania species sequenced, which contain a similar number of OG (Table 1).
Figure 2 compares the gene content of L. tarentolae to other Leishmania species with emphasis on L. major. Overall, 7331 OG are shared by L. tarentolae and L. major. Of these, 7225 OG (7662 genes in L. tarentolae and 7845 genes in L. major) have a similar copy number in L. tarentolae and at least another Leishmania species, 20 OG (32 genes in L. tarentolae and 131 genes in L. major) have a lower copy number in L. tarentolae than the three other species (Supplementary Table S3) and 86 OG (363 genes in L. tarentolae and 133 in L. major) have a higher copy number in L. tarentolae than the three other species (Supplementary Table S4). More than a third of the OGs with varying copy numbers have a putative function (see Figure 2 and Supplementary Tables S3 and S4).
Figure 2.
Differential distribution of genes and OGs of genes between L. tarentolae and L. major. aGene counts referring to L. major. bGene counts referring to L. tarentolae. Lists include the description of OGs of genes that have differential distribution between L. tarentolae and the three sequenced Leishmania pathogenic species. Counts of genes within the different OG are in parenthesis. The complete list of genes and OGs for selected categories is shown in Supplementary Tables S3–S6. Asterisk indicates genes with the highest copy number variability in L. tarentolae.
A total of 250 L. major genes distributed between 188 OGs were found to be absent from L. tarentolae (Figure 2 and Supplementary Table S5). Of these, 83 OG were shared by the three pathogenic species, 74 OG by L. major and L. infantum, 5 OG by L. major and L. braziliensis and 26 OG were unique to L. major (Figure 2).
A total of 73 OG (95 genes) were unique to L. tarentolae (Supplementary Table S6). From these, 31 OG had orthologs in other non-Leishmania species, including 29 in Trypanosoma spp. We also found 42 OG (65 genes) that were sequence orphans with no similarity to sequences found in databases.
Genes absent from L. tarentolae or present in lower copy number compared to the pathogenic Leishmania spp
Leishmania tarentolae lacks several genes coding for proteins implicated in trafficking. Indeed, the β1/β2-adaptins (LmjF11.0990; LmjF36.5595), μ-adaptin (LmjF31.3035) and the epsilon–adaptin (LmjF30.1545) (Figure 2 and Supplementary Table S5) were absent from L. tarentolae. Adaptins are involved in the formation of clathrin-associated adaptor protein (AP) complexes, which play a key role in the transport of proteins by regulating the formation of transport vesicles as well as cargo selection between the trans-Golgi network, endosomes, lysosomes and the plasma membrane (38,39). The calcium-dependent membrane binding proteins copines (LmjF28.1190) and Ras-like small GTP-binding proteins (LmjF36.1820), both involved in cell signalling and/or membrane trafficking/transport pathways and exocytosis, were also absent from L. tarentolae. The endosomal/lysosomal membrane-bound acid phosphatase (LmjF28.2650), potentially involved in intracellular trafficking (40), is also missing from L. tarentolae. Leishmania tarentolae also lacks the phosphatidylinositol 3-kinase 2 gene (LmjF14.0020) and the phosphatidylinositol-4-phosphate 5-kinase gene (LmjF26.2495) (Figure 2 and Supplementary Table S5) whose activities were linked to a diverse set of key cellular functions, including intracellular trafficking (41). The Tubby protein 1, that has been reported to bind phosphatidylinositol 4,5-bisphosphate on the plasma membrane and facilitate macrophage phagocytosis (42), is not present in L. tarentolae.
Leishmania tarentolae lacks three of the seven subunits of the Arp2/3 complex, notably the p40/ARPC1 (LmjF18.0920) p20/ARPC4 (LmjF02.0600) and p15/ARPC5 (LmjF05.0285) (Figure 2 and Supplementary Table S5). Only one Arp2/3 complex subunit coding gene was found and annotated in the L. tarentolae genome (not shown). The Arp2/3 complex is required for actin polymerization and reported to associate with several other cytoskeletal components, including microtubules (43,44). Interestingly, a number of microtubule-associated proteins (9 on chromosome 19) and a kinesin microtubule-associated protein (LmjF16.1580) are also missing from the L. tarentolae genome (Figure 2 and Supplementary Table S5).
Genes reported to play a role in the resistance to oxidative stress are absent from L. tarentolae. These include a subtilisin (LmjF28.2380) belonging to the S8A subfamily of proteases, shown recently to process the terminal peroxidases of the trypanothione reductase system in Leishmania (45); a Pfpi/DJ-1-like protein (LmjF35.3910) that defends cells against reactive oxygen species and mitochondrial damage (46); and a tyrosine/dopa decarboxylase (LmjF30.2500), a precursor of catecholamines, which may act as scavengers of free radicals (47) (Figure 2 and Supplementary Table S5). Collectively, these data suggest that L. tarentolae may deal less efficiently with oxidants than L. major. Interestingly, L. tarentolae strains were found to be >3.5-fold more sensitive to hydrogen peroxide than L. major and L. infantum. Indeed, the IC50 of L. tarentolae to H2O2 was 152 ± 7 µM but 514 ± 115 µM for L. major and 574 ± 121 µM for L. infantum.
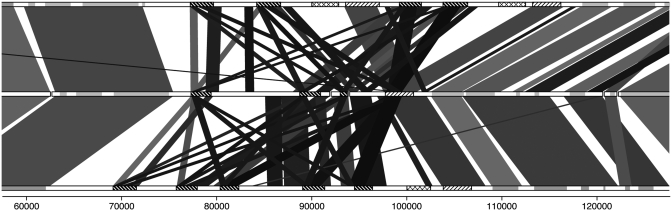
Gene content analysis also revealed differences in the glycoprotein content between L. tarentolae and the other pathogenic species. Genes involved in lipophosphoglycan (LPG) modifications were found to be either in lower copy number or absent from L. tarentolae. Chromosome 2 displayed important differences between L. tarentolae and the two other sequenced Old World species in terms of phosphoglycan transferases (Figure 3). This region extends on two de novo assembled scaffolds in L. tarentolae, providing reliable information on this locus. We found that phosphoglycan β 1,3 galactosyltransferase OG of genes necessary for the attachment of the side chain Gal residues to the LPG phosphoglycan (PG) repeats (48) are present in 6–8 copies in other Leishmania species but only in 2 copies in L. tarentolae (Supplementary Table S3). Within the same locus, phosphoglycan β 1,2 arabinosyltransferase (LmjF02.0180; LmjF02.0220), involved in the terminal capping oligosaccharide of LPG in L. major (49), is found in L. major and L. infantum but not in L. tarentolae (Supplementary Table S5). Leishmania tarentolae also lacks two glycosyltransferase genes (LmjF35.5250, LmjF29.2110), enzymes that catalyze the transfer of a monosaccharide unit to a glycosyl acceptor molecule. Moreover, L. tarentolae lacks calreticulin (LmjF31.2600), an endoplasmic reticulum (ER) chaperone ensuring the proper folding and quality control of newly synthesized glycoproteins destined for secretion or cell surface expression (50) (Supplementary Table S5).
Figure 3.
Differential distribution of genes involved in lipophosphoglycan and phosphoglycan modification in L. tarentolae (middle) as compared to L. major (top) and L. infantum (bottom). Phosphoglycan β 1,2 arabinosyltransferase are shaded with crosses, a first group of phosphoglycan β 1,3 galactosyltransferase is shaded with thin lines and a second group with bold lines. The other genes on chromosome tracks are grey. Leishmania tarentolae contig delimitation is in black in the middle lane. Shade of synteny blocks is proportional to sequence identity, the darker the more similar are the sequences. The scale represents nucleotide position on chromosome 2.
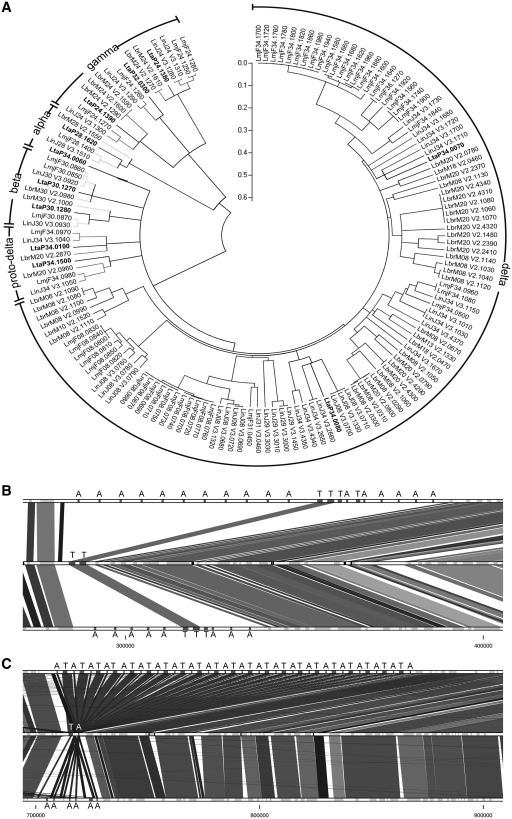
Amastins, one of the largest family of surface proteins in Leishmania (17) shown to be expressed primarily in the intracellular stage of the parasite (51,52), are underrepresented in L. tarentolae (Supplementary Table S3). Amastins, whose function has yet to be determined, are divided into four subfamilies based on their phylogeny and genomic positioning (53). We show that L. tarentolae contains the amastins of the α, β and γ subfamilies that are also shared with Trypanosoma or Crithidia, but lacks all but two of the amastins of the delta subfamily (Figure 4A). This subfamily is expressed preferentially in the intracellular stage of the parasite (51). No new amastins were discovered in L. tarentolae. Interestingly, tuzins, which are often associated on the same chromosomal locus with amastin genes in the pathogenic Leishmania species, were less diverse in L. tarentolae with only 4 tuzin genes compared to 6 and 28 genes in L. infantum and L. major, respectively. The amastin- and tuzin-rich region within chromosome 8 contains only two copies of the tuzin gene in L. tarentolae (Figure 4B). On L. tarentolae chromosome 34, amastin and tuzin genes are present in single copy as opposed to the situation in L. major where they are organized in tandem, suggesting co-expression (Figure 4C).
Figure 4.
Analysis of amastin-coding genes in pathogenic and non-pathogenic Leishmania spp. (A) Phylogenetic tree of the amastin genes in L. tarentolae, L. major, L. infantum and L. braziliensis. Labels refer to amastin subfamilies. L. tarentolae amastins are in bold. The evolutionary history was inferred using the Neighbor-Joining method, with a bootstrap test of 500 replicates. Phylogenetic analyses were conducted in MEGA4 (29). (B) Synteny of L. major (top), L. tarentolae (middle) and L. infantum (bottom) amastin/tuzin cluster located on chromosome 8. Amastins are marked with the letter A and tuzins with the letter T. (C) Synteny of L. major (top), L. tarentolae (middle) and L. infantum (bottom) amastin/tuzin cluster located on chromosome 34.
Gene family expansion in Leishmania tarentolae
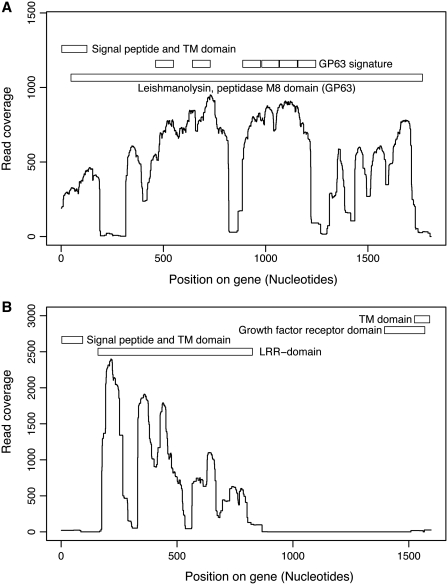
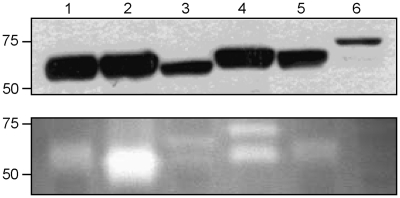
Genome comparative analysis between the L. tarentolae and the other sequenced Leishmania pathogenic species revealed that two surface-associated protein gene families were particularly enriched in the non-pathogenic L. tarentolae. The first family is leishmanolysin, a major surface zinc metalloprotease, also known as GP63; which is mostly, but not exclusively, expressed in the promastigote stage of several Leishmania species (54–56). The GP63 OG in L. tarentolae is highly expanded with 49 putative GP63 genes as compared to 29 in L. braziliensis, 7 in L. infantum and 5 in L. major (Supplementary Table S4). Due to the high copy number and sequence variability, assembly of the GP63 gene family was limited and resulted in several partial gene sequences. The density of read coverage for GP63 indicated portions of the gene that were well conserved in L. tarentolae while other regions were less represented, supporting a sequence diversity of GP63 in L. tarentolae (Figure 5A). Sequence diversity in the L. tarentolae GP63 gene may affect its function. Indeed, a gelatin zymography assay showed no protease activity in L. tarentolae, in contrast to the pathogenic Leishmania species (Figure 6).
Figure 5.
Density of read coverage for genes present in high copy number in L. tarentolae. For each position of the reference L. major genes, the number of corresponding reads were counted and plotted on the graph. Protein domains are indicated on the upper portion of each graph. (A) Leishmanolysin (GP63) gene; LmjF10.0480 is used as a reference. (B) Promastigote surface antigen PSA31C gene; LmjF31.1440 is used as a reference.
Figure 6.
Protease activity of GP63 in six Leishmania species. (A) Western blot using monoclonal antibody targetting GP63 shows the quantity of this protease in each sample. (B) Gelatin zymography assay determining the protease activity of GP63. No signal was observed for L. tarentolae, suggesting the absence of GP63 activity in this species. Lane 1, L. mexicana; lane 2, L. major; lane 3, L. donovani; lane 4, L. infantum; lane 5, L. amazonensis; and lane 6, L. tarentolae.
The second surface protein gene family that was expanded in L. tarentolae is distantly related to the promastigote surface antigen proteins (PSA) located on chromosome 31. PSA-2 was originally described in 1989 (57). The phylogenetic history of this gene family has recently been studied and members have been assigned to subfamilies based on their sequences and chromosomal locations (58). We observed that the L. tarentolae PSA genes are orthologous to PSA31C. There are 63 paralogs in L. tarentolae, while only a single copy is present in L. major and L. infantum and two copies in L. braziliensis (Supplementary Table S4). Leishmania tarentolae PSA proteins seem to contain only the leucine-rich repeated motif (LRR) (Figure 5B), a domain often implicated in the binding to other proteins or glycolipids. This gene is proposed to be involved in host–pathogen interactions (58).
Smaller copy number variations were observed for 84 additional OGs, which are shown in Supplementary Table S4.
Genes unique to Leishmania tarentolae
We found 73 OG (95 genes) unique to L. tarentolae (Figure 2 and Supplementary Table S6). Of these groups, 31 (42%) had OrthoMCL orthologs in species other than L. infantum, L. major and L. braziliensis, including Trypanosoma spp. (26 groups; 36%). Putative functions could be ascribed to 10 OG (14%). These include a C3HC4 finger protein, a trafficking protein particle complex subunit 2-like protein, a La domain-containing protein, a malate dehydrogenase, an OmpA family protein, a phosphoinositide kinase, two surface protein GP63 OGs, a zinc finger (Ran-binding) family protein and a poly(A) polymerase, which is different from the poly(A) polymerases found in the pathogenic Leishmania. The remaining OG are sequence orphans with no orthologs found in the orthoMCL database.
Experimental validation for gene content and copy number variations
We validated experimentally some of the changes highlighted from the genome sequence comparisons using comparative genomic hybridizations (CGH), Southern blot and read depth analysis. Full Leishmania genome microarrays designed for L. major and L. infantum were used to co-hybridize L. tarentolae with either the L. major or the L. infantum DNA to test whether we could identify genes specific to pathogenic species or genes with altered copy number. We found that 31–35% of the genes absent in L. tarentolae showed significantly higher signals with the DNA derived from the pathogenic species, which is statistically significant when compared to the overall ratio of 8% due to better hybridization of the DNAs derived from the pathogens (P < 0.001, one sample test for binomial proportion). Similarly, 20% of the L. tarentolae genes with lower copy number were validated by the CGH experiment (P < 0.001) confirming in part the sequencing data. These correlations are explained by the design of the microarray that contains several probes (45%) that have more than 10 mismatches with L. tarentolae, and by the fact that this microarray did not allow testing for the presence of L. tarentolae-specific genes. Considering these limitations, the CGH results allowed the validation of many differentially distributed genes between the Leishmania species. However, genes that are not validated by CGH may still be differentially distributed between the species. Genes confirmed by CGH are highlighted in Supplementary Tables S3 and S5.
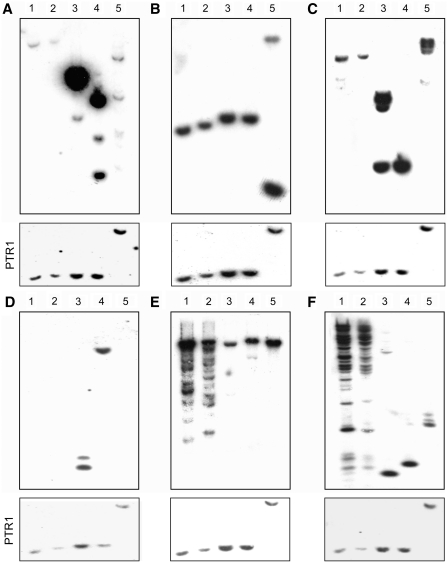
The correlation between sequence data and gene copy number was validated by Southern blot hybridizations for a few chosen genes (Figure 7). The delta-amastin subfamily is present in high copy number in L. major and L. infantum but not in L. tarentolae (Supplementary Table S3). The low copy number of delta-amastins in the TarII-Parrot L. tarentolae strain predicted from the sequencing data and their high copy number in the pathogenic species were confirmed by Southern blot hybridization (Figure 7A, lanes 3–5). Indeed the intense hybridization signals in the pathogenic species were due to the highly repetitive nature of a cluster of amastins giving rise to single restriction fragments (see Figure 7A). Interestingly, the copy number of delta-amastins was low not only in the TarII-Parrot strain (Figure 7A, lane 1) cultured for decades in the laboratory, but also in a more recent isolate from lizard with a limited history of in vitro culture passages (Figure 7A, lane 2). As a control, we used a probe recognizing the proto-delta amastin gene, which according to the sequence data is present in similar copy numbers in all species. Southern blots have indeed validated this assumption (Figure 7B).
Figure 7.
Southern blot hybridization of genes whose copy number varies between L. tarentolae and the other Leishmania pathogenic species. Total genomic DNA of Leishmania isolates was digested with XhoI, run on agarose gels, blotted and hybridized with a combination of PCR-specific probes derived from each species (see Supplementary Table S2 for primer sequences and probe details). (A) Delta-amastin; (B) Proto-delta-amastin (shared by the four species) as a control; (C) Phosphoglycan β 1,3 galactosyltransferase; (D) Phosphoglycan β 1,2 arabinosyltransferase; (E) GP63; and (F) Surface antigen protein PSA31C. DNA loading was estimated by hybridizing the same blot with the single copy gene PTR1 (shown in the lower portion of each panel). Lanes 1, L. tarentolae Parrott-TarII; lane 2, L. tarentolae S125; lane 3, L. major Friedlin; lane 4, L. infantum JPCM5; lane 5, L. braziliensis WHOM/BR/75/M2904.
In an independent set of experiments, Southern blot hybridizations corroborated sequencing data for the two genes involved in LPG side chain addition and LPG modification, indicating that phosphoglycan β 1,3 galactosyltransferase was present in lower copy number in L. tarentolae (Figure 7C) whereas phosphoglycan β 1,2 arabinosyltransferase was absent from this species (Figure 7D). Also, Southern blot analyses confirmed the expansion of the GP63 and PSA31C protein gene families in L. tarentolae as deduced from the sequencing data (Figure 7E and 7F). The heterogeneity in the hybridizing fragments gives some credence to the size diversity of GP63 (Figure 5A) and PSA31C (Figure 5B) genes, as suggested by the sequencing data. Moreover, both gene families have a high read coverage, 3413 reads per nucleotide for GP63 and 7280 reads per nucleotide for PSA31C, which supports the idea that these genes are in high copy number (Supplementary Table S4). Read depth analysis also supports the higher copy number of genes listed in Supplementary Table S4 and the lower copy number of genes listed in Supplementary Table S3. The mean read coverage for single-copy genes was 22.4 read/nucleotide.
Orthologous groups of genes varying in copy number or missing from L. tarentolae are developmentally regulated in pathogenic Leishmania species
We show here that some genes preferentially expressed in the amastigote stage, such as the amastins, are present in lower copy number or are absent from L. tarentolae, while genes reputed to have a higher expression in the insect stage (e.g. GP63 or PSA31C) are represented in higher copy number in L. tarentolae (Figure 7E and 7F). We performed additional bioinformatics analyses to test whether OGs of genes with a variable copy number between L. tarentolae and the pathogenic species were stage-regulated in the later. Specific expression of these groups to Leishmania's life cycle stages was based on published transcriptomics and proteomics studies (59–71). Overall, 23% of the 7694 OG were associated to one or the other developmental stage of the parasite with 890 OG associated to the promastigote stage and 1013 OG associated to the amastigote stage (Table 2). Association to either developmental stage is indicated in Supplementary Tables S3–S5. Comparison of each category to overall distribution was performed using the Fisher's exact test. Interestingly, 70% (16/23) of the OG with a lower copy number and ∼34% (29/86) of the OG with a higher copy number in L. tarentolae were differentially expressed in either life stage of the pathogenic Leishmania species (Table 2). Remarkably, OG absent from L. tarentolae were mainly associated to the amastigote life stage in other Leishmania (P < 0.001). Indeed, of the 66 OGs present in pathogenic Leishmania spp. but absent from L. tarentolae that were associated to one or the other developmental stage of the parasite, 18 genes were linked to the promastigote life stage, 52 to the amastigote stage and 4 to both life stages (Table 2).
Table 2.
Association of OGs of genes in L. tarentolae with differential expression in promastigote and/or amastigote life stages in pathogenic Leishmania species
| n | Promastigote | Amastigote | P-value* | |
|---|---|---|---|---|
| All OGs | 7694 | 890 | 1013 | NA |
| Higher copy number in L. tarentolae | 86 | 20 | 20 | 0.75 |
| Lower copy number in L. tarentolae | 23 | 11 | 11 | 0.83 |
| OGs absent from L. tarentolae | 278 | 18 | 52 | <0.001 |
*Promastigote versus amastigote comparison using two-tailed Fisher's exact test.
NA: Not Applicable.
Examples of genes that were absent from L. tarentolae but preferentially expressed in the amastigote stage of other Leishmania species are the delta-amastins and the hydrophilic surface protein gene family HASPA1, HASPA2, HASPB1, HASPB2 (Supplementary Table S5), known to be preferentially expressed in L. major, L. infantum and L. donovani amastigotes as determined by DNA microarray studies (68,69). This list also includes a 3,2-trans-enoyl-CoA isomerase, an ABC transporter-like protein, a class I nuclease-like protein, a D-isomer-specific 2-hydroxyacid dehydrogenase-protein, an Ef hand-like protein, a GTPase activator protein, a pteridine transporter and a tyrosine/dopa decarboxylase. The remaining amastigote OGs absent from L. tarentolae encode hypothetical proteins (Supplementary Table S5).
DISCUSSION
We have used next-generation DNA sequencing technologies to obtain a high-quality draft (72) of the genome sequence of L. tarentolae, the first non-human trypanosomatid pathogen sequenced. Genome sequence analysis revealed that L. tarentolae is syntenic to the three-sequenced pathogenic Leishmania species and that >90% of the approximately 8200 genes are shared by all the species. However, a number of genes shown to be either important for pathogenesis or preferentially expressed in the intracellular parasitic stage in the pathogenic species or more relevant to the insect stage were either absent from L. tarentolae or present at variable copy number, supporting the hypothesis that some of these genes may be responsible for the reduced capacity of L. tarentolae to live as an intracellular parasite and its diminished pathogenic potential to humans.
Among the genes absent from L. tarentolae, there was a significant bias for those expressed specifically in the amastigote intracellular stage in the pathogenic species (Table 2). Most of these genes correspond to hypothetical conserved or hypothetical proteins of unknown function (Supplementary Table S5) further highlighting the gaps in our understanding of the biology of intracellular parasites. Still, well-studied genes, such as the amastin gene family, especially the delta-amastins, are in low copy number in L. tarentolae (only 2 found, see Figure 4A) whereas high numbers of these genes (12–25 members) are found in the pathogenic species. The delta subfamily contains a diverse clade that is exclusive to Leishmania with one locus (proto-delta-amastins) found in other trypanosomatids (53). Although the function of amastins is still unknown, most of the delta-amastins undergo a stage-specific regulation and are preferentially expressed in amastigotes (51,52,68,73,74) and few in infective metacyclics (59). Given their specific expression in amastigotes, it has been argued that their expansion should be viewed as the adaptation of the parasite to a novel life stage after the acquisition of vertebrate parasitism (53). Leishmania tarentolae also lacks tuzins, conserved transmembrane proteins with unknown function, which are often contiguous with delta-amastins. Moreover, the majority of L. tarentolae genes displaying high variation in their copy number (e.g. GP63, PSA31C, phosphoglycan β 1,3 galactosyltransferase, ama1, etc) are developmentally regulated in the pathogenic Leishmania.
Two of the most studied abundant surface constituents of Leishmania promastigotes are the GPI-anchored molecules lipophosphoglycan (LPG) (75) and GP63 zinc metallo-protease (76). In contrast to the mammalian parasites, lizard Leishmania such as L. tarentolae and L. adleri seem to lack LPG (77). Sequencing data did not reveal any specific gene, absent or mutated, that could explain the lack of LPG. LPG has been implicated in many steps required for the establishment of initial macrophage infection, notably in the inhibition of phagolysosome biogenesis and in resistance to oxidants (75), but also in the development of innate immunity (78,79). Indeed, LPG-deficient L. major mutants showed attenuated virulence (80,81) although this was not the case for L. mexicana LPG-deficient mutants (82). In addition to LPG, L. tarentolae lacks enzymes that are involved in LPG modification like the phosphoglycan β 1,2 arabinosyltransferase, which is involved in the terminal capping of LPG in L. major (49), and has less copies of the β 1,3 galactosyltransferase gene, which adds the β 1,3 galactoside to the repeating PG of LPG (48) (Figure 3). These modifications play a role in Leishmania–sand fly interactions (48,49,83).
The major surface protease GP63, a well known virulence factor implicated in phagocytosis, parasite evasion of complement-mediated lysis, and induction of the host immune response (76,84), is highly expanded in L. tarentolae, as confirmed by Southern blot (Figure 7E) and read coverage analyses (Figure 5A). In the sand fly, GP63 is thought to be implicated in nutrient acquisition and attachment of promastigotes to the gut wall (76,85). Expansion of GP63 genes in L. tarentolae may thus facilitate nutrient acquisition, and in the absence of LPG, GP63 may be important for the parasite to maintain its attachment to the midgut. More recently, it was shown that during Leishmania–macrophage interaction, GP63 is internalized by macrophages, where it interacts and cleaves several intracellular macrophage proteins, including actin cytoskeleton regulators, protein tyrosine phosphatases, and transcription factors (86–89). Collectively, these cleavage-dependent activation events lead to the down-regulation of IFN-gamma signalling and downstream macrophage activation, including NO production (90). Interestingly, L. tarentolae was shown to lack protease activity (91), as also validated here by gelatin zymography (Figure 6). While GP63 genes were difficult to assemble, read analysis showed that some portions of the gene, especially the N-terminal and C-terminal regions, were underrepresented or had increased sequence variability while other portions were conserved (Figure 5A). It has been reported previously that the C-terminus of GP63 contains a GPI membrane anchor (92). Moreover, it has been shown recently that deletion of 180–211 amino acids from the C-terminal domain of GP63 resulted in 50% loss of its catalytic activity (93). Our data (Figures 5A and 6) and published reports (86,91) support the possibility that the high sequence variability of the L. tarentolae GP63 gene may affect GP63 protease's activity.
In addition to GP63 genes, L. tarentolae has expanded the promastigote surface antigen PSA31C for which only a single copy is present in the pathogenic Leishmania (58). The PSA proteins are part of eight subfamilies that have usually a signal peptide, a cysteine-rich region, leucine-rich repeats, a threonine/serine-rich region and a domain possibly acting as a GPI anchor (58). PSA were found to be either membrane-bound, secreted or soluble but little is known about their function (58). The expression of some PSA genes was found to be increased in metacyclic parasites (94). In L. tarentolae, the only recognizable feature in the PSA31C subfamily is the leucine-rich repeats domain (Figure 5B). The expansion of this subfamily in L. tarentolae (Figure 7F) is striking and warrants further analysis. One hypothesis is that the PSA31C gene has expanded in lizard parasites to facilitate promastigote survival either in the insect vector or in the lizard.
Another interesting feature of the L. tarentolae genome is the lack of a number of genes encoding key functions of vesicular protein trafficking. In fact, L. tarentolae lacks the large adaptin subunits (β1/β2) and the medium-sized μ-adaptins that are part of the heterotetrameric AP-1 complex, which is involved in the formation of clathrin-coated vesicles (CCVs) mediating protein transport between the trans-Golgi network and endosomes (38). Leishmania mutants lacking AP-1 subunits showed significant defects in Golgi structure, endocytosis or exocytic transport and displayed reduced rates of endosome-to-lysosome transport (95). Moreover, it was shown that Sigma 1- and μ 1-adaptin homologues of L. mexicana are required for parasite survival in the infected host (96). In addition, the absence of membrane-bound acid phosphatase localizing to endosomal/lysosomal compartments (40) and of copines, soluble, calcium-dependent membrane-binding proteins with C2 domains known to be involved in cell signalling and/or membrane trafficking pathways and exocytosis (97) further suggests that L. tarentolae may have a deficient vesicular protein transport. In line with this, L. tarentolae lacks members of the Ras-like small GTP-binding proteins that have emerged as master regulators of cellular membrane transport by interacting with C2 protein domains (98). Also, L. tarentolae lacks three of the seven subunits of the Arp2/3 complex, which has been shown to function in cellular processes ranging from cell motility, cytokinesis and endocytosis to trafficking and cell–cell communication (44). Arp2/3 complex is a key player in initiating actin polymerization at sites of endocytosis (99). Since in the absence of ARPC1, ARPC4 and ARPC5, the Arp2/3 complex cannot be formed (44,100), it is possible that L. tarentolae shows defects in endocytosis, which may impact the capacity of the parasite to communicate efficiently with the intracellular environment in the macrophage phagolysosome.
Some other genes known to play a role in virulence were also missing from L. tarentolae. Indeed, L. tarentolae lacks one of the two subtilisin (SUB) proteases found to process the terminal peroxidases of the trypanothione reductase system (45). Subtilisin promotes survival of Leishmania amastigotes by serving as a maturase for the trypanothione reductase system, which is essential to maintain redox homeostasis in the host macrophage and to protect the parasite against oxidative damage (101). Interestingly, SUB-deficient Leishmania showed increased sensitivity to hydroperoxides and reduced viability in mouse infection models (45). In addition, L. tarentolae lacks DJ-1, a multifunctional oxidative stress response protein that defends cells against reactive oxygen species and mitochondrial damage (46). This is consistent with the observed decrease of L. tarentolae survival in the presence of H2O2.
Here, we provide a high-quality draft of the genome sequence of the lizard L. tarentolae using next-generation sequencing (NGS) technologies. Limitations of NGS technologies are well known (102) and similar problems were observed with the L. tarentolae genome assembly. The size of the L. tarentolae genome is somewhat smaller (∼5%) than that of the other sequenced Leishmania spp. due to collapse of regions with identical nucleotide sequence. Also, repeated genes such as GP63 or PSA31C had a tendency to be fragmented in the assembly. However, these problems were attenuated by read number analysis. Homopolymers are an important cause of sequencing errors when using the 454 platform and can lead to frame shifts that make it difficult to differentiate actual sequencing errors from pseudogenes. In most cases, comparison of these specific regions to reads obtained by Illumina sequencing validated the presence or absence of genes. Most importantly, the current sequence draft provides additional research avenues to investigate Leishmania pathogenesis. The absence of LPG, acid phosphatase and delta-amastin glycoproteins on the surface of L. tarentolae combined to GP63 lacking most likely its protease activity could lead to more vulnerable parasites, which may be more sensitive to complement-mediated lysis with a diminished capacity of survival within the host macrophage. In addition, a possible defect in the vesicular trafficking of glycoproteins, plasma-membrane proteins and secreted proteins may have important consequences on the ability of L. tarentolae to survive as an intracellular parasite. Furthermore, L. tarentolae lacks several proteins of unknown functions (including the delta-amastins) shown to be expressed preferentially in the amastigote stage. The absence of these proteins may explain why L. tarentolae cannot replicate efficiently in mammalian macrophages. In summary, this study provides insights into the reconstruction of the steps leading to increase adaptation for intracellular parasitism and suggests a number of hypothesis that can be experimentally tested.
ACCESSION NUMBER
The genome sequence of L. tarentolae has been deposited in tritrypdb.org. Upon inclusion, the sequence will be transferred to Genbank.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6, Supplementary Figures 1–36.
FUNDING
Canadian Institutes of Health Research group grant (GR14500 to M.Ou., B.P., J.C., M.J.T. and M.Ol.). Funding for open access charge: Canada research chair in medical genomics (J.C.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Arno Velds and Ron Kerkhoven from the Netherlands Cancer Institute for providing Illumina runs and Dr. Ken Dewar at the McGill University and Genome Quebec Innovation Centre for generating the Roche/454 sequencing data included in the genome assembly. We thank also Prof. Jean-Pierre Dedet and Prof. Francine Pratlong at Université de Montpellier, France, for sending us the L. tarentolae S125 isolate from the ‘Leishmania Biological Resource Centre and Centre National de Références des Leishmania’ and for useful discussions on lizard Leishmania. We are grateful to Prof. Steve Beverley (Washington University School of Medicine, St. Louis, Missouri, USA) for discussions on NGS and for suggestions on kinetoplast DNA removal, and Dr. Sachiko Sato (Université Laval) for discussions on LPG. FR was the recipient of a CIHR doctoral award. SB is recipient of a CIRH doctoral award. JC, MJT and MO are holders of Tier 1 Canada Research Chairs.
REFERENCES
- 1.Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet. 2005;366:1561–1577. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- 2.Bates PA. Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int. J. Parasitol. 2007;37:1097–1106. doi: 10.1016/j.ijpara.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noyes HA, Chance ML, Croan DG, Ellis JT. Leishmania (sauroleishmania): A comment on classification. Parasitol. Today. 1998;14:167. doi: 10.1016/s0169-4758(97)01196-4. [DOI] [PubMed] [Google Scholar]
- 4.Simpson L, Holz G. The status of Leishmania tarentolae/Trypanosoma platydactyli. Parasitol. Today. 1988;4:115–118. doi: 10.1016/0169-4758(88)90043-9. [DOI] [PubMed] [Google Scholar]
- 5.Elwasila M. Leishmania tarentolae Wenyon, 1921 from the gecko Tarentola annularis in the Sudan. Parasitol. Res. 1988;74:591–592. doi: 10.1007/BF00531640. [DOI] [PubMed] [Google Scholar]
- 6.Wilson V, Southern B. Lizard Leishmania. In: Lumsden W, Evans D, editors. Biology of Kinetoplastida. New York: Academic press; 1979. pp. 242–268. [Google Scholar]
- 7.Killick-Kendrick R, Lainson R, Rioux J-A, Saf'janova VM. The taxonomy of Leishmania-like parasites in reptiles. In: Rioux JA, editor. 1986. Leishmania. Taxonomie et phylogenèse. Application Éco-epidemiologiques (Colloque International du CNRS/INSERM, 1984), IMEE, Montpellier, pp. 143–148. [Google Scholar]
- 8.Taylor VM, Muñoz DL, Cedeño DL, Vélez ID, Jones MA, Robledo SM. Leishmania tarentolae: utility as an in vitro model for screening of antileishmanial agents. Parasitol. Res. 2010;126:471–475. doi: 10.1016/j.exppara.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 9.Breton M, Tremblay MJ, Ouellette M, Papadopoulou B. Live nonpathogenic parasitic vector as a candidate vaccine against visceral leishmaniasis. Infect. Immun. 2005;73:6372–6382. doi: 10.1128/IAI.73.10.6372-6382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breton M, Zhao C, Ouellette M, Tremblay MJ, Papadopoulou B. A recombinant non-pathogenic Leishmania vaccine expressing human immunodeficiency virus 1 (HIV-1) Gag elicits cell-mediated immunity in mice and decreases HIV-1 replication in human tonsillar tissue following exposure to HIV-1 infection. J. Gen. Virol. 2007;88:217–225. doi: 10.1099/vir.0.81995-0. [DOI] [PubMed] [Google Scholar]
- 11.Ouellette M, Hettema E, Wüst D, Fase-Fowler F, Borst P. Direct and inverted DNA repeats associated with P-glycoprotein gene amplification in drug resistant Leishmania. EMBO J. 1991;10:1009–1016. doi: 10.1002/j.1460-2075.1991.tb08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrillo-Peixoto ML, Beverley SM. Amplified DNAs in laboratory stocks of Leishmania tarentolae: extrachromosomal circles structurally and functionally similar to the inverted-H-region amplification of methotrexate-resistant Leishmania major. Mol. Cell. Biol. 1988;8:5188–5199. doi: 10.1128/mcb.8.12.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White TC, Fase-Fowler F, van Luenen H, Calafat J, Borst P. The H circles of Leishmania tarentolae are a unique amplifiable system of oligomeric DNAs associated with drug resistance. J. Biol. Chem. 1988;263:16977–16983. [PubMed] [Google Scholar]
- 14.Simpson L, Aphasizhev R, Gao G, Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basile G, Peticca M. Recombinant protein expression in Leishmania tarentolae. Mol. Biotechnol. 2009;43:273–278. doi: 10.1007/s12033-009-9213-5. [DOI] [PubMed] [Google Scholar]
- 16.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream M-A, Adlem E, Aert R, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, Peters N, Adlem E, Tivey A, Aslett M, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch EF, Maniatis T. Sambrook J E F Fritsch and T Maniatis Molecular Cloning A Laboratory Manual. 2nd edn. Vol. 1, 2 and 3. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. Molecular cloning: a laboratory manual, second edition, Vols. 1, 2 and 3; p. 999. (USA Illus Paper) [Google Scholar]
- 19.Zerbino DR, Birney E, Spring C. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boisvert S, Laviolette F, Corbeil J. Ray: Simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J. Comput. Biol. 2010;17:1519–1533. doi: 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sommer DD, Delcher AL, Salzberg SL, Pop M. Minimus: a fast, lightweight genome assembler. BMC Bioinf. 2007;8:64. doi: 10.1186/1471-2105-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19:215–225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 24.Stanke M, Schöffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinf. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bateman A, Birney E, Durbin R, Eddy SR, Finn RD, Sonnhammer EL. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogh A, Sj lander K, Brown M, Mian IS, Haussler D. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- 28.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 30.Chen F, Mackey AJ, Stoeckert CJ, Roos DS. OrthoMCL-DB: Querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ubeda J-M, Légaré D, Raymond F, Ouameur AA, Boisvert S, Rigault P, Corbeil J, Tremblay MJ, Olivier M, Papadopoulou B, et al. Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 2008;9:R115. doi: 10.1186/gb-2008-9-7-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 36.Rovai L, Tripp C, Stuart K, Simpson L. Recurrent polymorphisms in small chromosomes of Leishmania tarentolae after nutrient stress or subcloning. Mol. Biochem. Parasit. 1992;50:115–125. doi: 10.1016/0166-6851(92)90249-j. [DOI] [PubMed] [Google Scholar]
- 37.Wincker P, Ravel C, Blaineau C, Pages M, Jauffret Y, Dedet JP, Bastien P. The Leishmania genome comprises 36 chromosomes conserved across widely divergent human pathogenic species. Nucleic Acids Res. 1996;24:1688–1694. doi: 10.1093/nar/24.9.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mousavi SA, Malerød L, Berg T, Kjeken R. Clathrin-dependent endocytosis. Biochem. J. 2004;377:1–16. doi: 10.1042/BJ20031000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohno H. Physiological roles of clathrin adaptor AP complexes: lessons from mutant animals. J. Biochem. 2006;139:943–948. doi: 10.1093/jb/mvj120. [DOI] [PubMed] [Google Scholar]
- 40.Weise F, Thilo L, Engstler M, Wiese M, Benzel I, Kühn C, Bühring H-J, Overath P. Binding affinity and capacity of putative adaptor-mediated sorting of a Type I membrane protein in Leishmania mexicana. Mol. Biochem. Parasit. 2005;142:203–211. doi: 10.1016/j.molbiopara.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 41.Cantrell DA. Phosphoinositide 3-kinase signalling pathways. J. Cell Sci. 2001;114:1439–1445. doi: 10.1242/jcs.114.8.1439. [DOI] [PubMed] [Google Scholar]
- 42.Caberoy NB, Zhou Y, Li W. Tubby and tubby-like protein 1 are new MerTK ligands for phagocytosis. EMBO J. 2010;29:3898–3910. doi: 10.1038/emboj.2010.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welch MD, Mullins RD. Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 2002;18:247–288. doi: 10.1146/annurev.cellbio.18.040202.112133. [DOI] [PubMed] [Google Scholar]
- 44.Pollard TD, Beltzner CC. Structure and function of the Arp2/3 complex. Curr. Opin. Struct. Biol. 2002;12:768–774. doi: 10.1016/s0959-440x(02)00396-2. [DOI] [PubMed] [Google Scholar]
- 45.Swenerton RK, Knudsen GM, Sajid M, Kelly BL, McKerrow JH. Leishmania subtilisin is a maturase for the trypanothione reductase system and contributes to disease pathology. J. Biol. Chem. 2010;285:31120–31129. doi: 10.1074/jbc.M110.114462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson MA. The role of cysteine oxidation in DJ-1 function and dysfunction. Antioxid. Redox Signal. 2011;15:111–112. doi: 10.1089/ars.2010.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jodko K, Litwinienko G. [Oxidative stress in the neurodegenerative diseases-potential antioxidant activity of catecholamines] Postepy biochem. 2010;56:248–259. [PubMed] [Google Scholar]
- 48.Butcher BA, Turco SJ, Hilty BA, Pimenta PF, Panunzio M, Sacks DL. Deficiency in β1,3-galactosyltransferase of a Leishmania major lipophosphoglycan mutant adversely influences the Leishmania-sand fly interaction. J. Biol. Chem. 1996;271:20573–20579. doi: 10.1074/jbc.271.34.20573. [DOI] [PubMed] [Google Scholar]
- 49.Dobson DE, Mengeling BJ, Cilmi S, Hickerson S, Turco SJ, Beverley SM. Identification of genes encoding arabinosyltransferases (SCA) mediating developmental modifications of lipophosphoglycan required for sand fly transmission of leishmania major. J. Biol. Chem. 2003;278:28840–28848. doi: 10.1074/jbc.M302728200. [DOI] [PubMed] [Google Scholar]
- 50.Williams DB. Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J. Cell Sci. 2006;119:615–623. doi: 10.1242/jcs.02856. [DOI] [PubMed] [Google Scholar]
- 51.Rochette A, McNicoll F, Girard J, Breton M, Leblanc E, Bergeron MG, Papadopoulou B. Characterization and developmental gene regulation of a large gene family encoding amastin surface proteins in Leishmania spp. Mol. Biochem. Parasit. 2005;140:205–220. doi: 10.1016/j.molbiopara.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 52.Wu Y, El Fakhry Y, Sereno D, Tamar S, Papadopoulou B. A new developmentally regulated gene family in Leishmania amastigotes encoding a homolog of amastin surface proteins. Mol. Biochem. Parasit. 2000;110:345–357. doi: 10.1016/s0166-6851(00)00290-5. [DOI] [PubMed] [Google Scholar]
- 53.Jackson AP. The evolution of amastin surface glycoproteins in trypanosomatid parasites. Mol. Biol. Evol. 2010;27:33–45. doi: 10.1093/molbev/msp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider P, Rosat JP, Bouvier J, Louis J, Bordier C. Leishmania major: Differential regulation of the surface metalloprotease in amastigote and promastigote stages. Parasitol. Res. 1992;75:196–206. doi: 10.1016/0014-4894(92)90179-e. [DOI] [PubMed] [Google Scholar]
- 55.Ilgoutz SC, McConville MJ. Function and assembly of the Leishmania surface coat. Int. J. Parasitol. 2001;31:899–908. doi: 10.1016/s0020-7519(01)00197-7. [DOI] [PubMed] [Google Scholar]
- 56.Yao C, Donelson JE, Wilson ME. The major surface protease (MSP or GP63) of Leishmania sp. Biosynthesis, regulation of expression, and function. Mol. Biochem. Parasit. 2003;132:1–16. doi: 10.1016/s0166-6851(03)00211-1. [DOI] [PubMed] [Google Scholar]
- 57.Murray PJ, Spithill TW, Handman E. The PSA-2 glycoprotein complex of Leishmania major is a glycosylphosphatidylinositol-linked promastigote surface antigen. J. Immunol. 1989;143:4221–4226. [PubMed] [Google Scholar]
- 58.Devault A, Bañuls A-L. The promastigote surface antigen gene family of the Leishmania parasite: differential evolution by positive selection and recombination. BMC Evol. Biol. 2008;8:292. doi: 10.1186/1471-2148-8-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alcolea PJ, Alonso A, Sánchez-Gorostiaga A, Moreno-Paz M, Gómez MJ, Ramos I, Parro V, Larraga V. Genome-wide analysis reveals increased levels of transcripts related with infectivity in peanut lectin non-agglutinated promastigotes of Leishmania infantum. Genomics. 2009;93:551–5064. doi: 10.1016/j.ygeno.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 60.Alcolea PJ, Alonso A, Gómez MJ, Moreno I, Domínguez M, Parro V, Larraga V. Transcriptomics throughout the life cycle of Leishmania infantum: high down-regulation rate in the amastigote stage. Int. J. Parasitol. 2010;40:1497–1516. doi: 10.1016/j.ijpara.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 61.Brotherton M-C, Racine G, Foucher AL, Drummelsmith J, Papadopoulou B, Ouellette M. Analysis of stage-specific expression of basic proteins in Leishmania infantum. J. Proteome Res. 2010;9:3842–3853. doi: 10.1021/pr100048m. [DOI] [PubMed] [Google Scholar]
- 62.Depledge DP, Evans KJ, Ivens AC, Aziz N, Maroof A, Kaye PM, Smith DF. Comparative expression profiling of Leishmania: modulation in gene expression between species and in different host genetic backgrounds. PLoS Neglected Trop. Dis. 2009;3:e476. doi: 10.1371/journal.pntd.0000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holzer TR, McMaster WR, Forney JD. Expression profiling by whole-genome interspecies microarray hybridization reveals differential gene expression in procyclic promastigotes, lesion-derived amastigotes, and axenic amastigotes in Leishmania mexicana. Mol. Biochem. Parasit. 2006;146:198–218. doi: 10.1016/j.molbiopara.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 64.Leifso K, Cohen-Freue G, Dogra N, Murray A, McMaster WR. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: The Leishmania genome is constitutively expressed. Mol. Biochem. Parasit. 2007;152:35–46. doi: 10.1016/j.molbiopara.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 65.McNicoll F, Drummelsmith J, Müller M, Madore E, Boilard N, Ouellette M, Papadopoulou B. A combined proteomic and transcriptomic approach to the study of stage differentiation in Leishmania infantum. Proteomics. 2006;6:3567–3581. doi: 10.1002/pmic.200500853. [DOI] [PubMed] [Google Scholar]
- 66.Paape D, Barrios-Llerena ME, Le Bihan T, Mackay L, Aebischer T. Gel free analysis of the proteome of intracellular Leishmania mexicana. Mol. Biochem. Parasit. 2010;169:108–114. doi: 10.1016/j.molbiopara.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 67.Rochette A, Raymond F, Corbeil J, Ouellette M, Papadopoulou B. Whole-genome comparative RNA expression profiling of axenic and intracellular amastigote forms of Leishmania infantum. Mol. Biochem. Parasitol. 2009;165:32–47. doi: 10.1016/j.molbiopara.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 68.Rochette A, Raymond F, Ubeda J-M, Smith M, Messier N, Boisvert S, Rigault P, Corbeil J, Ouellette M, Papadopoulou B. Genome-wide gene expression profiling analysis of Leishmania major and Leishmania infantum developmental stages reveals substantial differences between the two species. BMC Genomics. 2008;9:255. doi: 10.1186/1471-2164-9-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saxena A, Lahav T, Holland N, Aggarwal G, Anupama A, Huang Y, Volpin H, Myler PJ, Zilberstein D. Analysis of the Leishmania donovani transcriptome reveals an ordered progression of transient and permanent changes in gene expression during differentiation. Mol. Biochem. Parasit. 2007;152:53–65. doi: 10.1016/j.molbiopara.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Srividya G, Duncan R, Sharma P, Raju BVS, Nakhasi HL, Salotra P. Transcriptome analysis during the process of in vitro differentiation of Leishmania donovani using genomic microarrays. Parasitology. 2007;134:1527–1539. doi: 10.1017/S003118200700296X. [DOI] [PubMed] [Google Scholar]
- 71.Saxena A, Worthey EA, Yan S, Leland A, Stuart KD, Myler PJ. Evaluation of differential gene expression in Leishmania major Friedlin procyclics and metacyclics using DNA microarray analysis. Mol. Biochem. Parasit. 2003;129:103–114. doi: 10.1016/s0166-6851(03)00100-2. [DOI] [PubMed] [Google Scholar]
- 72.Chain PSG, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, Ali J, Birren B, Bruce DC, Buhay C, et al. Genomics. Genome project standards in a new era of sequencing. Science. 2009;326:236–237. doi: 10.1126/science.1180614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McNicoll F, Müller M, Cloutier S, Boilard N, Rochette A, Dubé M, Papadopoulou B. Distinct 3′-untranslated region elements regulate stage-specific mRNA accumulation and translation in Leishmania. J. Biol. Chem. 2005;280:35238–35246. doi: 10.1074/jbc.M507511200. [DOI] [PubMed] [Google Scholar]
- 74.Boucher N, Wu Y, Dumas C, Dube M, Sereno D, Breton M, Papadopoulou B. A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3′-untranslated region element. J. Biol. Chem. 2002;277:19511–19520. doi: 10.1074/jbc.M200500200. [DOI] [PubMed] [Google Scholar]
- 75.Lodge R, Descoteaux A. Leishmania invasion and phagosome biogenesis. Subcell. Biochem. 2008;47:174–181. doi: 10.1007/978-0-387-78267-6_14. [DOI] [PubMed] [Google Scholar]
- 76.Yao C. Major surface protease of trypanosomatids: one size fits all? Infect. Immun. 2010;78:22–311. doi: 10.1128/IAI.00776-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Previato JO, Jones C, Wait R, Routier F, Saraiva E, Mendonça-Previato L. Leishmania adleri, a lizard parasite, expresses structurally similar glycoinositolphospholipids to mammalian Leishmania. Glycobiology. 1997;7:687–695. doi: 10.1093/glycob/7.5.687. [DOI] [PubMed] [Google Scholar]
- 78.Becker I, Salaiza N, Aguirre M, Delgado J, Carrillo-Carrasco N, Kobeh LG, Ruiz A, Cervantes R, Torres AP, Cabrera N, et al. Leishmania lipophosphoglycan (LPG) activates NK cells through toll-like receptor-2. Mol. Biochem. Parasit. 2003;130:65–74. doi: 10.1016/s0166-6851(03)00160-9. [DOI] [PubMed] [Google Scholar]
- 79.Aebischer T, Bennett CL, Pelizzola M, Vizzardelli C, Pavelka N, Urbano M, Capozzoli M, Luchini A, Ilg T, Granucci F, et al. A critical role for lipophosphoglycan in proinflammatory responses of dendritic cells to Leishmania mexicana. Eur. J. Immunol. 2005;35:476–486. doi: 10.1002/eji.200425674. [DOI] [PubMed] [Google Scholar]
- 80.Späth GF, Lye L-F, Segawa H, Sacks DL, Turco SJ, Beverley SM. Persistence without pathology in phosphoglycan-deficient Leishmania major. Science. 2003;301:1241–1243. doi: 10.1126/science.1087499. [DOI] [PubMed] [Google Scholar]
- 81.Späth GF, Garraway LA, Turco SJ, Beverley SM. The role(s) of lipophosphoglycan (LPG) in the establishment of Leishmania major infections in mammalian hosts. Proc. Natl. Acad. Sci. USA. 2003;100:9536–9541. doi: 10.1073/pnas.1530604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ilg T. Lipophosphoglycan is not required for infection of macrophages or mice by Leishmania mexicana. EMBO J. 2000;19:1953–1962. doi: 10.1093/emboj/19.9.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dobson DE, Scholtes LD, Valdez KE, Sullivan DR, Mengeling BJ, Cilmi S, Turco SJ, Beverley SM. Functional identification of galactosyltransferases (SCGs) required for species-specific modifications of the lipophosphoglycan adhesin controlling Leishmania major-sand fly interactions. J. Biol. Chem. 2003;278:15523–15531. doi: 10.1074/jbc.M301568200. [DOI] [PubMed] [Google Scholar]
- 84.Joshi PB, Kelly BL, Kamhawi S, Sacks DL, McMaster WR. Targeted gene deletion in Leishmania major identifies leishmanolysin (GP63) as a virulence factor. Mol. Biochem. Parasit. 2002;120:33–40. doi: 10.1016/s0166-6851(01)00432-7. [DOI] [PubMed] [Google Scholar]
- 85.Button LL, McMaster WR. Molecular cloning of the major surface antigen of Leishmania. J. Exp. Med. 1988;167:724–729. doi: 10.1084/jem.167.2.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hallé M, Gomez MA, Stuible M, Shimizu H, McMaster WR, Olivier M, Tremblay ML. The Leishmania surface protease GP63 cleaves multiple intracellular proteins and actively participates in p38 mitogen-activated protein kinase inactivation. J. Biol. Chem. 2009;284:6893–6908. doi: 10.1074/jbc.M805861200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gomez MA, Contreras I, Hallé M, Tremblay ML, McMaster RW, Olivier M. Leishmania GP63 alters host signaling through cleavage-activated protein tyrosine phosphatases. Sci. Signal. 2009;2:ra58. doi: 10.1126/scisignal.2000213. [DOI] [PubMed] [Google Scholar]
- 88.Contreras I, Gómez MA, Nguyen O, Shio MT, McMaster RW, Olivier M. Leishmania-induced inactivation of the macrophage transcription factor AP-1 is mediated by the parasite metalloprotease GP63. PLoS pathogens. 2010;6:e1001148. doi: 10.1371/journal.ppat.1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gregory DJ, Godbout M, Contreras I, Forget G, Olivier M. A novel form of NF-kappaB is induced by Leishmania infection: Involvement in macrophage gene expression. Eur. J. Immunol. 2008;38:1071–1081. doi: 10.1002/eji.200737586. [DOI] [PubMed] [Google Scholar]
- 90.Olivier M, Gregory DJ, Forget G. Subversion mechanisms by which Leishmania parasites can escape the host immune response: a signaling point of view. Clin. Microbiol. Rev. 2005;18:293–305. doi: 10.1128/CMR.18.2.293-305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Campbell DA, Kurath U, Fleischmann J. Identification of a gp63 surface glycoprotein in Leishmania tarentolae. FEMS Microbiol. Lett. 1992;75:89–92. doi: 10.1016/0378-1097(92)90461-v. [DOI] [PubMed] [Google Scholar]
- 92.Voth BR, Kelly BL, Joshi PB, Ivens AC, McMaster WR. Differentially expressed Leishmania major gp63 genes encode cell surface leishmanolysin with distinct signals for glycosylphosphatidylinositol attachment. Mol. Biochem. Parasit. 1998;93:31–41. doi: 10.1016/s0166-6851(98)00013-9. [DOI] [PubMed] [Google Scholar]
- 93.Mazumder S, Ganguly A, Ali N. The effect of C-terminal domain deletion on the catalytic activity of Leishmania donovani surface proteinase GP63: Role of Ser446 in proteolysis. Biochimie. 2010;92:1876–1885. doi: 10.1016/j.biochi.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 94.Beetham JK, Donelson JE, Dahlin RR. Surface glycoprotein PSA (GP46) expression during short- and long-term culture of Leishmania chagasi. Mol. Biochem. Parasit. 2003;131:109–117. doi: 10.1016/s0166-6851(03)00197-x. [DOI] [PubMed] [Google Scholar]
- 95.Vince JE, Tull DL, Spurck T, Derby MC, McFadden GI, Gleeson PA, Gokool S, McConville MJ. Leishmania adaptor protein-1 subunits are required for normal lysosome traffic, flagellum biogenesis, lipid homeostasis, and adaptation to temperatures encountered in the mammalian host. Eukaryotic Cell. 2008;7:1256–1267. doi: 10.1128/EC.00090-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gokool S. Sigma 1- and mu 1-Adaptin homologues of Leishmania mexicana are required for parasite survival in the infected host. J. Biol. Chem. 2003;278:29400–29409. doi: 10.1074/jbc.M304572200. [DOI] [PubMed] [Google Scholar]
- 97.Tomsig JL, Creutz CE. Copines: A ubiquitous family of Ca(2+)-dependent phospholipid-binding proteins. Cell. Mol. Life Sci. 2002;59:1467–1477. doi: 10.1007/s00018-002-8522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pannekoek W-J, Kooistra MRH, Zwartkruis FJT, Bos JL. Cell-cell junction formation: The role of Rap1 and Rap1 guanine nucleotide exchange factors. Biochim. Biophys. Acta. 2009;1788:790–796. doi: 10.1016/j.bbamem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 99.Martin AC, Welch MD, Drubin DG. Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat. Cell Biol. 2006;8:826–833. doi: 10.1038/ncb1443. [DOI] [PubMed] [Google Scholar]
- 100.Winter DC, Choe EY, Li R. Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc. Natl. Acad. Sci. USA. 1999;96:7288–7293. doi: 10.1073/pnas.96.13.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Krauth-Siegel RL, Comini MA. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochim. Biophys. Acta. 2008;1780:1236–1248. doi: 10.1016/j.bbagen.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 102.Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nature Met. 2010;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.