Abstract
Despite the theoretical bases for the association of topoisomerases and supercoiling changes with transcription and replication, our knowledge of the impact of topological constraints on transcription and replication is incomplete. Although mutation of topoisomerases affects expression and stability of the rDNA region it is not clear whether the same is the case for RNAPII transcription and genome integrity in other regions. We developed new assays in which two convergent RNAPII-driven genes are transcribed simultaneously. Plasmid-based systems were constructed with and without a transcription terminator between the two convergent transcription units, so that the impact of transcription interference could also be evaluated. Using these assays we show that Topos I and II play roles in RNAPII transcription in vivo and reduce the stability of RNAPII-transcribed genes in Saccharomyces cerevisiae. Supercoiling accumulation in convergent transcription units impairs RNAPII transcription in top1Δ strains, but Topo II is also required for efficient transcription independent of Topo I and of detectable supercoiling accumulation. Our work shows that topological constraints negatively affect RNAPII transcription and genetic integrity, and provides an assay to study gene regulation by transcription interference.
INTRODUCTION
DNA in its natural form in the cell is supercoiled. In eukaryotes this state facilitates its organization into chromatin around nucleosomes. DNA topoisomerases control supercoiling and their action is required in replication and transcription as well as in chromosome condensation and segregation (1,2). There are two types of DNA topoisomerases (Topo), I and II, which differ in their catalytic way of action, the first relying on a single-strand DNA cleavage, the second on a double-strand break (DSB). Topoisomerases I and II are the most representative of each type in all organisms. They are functionally interchangeable in eukaryotes, but not in bacteria.
During replication DNA topoisomerases can act at different stages. In Escherichia coli reconstituted replication systems require DNA supercoiling for replication initiation and the action of DNA topoisomerases (3). However, it is during elongation of replication that topoisomerases are expected to have a key role. They are required to remove the positive supercoiling that are generated by the continuous opening of the template strands. The intertwined daughter DNA molecules resulting after replication completion require the action of type II topoisomerases for their segregation. This late function in replication cannot be performed by type I topoisomerases but only by type II (4,5).
In contrast to replication, transcription does not involve a continuous separation of DNA strands. However, RNA polymerase progression through the DNA template would lead to the accumulation of local positive supercoiling ahead of the RNA polymerase and negative supercoiling behind. The twin-supercoiled-domain model was proposed to explain removal of this local supercoiling accumulation by topoisomerases during transcription (6). Evidence for transcriptional supercoiling associated with transcription has been provided in E. coli (6,7) and yeast (8,10). The relaxation of transcription-dependent supercoiling is generally considered to be the main function of Topo I, which is considered to play a swivel role in transcription (1,2). In eukaryotes such as the yeast Saccharomyces cerevisiae, the fact that top1Δ mutants are viable suggests that Topo II can replace the function of Topo I; whereas the opposite is not the case, since Topo I cannot replace Topo II at the end of replication during segregation.
Despite the theoretical bases for the association of topoisomerases and supercoiling changes with transcription, experimental evidence for the impact of topoisomerases deficiency and topological constraints in transcription is incomplete. In bacteria, the importance of Topo I has been well documented in rDNA transcription (11) or transcription from operons involved in amino acid metabolism (12,13). In yeast, an effect of topoisomerase activity has been reported on transcription initiation. This may be related to the need of negatively supercoiled DNA at promoter regions to facilitate transcription initiation (14). The effect of DNA topoisomerases on transcription elongation has been established for rDNA, in which the rate of RNAPI transcription is reduced when both DNA topoisomerases are inactivated in yeast (8,15). Instead, none or a poor effect has been reported for mRNA transcription as tested in endogenous and lacZ reporters fused to GAL promoters in vivo (10,16).
In addition to the effect of Topo mutations in cell growth, transcription and replication, they have an important impact on genome integrity. Stability of tandem rDNA repeats in yeast has been shown to be strongly dependent on DNA topoisomeases I and II, whereas this was not the case for direct repeats outside of the rDNA region (17). Hyper-recombination has been shown in plasmid-borne direct-repeats in double top1 top2 mutants (18). It has been proposed that this is mainly the result of the failure to relax the negative rather than the positive supercoiling of DNA (18). Nevertheless, impairment of replication fork progression by obstacles or DNA lesions can lead to replication fork collapse that in turn could trigger recombination as a mechanism of replication restart (19). Thus, hyper-recombination of yeast topoisomerase mutants could be linked to their role in replication, but the functional relevance of Topo I in rDNA transcription and its large accumulation in the nucleolus, makes plausible that hyper-recombination could also be linked to transcription. Indeed, rDNA genes contain the RNAPI transcription-dependent HOT1 recombination hotspots (20,21), and one key function of the Fob1 barrier to DNA replication is to avoid replication–transcription collisions that could potentially compromise genome integrity (22).
In this sense, one intriguing class of hyper-recombination mutations in yeast is composed of those of the THO complex. THO is a conserved protein complex composed of stoichiometric amounts of Hpr1, Tho2, Mft1 and Thp2, with a role in transcription elongation, mRNP biogenesis and RNA export (23). THO mutants, as well as mutants related to mRNP biogenesis factors such as the THSC/TREX-2 complex, cause a strong hyper-recombination between direct repeats that are transcription-dependent. This phenotype is linked to the co-transcriptional formation of R-loops that can impair the progression of replication (24,25). Interestingly, in E. coli mutants lacking topoisomerase I, an excess of negative supercoiling during transcription can lead to R-loops (26). Furthermore, THO and THSC mutants show synthetic growth defects in combination with top1 and top2 mutations (27). All this evidence raises the question of whether hyper-recombination in the rDNA region in topoisomerases mutants could be linked to transcription.
Thus, although topological constraints and topoisomerase mutations have a clear impact in expression and stability of the rDNA locus this is not that clear for RNAPII transcription and genome integrity at regions others than rDNA. To investigate this, new and highly sensitive in vivo assays were developed based on plasmid-borne DNA sequences of increasing lengths that are transcribed form strong promoters with or without convergent transcription coming from a second promoter. We show that both top1 and top2 mutations drastically reduce RNAPII transcription, a defect that is strengthened as the length of the transcribed DNA sequence increases and by convergent transcription. Importantly, top2-1 mutation causes a significant increase in direct-repeat recombination in a top1Δ mutant background; however, this hyper-recombination was predominantly independent of transcription of the direct-repeat system itself. Therefore, this study provides new evidence for the in vivo relevance of Topos I and II in RNAPII transcription and genome integrity. We propose that DNA topoisomerases has a non-interchangeable role in the maintenance of integrity of RNAPII genes providing evidence for the negative effect of topological constraints in transcription and genome stability in DNA regions beyond the rDNA. Although a similar study would need to be performed in chromosome-borne constructs, we believe that these results may also be valid for chromosomal regions, provided that local supercoiling dynamics follow universal rules.
MATERIALS AND METHODS
Strains and plasmids
Yeast strains and plasmids are described in Supplementary Tables S1 and S2, respectively.
Chromatin immunoprecipitation analysis
For ChIP experiments, strains were grown in synthetic medium (SC) 2% glycerol-2% lactate to an OD660 of 0.5 (for GAL1). For GAL1 ChIPs the culture was split in two, and one half was supplemented with 2% glucose (repressed transcription) and the other with 2% galactose (activated transcription). For Ptet-based systems, yeast strains were grown in synthetic medium (SC), the culture was split in two, and one half was supplemented with doxycycline. Samples were then taken after 4 h of induction and ChIP assays were performed as described (28). Polyclonal anti-PolII (N-20) (Santa Cruz Biotechnology) and Protein A–Sepharose were used for RNAPII immunoprecipitation. The GFX purification system (Amersham) was used for the last DNA purification step. We used the PCR of the intergenic region at positions 9716–9863 of chromosome V as a negative control. Real-time quantitative PCR and calculations of the relative abundance of each DNA fragment was performed as described (29). For each experiment, the DNA ratios in the different regions were calculated from the DNA amount of these regions relative to the intergenic region. Median and Standard Deviation (SD) of three independent experiments are shown.
Nucleic acid isolation
For DNA isolation, pellets were re-suspended to a final concentration of 1.5–2 × 109/ml in NIB pH 7.2 (17% glycerol, 50 mM MOPS, 150 mM KAc, 2 mM MgCl, 0.5 mM spermidine and 0.15 mM spermine) with 3 mg/ml Zymoliase-20T and incubated at room temperature for 25 min. Cellular suspensions were diluted seven times with chilled water and centrifuged to collect nuclei. Pellets were re-suspended in 720 µl of TE (50 mM Tris–HCl, pH 8, 20 mM EDTA) and mixed with 80 µl of 10% SDS. After phenol–chloroform–isoamylalcohol (25:24:1) extraction and ethanol precipitation, DNA was finally re-suspend in 20–30 µl of TE.
Chloroquine gel electrophoresis
Electrophoresis was performed at room temperature on horizontal slab gels (0.7% agarose) containing TPE buffer (50 mM Tris–phosphate [pH 7.2], 1 mM EDTA and 25 mM phosphoric acid). Chloroquine (Sigma) was added to the melted agarose from a 10 mg/ml stock prior to casting the gel and was present in the electrophoresis buffer. The buffer was re-circulated continuously during electrophoresis. The concentration of chloroquine was 4µg/ml. Each lane had 20 µg of total nucleic acids. The gels were run at 40 V for 48 h.
Miscellaneous
Yeast methods, α-32P-labeled DNA probes, northern and Southern analyses were performed according to standard procedures. RNA was isolated from mid-log phase cells.
Recombination frequencies were obtained by fluctuation tests as the median value of six independent colonies isolated from SC plates. The final frequency given for each strain and condition is the mean and SD of three to four median values as described (30). Recombinants Leu+ were selected in SC- Leu plates.
RESULTS
Convergent transcription in systems with or without terminator sequences
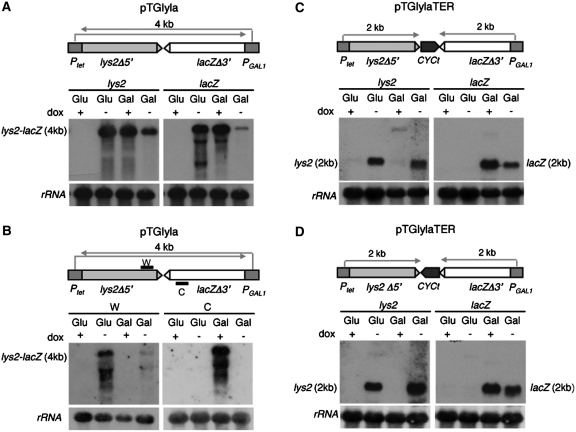
For a fine assessment of the possible effect of topological constraints in transcription, a plasmid-based system was constructed in which a 4-kb sequence was transcribed from two convergent and differentially regulated promoters located on each side. For this construction, we fused a 2-kb lacZ fragment under the GAL1 promoter to a 2-kb lys2 fragment under the quimeric tet promoter so that transcription driven from the two promoters was convergent (Figure 1). Two versions of this system were constructed in monocopy CEN-based plasmids: one with a 0.4-kb CYC1 terminator between the lys2 and lacZ sequences (pTGlylaTER) and the other without transcription terminator (pTGlyla). The CYC1 terminator sequence contains two converging terminators so that it acts in both orientations. As expected, whereas in the pTGlyla system without terminator the 4-kb lys2-lacZ mRNA was produced when cells were cultivated either with 2% galactose (GAL1 promoter activated) or without doxycycline (tet promoter activated), but not in 2% glucose + doxycycline (GAL1 and tet promoters repressed) (Figure 1A). To confirm that the band detected in 2% galactose was driven from the GAL1 promoter and the band detected in the absence of doxycycline was driven from the tet promoter northern analysis was performed in which hybridization was made with specific ssDNA probes. As expected, the 4-kb mRNA band was specific to a DNA strand. It was detected by the Watson-chain-specific (W) probe in the absence of doxycycline (Ptet active), and with the Crick-chain-specific (C) probe in galactose (PGAL1 active), according to the promoter from which each transcript was transcribed (Figure 1B). Instead, in the pTGlylaTER system either the 2-kb lacZ or the 2-kb lys2 mRNAs were detected in galactose regardless of the presence of doxycycline, or in doxycycline regardless of the carbon source, respectively (Figure 1C and D).
Figure 1.
Transcription analyses of the lys2-CYCt-lacZ and lys2-lacZ fusion constructs. (A) Northern analyses of lys2-lacZ-containing mRNAs driven from the tet and GAL1 promoters. The 3-kb XbaI-EcoRV lacZ and a 589-bp 25 S rDNA internal fragments obtained by PCR (rRNA) were used as DNA probes. (B) Northern analyses of lys2-lacZ. The oligos C and W were used as single-stranded DNA probes. (C), (D) Northern analyses of pTGlylaT-1 and pTGlylaT-5 plasmids. Arrows refers to the mRNA being produced from the specified promoter. No arrow indicates that there is no transcription from that promoter. All experiments are made in the absence of doxycycline, so that the tet promoter is permanently active. A total of 2% glucose (Glu) or galactose (Gal) are used to either repress or activate the GAL1 promoter. Other details as described in ‘Materials and Methods’ section.
Importantly, in the pTGlyla system, the level of 4-kb lys2-lacZ transcripts was lower when both the PGAL1 and Ptet promoters were simultaneously active than when only one was active, as observed with either the lys2 or the lacZ probe (Figure 1A and B). This result is consistent with transcription interference either at the promoter or transcription stalling provided by RNAPII collisions. Since the 4-kb transcript driven from either the tet or GAL1 promoters reaches the downstream converging promoter, the latter may undergo transcription interference. When the transcripts were analyzed with specific ssDNA probes it was found that after galactose activation the Ptet-driven 4-kb transcript, as detected with the W probe, was reduced, whereas the PGAL1-driven transcript was below detection levels (Figure 1B). This indicates that since the Ptet-driven RNAPII reaches PGAL1, it causes promoter interference impeding PGAL1 activation, even though transcriptional stalling cannot be excluded either. Similarly, PGAL1-driven transcription seems to reduce the efficiency of Ptet-driven transcription, although to a lesser extent.
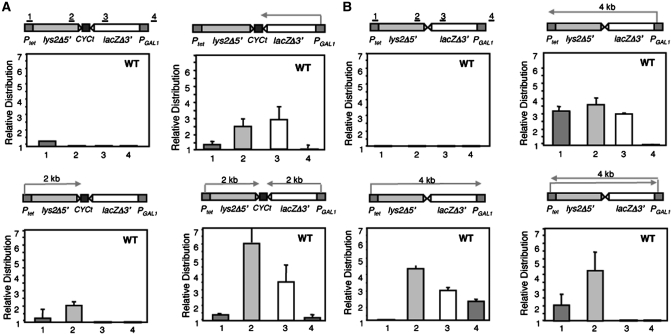
To assay whether the northern results could be interpreted in terms of RNAPII progression, a chromatin–immunoprecipitation (ChIP) analysis was performed with anti-Rpb1 N-ter antibodies in both the pTGlylaTER and pTGlyla systems under all possible conditions (with either none, only one or both promoters activated). As can be seen in Figure 2, RNAPII recruitment was not observed in either of the regions analyzed when both promoters were repressed in either system, as expected. In the pTGlylaTER system RNAPII was detected in lys2 when only Ptet was active and in both lys2 and lacZ when only PGAL1 was active (Figure 2A). The latter was consistent with transcription termination read-through observed for transcription initiated at the strong PGAL1 promoter, as observed by northern (Figure 1). In the pTGlyla system, RNAPII recruitment was observed at both the lys2 and lacZ regions when either Ptet or PGAL1 were the only active promoters, whereas recruitment to one promoter was only detected when transcription was driven from the opposite promoter (Figure 2B), implying that such polymerases corresponded to elongating RNAPIIs. When both promoters were active, the lys2 and lacZ signals were detected, as expected, in the pTGlylaTER system, whereas only the lys2 signal was detected in pTGlyla. This confirms that the northern analysis used can be interpreted in terms of RNAPII capacity to transcribe the different regions analyzed.
Figure 2.
Distribution of RNAPII along the lys2-lacZ transcription units of plasmids pTGlyla and pTGlylaTER. ChIP analyses in the wild-type strain (HRN1-4A) carrying the plasmids pTGlylaTER (A) or TGlyla (B). Schemes of the gene and the PCR-amplified fragments are shown. DNA ratios in regions 1–4 were calculated from the amounts obtained for these regions relative to the amounts of the intergenic region. ChIPs were performed from three independent cultures, and quantitative PCRs were repeated three times for each culture. SDs are indicated as error bars.
Transcription elongation is sensitive to Topos I and II inactivation
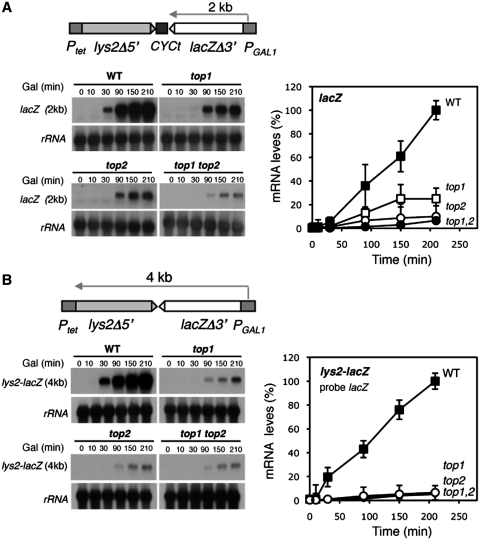
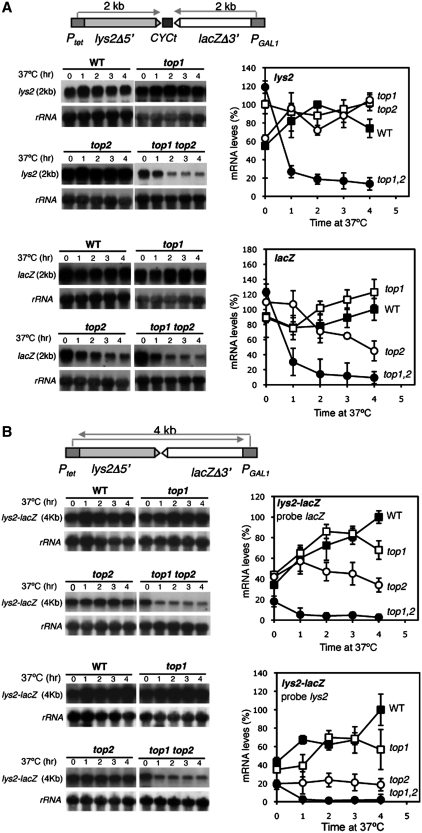
Next, the two constructs were used to analyze transcription in the presence of doxycycline, so that the Ptet promoter was inactivated and transcription was only driven from PGAL1. Under these conditions kinetics experiments were performed of PGAL1-driven transcription with 2% galactose to determine the efficiency of transcription of the 4-kb lacZ-lys2 transcript in pTGlyla and the 2-kb LacZ transcript in pTGlylaTER. As can be seen in Figure 3, transcription of LacZ was clearly reduced in top1Δ mutants, more reduced in top2-1 mutants and further in the double top1Δ top2-1 mutants. Transcription of the 4-kb lys2-lacZ was dramatically decreased in all single and double top mutants. This result is consistent with a defect in transcription elongation so that the longer the RNA is transcribed the stronger the effect. The double mutant showed the strongest effect and a similar dramatic decrease in the accumulation of both of the 2- and 4-kb transcripts.
Figure 3.
Transcription analyses of lys2-CYCt-lacZ and lys2-lacZ fusion transcript driven form the GAL1 promoter. (A) Northern analyses of lys2-CYCt-lacZ containing mRNAs driven from the GAL1 promoter in wild-type (HRN1-4A), top1Δ (TOHR-11A), top2-1 (TORH-6A) and top1Δ top2-1 (TOHR-13C) strains. Mid-log phase plasmid-transformed cells were diluted in 3% glycerol–2%lactate synthetic complete (SC)-Trp medium plus doxycycline and diluted into identical fresh media to an OD600 of 0.4 and incubated for 16 h. Galactose was then added and samples were taken for northern analyses at different times. RNA levels in arbitrary units were obtained by quantification of signals intensities in a FUJI FLA 5000 and normalized with respect to rRNA levels of each sample. Wild-type mRNA levels were taken as 100%. (B) Northern analyses of lys-lacZ-containing mRNAs driven from the GAL1 promoter using the same strains as in A.
Different studies have revealed that yeast mutations impairing transcription elongation causes a reduction in mRNA accumulation that in most cases increases with the length of the transcript unit analyzed (23). Using plasmid-borne PGAL1::PHO5 and PGAL1::lacZ constructs containing the 1.2-kb and 3.1-kb yeast PHO5 and bacterial lacZ ORFs, respectively, under the same GAL1 promoter, the negative effect of topoisomerase mutations was confirmed in transcription of long DNA sequences. The single and double Topo mutants have a significant effect on the efficiency of transcription of the PHO5 ORF, the reduction in the accumulation of the lacZ mRNA being stronger in the single top2-1 and in the double thp1Δ top2-1 (Supplementary Figure S1). Given that transcription in both constructs is driven from the same GAL1 promoter, this result excludes the possibility that the major transcription defects observed in top mutants occur at transcription initiation, but at elongation. In addition, consistent with the idea that eukaryotic Topos I and II can partially replace each other, we showed that overexpression of Top1 can partially suppress the lacZ mRNA accumulation defect of top2-1 mutants (Supplementary Figure S2).
Convergent transcription is impaired under Topos I and II inactivation as a function of supercoiling accumulation
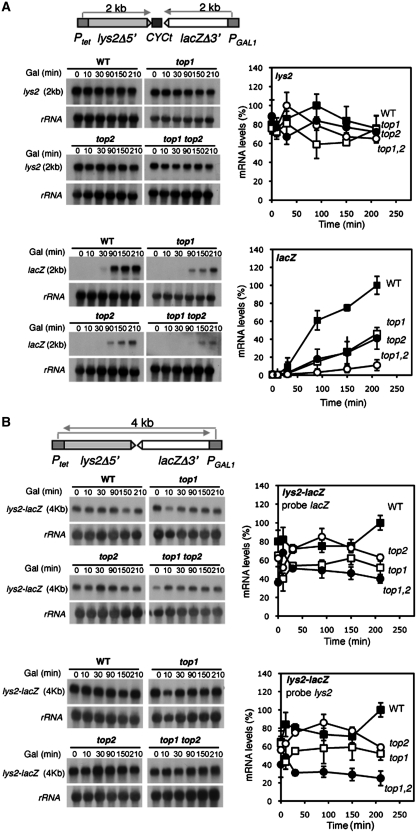
Next, we assayed whether or not convergent transcription affected transcription efficiency. This analysis of convergent transcription was made in the absence of doxycycline to allow continuous Ptet-driven transcription. Time-course kinetics was performed for the PGAL1-driven transcripts after galactose addition. As can be seen in Figure 4, in the pTGlylaTER system with the transcription terminator the overall levels of the Ptet-driven lys2 transcript did not show a significant change after activation of the converging PGAL1. At 200 min of PGAL1 activation the overall levels of lys2 transcripts were similar to the levels before activation. At intermediate times some differences between wild-type and mutant strains were evident, but these were minor. This may be a response to cell adaptation to new conditions. Importantly, at 200 min of activation the lys2 transcript levels were the same in wild-type and mutant strains, with a slight decrease in top1Δ top2-1 mutants. Instead, transcription from the GAL1 promoter, as determined by accumulation of the 2-kb lacZ transcript, was clearly affected in top1Δ and top2-1 mutants (38–40% of wild-type levels) and much more in the double top1Δ top2-1 mutants (10% of wild-type levels) (Figure 4A). The clear impairment in transcription of the 2-kb lacZ transcript caused by topoisomerase mutations as compared to the poor or no effect on the 2-kb lys2 transcript (both transcripts are of the same size but are driven from promoters of different strengths), indicates that the higher the transcription rate the higher the requirement of topoisomerases.
Figure 4.
Northern analyses of convergent transcription in the lys2-CYCt-lacZ and lys2-lacZ fusion constructs. (A) Northern analyses of lys2-CYCt-lacZ mRNAs driven from the tet and GAL1 promoters in wild-type (HRN1-4A), top1Δ (TOHR-11A), top2-1 (TORH-6A) and top1Δ top2-1 (TOHR-13C) strains. The 3-Kb XbaI-EcoRV lacZ internal fragment were used as DNA probe. (B) Northern analyses of lys2-lacZ mRNAs driven from the Ptet and PGAL1 promoters. Other details as described for Figure 3.
In the pTGlyla system there is little change in the total amount of 4-kb transcripts (Figure 4B) during the kinetics performed. As shown above, these results indicate that pTGlyla, which contains no terminator sequence between the converging promoters, undergoes promoter interference caused at PGAL1 by the RNAPII coming from Ptet. However, despite the interference phenomenon, transcripts were moderately low in single top1 and top2 mutants (60–65% of wild-type levels after 200 min activation) and lower in double top1 top2 mutants (38–45%) as compared to wild-type levels. This is consistent with the importance of topoisomerase activities in converging transcription. It is worth noting the different topoisomerase requirements of transcription of the 4-kb lys2-lacZ and 2-kb lys2 fragments in the pTGlyla and pTGlylaTER systems (compare Figures 4A. up and D, bottom). This is due to the longer length of the lacZ-lys2 fragment, consistent with the conclusion that the longer the transcript the higher the dependency of transcription on topoisomerase activity (Figure 1). Altogether, the results indicate that topoisomerases I and II are required for efficient RNAPII transcription elongation.
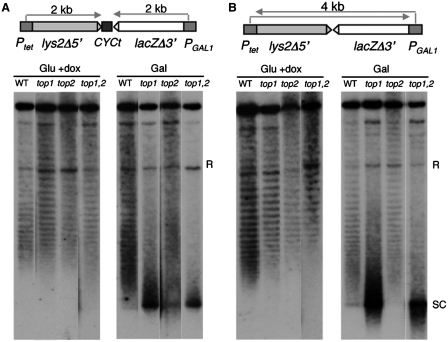
To assay whether the transcription defect caused by top mutations was linked to the incapacity to resolve supercoiling, rather than to a specific function of any of the topoisomerases at specific target DNA sites, supercoiling of the plasmids was analyzed carrying both the pTGlylaTER and pTGlyla systems under conditions of transcription activation of both promoters. As can be seen in Figure 5, a large proportion of supercoiled DNA is accumulated in top mutants, in particular in top1 and top1 top2 when both promoters are active, but not when they were inactive. Experiments were performed similarly to previous reports using chloroquine concentrations in which all topoisomers show negative supercoiling (9). Importantly, accumulation of the supercoiled plasmid band is clearly stronger in pTGlyla versus pTGlylaTER, consistent with the conclusion that the longer the length of the transcription unit, the higher the change in supercoiling produced by transcription and the stronger the requirement of topoisomerases. Even though on a theoretical basis the impact of transcription on DNA supercoiling is local, TOP1 inactivation has a global impact on plasmid supercoiling as has been previously reported (9). As previously shown (9) accumulation of global supercoiling was not evident in top2-1 mutants.
Figure 5.
Analysis of the effect of top1Δ and top2-1 on DNA supercoiling. (A) Chloroquine gel electrophoresis analysis of plasmid pTGlylaTER in wild-type (HRN1-4A), top1Δ (TOHR-11A), top2-1 (TORH-6A) and top1Δ top2-1 (TOHR-13C). Strains were cultured under non-transcription conditions in glucose plus doxycycline (Glu +dox) and under transcription conditions in galactose without doxycycline (Gal) at 30°C. Electrophoresis was carried out in the presence of 4 µg/ml chloroquine, and hybridization was performed with a labelled LYS2 DNA probe. Southern reveals bands representing topoisomers differing in linking number by steps of one. With the chloroquine concentration used, all topoisomers are negatively supercoiled, with those of increasing negative superhelicity migrating faster in the gel. The band marked as SC consists of the most negatively supercoiled species that are not resolved by the concentration of chloroquine used here. (B) Chloroquine gel electrophoresis analysis of plasmid pTGlyla.
Efficient RNAPII-driven convergent transcription requires Topos I and II
So far, all our previous experiments were performed at 30°C, a semi-permissive temperature to allow growth of top2-1 thermosensitive mutants. Yet, it was possible to see a synergistic effect on double top1 top2 mutants suggesting that Topo II has a role in transcription in vivo. This conclusion was confirmed by assaying the effect on complete Top II inactivation. These experiments were repeated on convergent transcription in both the pTGlyla and pTGlylaTER systems after shifting cells to the restrictive temperature of 37°C. As can be seen in Figure 6A, in the pTGlylaTER system in which both convergent promoters are activated, whereas lys2 and lacZ transcript levels show little effect on top1Δ and top2-1 single mutants, which were dramatically reduced in the double top1Δ top2-1 mutants (20% of wild-type levels after 2 h at 37°C). In the pTGlyla system, whereas in wild-type and, to a lesser extent, in the top1Δ mutant, the 4-kb transcript levels increased with time at 37°C consistent with a faster metabolism at this temperature, they did not change in top2-1 mutants and dropped dramatically in top1Δ top2-1 mutants (Figure 6B). These results were confirmed by analyzing the effect of top2-1 at different temperatures, from 30°C to 33°C in which it could be seen a dramatic decrease in the accumulation of lacZ mRNA in the top1Δ background as the temperature increased (Supplementary Figure S2). Altogether, the results indicate that Topo II plays a critical role in RNAPII transcription in vivo, which indeed is quantitatively more important than that of Topo I.
Figure 6.
Effect of top1 and top2 mutations on transcription of lys-CYCt-lacZ and lys-lacZ fusion constructs at 37°C. (A) Northern analyses of lys-CYCt-lacZ-containing mRNAs driven from the tet and GAL1 promoters in wild-type (HRN1-4A), top1Δ (TOHR-11A), top2-1 (TORH-6A) and top1Δ top2-1 (TOHR-13C) strains at 37°C. Mid-log phase plasmid-transformed cells were diluted to an OD600 of 0.4 synthetic complete (SGal)-Trp medium at 26°C and shifted to 37°C, samples were taken for northern analyses at different times. (B) Northern analyses of lys-lacZ mRNAs driven from the Ptet and PGAL1 promoters. Other details as described in Figure 3.
top1Δ and top2-1 confer a general hyper-recombination phenotype
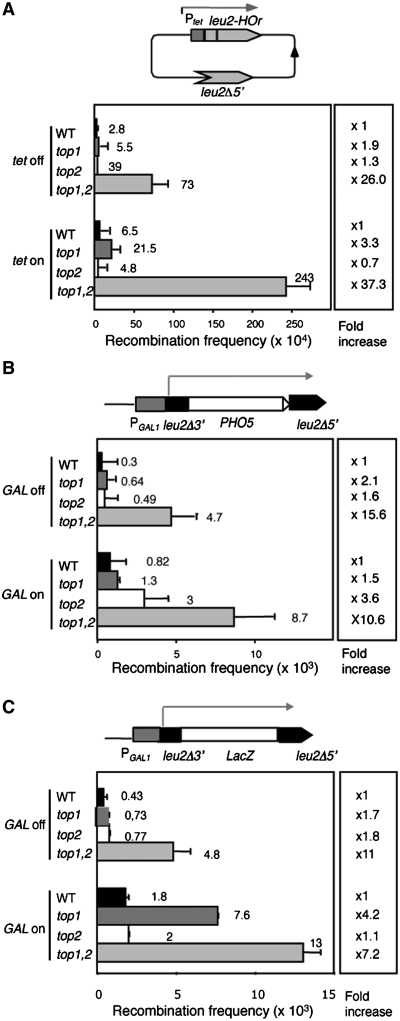
It is known that top1Δ mutants confer a strong hyper-recombination at the rDNA locus (17). To test whether this lack of effect was general for any DNA region and for different types of recombination events and whether it was related to transcription, the effect of top mutations on recombination was analyzed in different and highly sensitive plasmid-borne recombination systems. First we assayed recombination in the pTINV system based on two inverted repeats of the LEU2 gene, one copy of which is a 5′-end truncation (leu2Δ5′) and the other a 21-bp insertion mutation under the control of the regulated tet promoter (Ptet::leu2-HOr) (31). This system allows detection of gene conversion events of the leu2-HOr allele with or without crossovers as a function of transcription, which leads to Leu+ recombinants that are scored on SC-leu plates (Figure 7A). As can be seen in Figure 7A, whereas Leu+ recombination levels in top1Δ and top2-1 mutants were similar to the wild-type both with or without transcription, recombination was increased 26- and 37-fold without or with transcription. Next, we tested the effect on recombination in the pGL-PHO5 system carrying two 0.6-kb leu2 direct repeats under the control of the GAL1 promoter and the PHO5 ORF located between the repeats (32).This system is used to study deletions occurring by recombination between the two leu2 direct repeats that are scored as Leu+ colonies (Figure 7B). It can be used either with active transcription of the direct-repeat construct in media supplemented with 2% galactose or without transcription in media with 2% glucose. Leu+ deletions occurred at similar levels in top1Δ and top2-1 mutants as in the wild-type, whereas in top1Δ top2-1 mutants there was a significant increase in recombination both with or without transcription (15- and 10-fold increase, respectively) (Figure 7B). These data indicate that only a simultaneous defect of topoisomerases I and II lead to a general increase in recombination, which is primarily independent of transcription of the DNA sequences involved in the recombination event. Similar results were observed with the pGL-lacZ system (32), the same as pGL-PHO5 with LacZ between the leu2 repeats (Figure 7C). A clear increase in recombination was observed in top1 top2 mutants with or without transcription (7.2- and 11-fold increase). However, it is worth noting that a significant increase (4.2-fold) was also observed in single top1 mutants but only under transcription conditions, suggesting that the torsional stress caused by transcription elongation through the lacZ sequence is recombinogenic in the absence of Topo I.
Figure 7.
Recombination analyses of wild-type, top1Δ, top2-1 and top1Δ top2-1 strains. (A) Recombination between inverted repeat in plasmid pRS316-TINV carrying two inverted leu2 sequences. One copy is the leu2-HOr allele under the tet promoter and the other is a 5′-end truncated allele (leu2Δ5′). Transcription of leu2 is driven from tet promoter. Leu+ recombinants can arise by gene conversion of leu2-HOr without an associated inversion or by crossover occurring upstream of the HO site, whether or not associated with gene conversion. (B) Recombination between directs repeats in the system GL::PHO5 carrying a 600-bp truncated leu2 repeat under the PGAL1 promoter. Leu+ recombinants can arise by the deletion of the PHO5 sequence. (C) Recombination between directs repeats in the system GL::LacZ carrying 600-bp truncated leu2 repeats under the PGAL1 promoter. Leu+ recombinants arise by the deletion of the LacZ sequence.
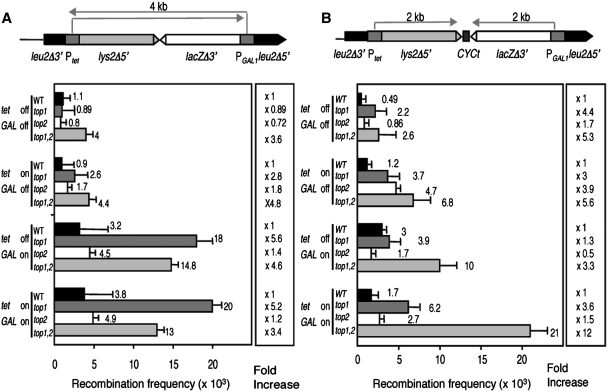
To test whether hyper-recombination could also be observed in direct-repeat systems containing the TGlyla and TGlylaTER converging transcription systems, new plasmid-borne recombination systems were constructed by inserting the TGlyla and TGlylaTER constructs between the two 0.6-kb leu2 direct-repeats of the plasmid-borne L repeat construct previously published (33) to create the recombination systems L-lyla and L-lylaTER (Figure 8). The frequency of Leu+ deletions was determined under conditions in which none (media with glucose and doxycycline), only one (with galactose and doxycycline or glucose without doxycycline) or both (with galactose without doxycycline) promoters were activated. As can be seen in Figure 8A, a 3.4- to 5.6-fold increase in Leu+ deletions was observed in the double top1Δ top2-1 mutants under the four different conditions of transcription tested as well as in the single top1Δ mutant when GAL1p-driven transcription was active, regardless of transcription from the tet promoter. Similar results were observed in the L-lylaTER (Figure 8B). In these cases, a 3.3- to 12-fold increase in recombination was observed in the double mutant top1Δ top2-1 in the four conditions, the major effect being observed when both promoters were active. A weaker effect could be seen in single top1Δ mutants. Altogether, these results indicate that only a simultaneous defect of topoisomerases I and II lead to a general increase in recombination, and this effect is primarily independent of transcription of the DNA repeats and their intervening DNA sequences involved in the recombination event.
Figure 8.
Recombination analyses of wild-type, top1Δ, top2-1 and top1Δ top2-1 strains in DNA substrates undergoing converging transcription. (A) Recombination frequencies were determined in wild-type (HRN1-4A), top1Δ (TOHR-11A), top2-1 (TORH-6A) and top1Δ top2-1 (TOHR-13C) strains transformed with plasmid pLlyla carrying the direct-repeat system L-Llyla in which transcription is under the PLEU2 promoter. (B) Recombination frequencies in wild-type (HRN1-4A), top1Δ (TOHR-11A), top2-1 (TORH-6A) and top1Δ top2-1 (TOHR-13C) strains transformed with plasmid pLlylaTER carrying the direct-repeat system L-LlylaTER. For more details see ‘Materials and Methods’ section.
In this sense, mutations in the THO complex, such as hpr1Δ, cause a strong increase in direct-repeat recombination that is linked to transcription. As Topo mutations lead to weaker hyper-recombination that is not restricted to direct-repeat systems and mainly in a transcription-independent manner, it seemed reasonable that both type of mutations may affect recombination via different pathways. To test this possibility recombination in the L-lyla system was analyzed in double mutant combinations of hpr1Δ with top1Δ and top2-1 mutations. As can be seen in Supplementary Figure S3, hyper-recombination was clearly observed in hpr1Δ mutants under Ptet or PGAL1-driven transcription (12- to 15-fold), but to a lesser extent when both the tet and GAL1 promoters were inactive. The triple mutant hpr1Δ top1Δ top2-1 shows a synergistic effect on hyper-recombination that reached 47- to 71-fold increase when only one promoter was active, and 123-fold when both promoters were active. Although we cannot exclude that transcription outside of the DNA recombination construct in the plasmid could influence the general levels of recombination due to its global impact on plasmid supercoiling, our result shows that, in contrast to THO–complex mutations, the effect of Topos I and II mutations in non-rDNA recombination is primarily not linked to transcription of the recombination system.
DISCUSSION
Here we show that Topos I and II play a role in eukaryotic RNAPII-driven transcription and genome stability in the yeast S. cerevisiae. RNAPII transcription requires Topos I and II at the elongation step, this being the role of Topo II in elongation independent of Topo I and of supercoiling accumulation. Notably, the impact of supercoiling accumulation in genome instability is not related to transcription of the direct-repeat in which instability is measured. Consequently, our work reveals independent effects of the overlapping functions of Topos I and II in transcription and genome integrity. In addition, we show that a promoter cannot be efficiently activated if the anti-sense DNA strand is transcribed, a result that suggests new possibilities to explain regulation of gene expression by promoter interference.
RNAPII transcription elongation impairment by topological constraints in vivo
The relevance of topoisomerases and supercoiling constraint in transcription was anticipated and demonstrated in the past in prokaryotic and eukaryotic cells (2,5). Different reports have shown that the levels of DNA supercoiling have an impact in RNA synthesis in E. coli (34,35,36). Requirement of DNA topoisomerases in transcription can be explained by the twin-supercoiled-domain model (6). The negative impact of failure to control DNA supercoiling in transcription is believed to be linked to the transcription elongation step. Evidence for this has been provided in the rDNA regions, as in yeast (2,14). Other studies of transcription at the rDNA region in human cells are consistent with this view (37). In yeast, it has also been shown that positive supercoiling diminishes mRNA transcription, probably at the initiation step (15). In human cells it has been shown that DNA topoisomearse I is involved in both repression and activation of transcription (38), providing evidence that DNA supercoiling can also control transcription initiation. All these data plus the observation that yeast topoisomerase mutants accumulate transcription-dependent supercoiling (9) indicates that this supercoiling needs to be removed by the action of DNA topoisomerases to allow transcription. Whereas the transcriptional role of Topo I has been previously documented, no evidence has been reported for a role of Topo II in RNAPII transcription in vivo, despite the fact that Topos I and II are redundant in eukaryotes. Evidence for a role of Topo II in RNAPII transcription has been shown in chromatin templates in vitro (39). In addition, despite the expectation that transcription elongation should be sensitive to supercoiling, according to the twin-supercoiled-domain model, experimental evidence for the negative effect that supercoiling may have in RNAPII-driven transcription elongation was lacking.
Our study on RNAPII transcription has been based on two convergent transcription units. Convergent transcription should create a local accumulation of positive supercoiling ahead of each polymerase that would quickly accumulate in topoisomerases mutants. Our work shows that, indeed, convergent transcription is reduced when both promoters are active in both single top1 and top2 mutants. Such a reduction in transcription efficiency is synergistic in double mutants top1 top2, in which transcription is reduced to almost undetectable levels. The results reveal a role for Topo II in transcription. It is likely that Topo II function has a minor impact in wild-type cells expressing Topo I. Nevertheless, our result suggests that Topo2 can replace Topo I in transcription when the latter is absent in the cell. This negative effect of Topo mutations on transcription is seen when the length of the mRNA is 4 or 2 kb, which likely indicates that 2-kb of transcription run is already sufficient to see the effect of supercoiling accumulation in transcription. Interestingly, transcription in simple systems in which transcription is only unidirectional, no effect of topoisomerases mutations is observed for 1.2-kb transcript units, but only in 3-kb long transcription units, even though this effect is clearly lower than for convergent transcription. This is consistent with a major impact of convergent transcription on local DNA supercoiling and with the idea that the longer the DNA sequence transcribed the higher the supercoiling. The impact in supercoiling has been shown indeed by chloroquine gels, as expected from previous work (9) (Figure 5). Although we cannot exclude the possibility that the reduction in transcription efficiency could be determined at the level of transcription initiation, altogether the data indicate that transcription elongation is a major step impaired by supercoiling accumulation. Accordingly with, we have been able to observe a synergistic effect on transcription efficiency in a top1 top2 mutant carrying a deletion of the SPT4 elongation factor gene in the 1.2-kb transcript. This indicates that under impaired transcription elongation, supercoiling accumulation has a negative impact on transcription elongation (Supplementary Figure S4).
Therefore, our work shows that mRNA transcription is strongly impaired by topoisomerase inactivation and supercoiling accumulation. Both Top2 and Top1 play important roles in removing superhelicity during transcription. Therefore, Top2 not only substitutes for Top1 to control supercoiling accumulation during transcription elongation, but it also plays a function in wild-type cells, consistent with its reported roles in RNAPII transcription in chromatin templates (39). Interestingly it has recently been shown that Topo2 is required to relax nucleosomal DNA (40) and for rDNA transcription in yeast (2). Further investigation would be necessary to define how Top2 acts during transcription, and whether its role is only related to supercoiling accumulation, given the fact that under the conditions studied no strong accumulation of DNA supercoiling is observed in top2 mutants in contrast to top1 mutants (Figure 5).
It is worth noting that our assays provide an example in which promoter interference has a negative impact on the ability of a promoter to be activated. Thus, when the GAL1 promoter was active, in which case the anti-sense DNA strand of the tet promoter was transcribed, the tet promoter could not be efficiently activated. This suggests that promoter transcription interferes with its activity, opening the possibility that promoter inactivation by CUTs or anti-sense RNAs (41,42) might also be caused by transcription, which could presumably disrupt the proteins and architecture of a functional promoter.
Direct-repeat hyper-recombination in topoisomerase-defective cells
Previous evidence has shown that rDNA stability was highly dependent on DNA topoisomerases I and, to a lesser extent, on Topo II (17). Short circular plasmids carrying non-rDNA regions have been shown to suffer frequent multimerization in yeast Topo mutants (43). Although multimerization is attributed to recombination, no study has been performed on whether such multimerization could be due to replication impairment leading to re-initiation and/or sigma-like type of replication due to enhanced superhelicity. The fact that this is observed in short circles indicate a specific structural demand, presumably associated with changes in supercoiling that may not be related only to recombination. The increased in recombination observed in our systems is in any case lower and not accompanied by multimerization.
A different study using direct-repeat systems such as those used in this study, revealed increases in direct-repeat recombination of 40- to 100-fold only in top1 top2 mutants (18). An important difference between those direct-repeats did not contain DNA sequences between the tandem direct repeats whereas the systems used in this study contained between 2 and 4 kb. This may explain why in our case at most we get a 10-fold increase in recombination. We previously showed that the distance between the repeats has an impact on recombination: the longer the intervening sequence the higher the probability that recombination events are initiated there (33). Long repeats may have a steric impediment that would interfere with a proper pairing of the two repeats along their length, whereas such steric limitations may not exist when the distance between the repeats is longer than the repeat units. Failure to relax negative supercoiling might limit pairing of both DNA repeats. Interestingly, however, our study reveals that topoisomerase depletion strongly increases gene conversion (37-fold). As it seems plausible that gene conversion is mainly initiated by DSBs, our study suggests therefore a general impact of negative supercoiling on genome fragility linked to DNA breaks.
Given the impact of Topo depletion on transcription in general and transcription of the convergent constructs used in this study in particular, it is notable that the increase in recombination is not linked to the increase in the levels of transcription of the direct repeat itself. Even when convergent transcription is activated between the two direct repeats, with or without a transcription terminator in between, hyper-recombination is not significantly affected. This implies that the hyper-recombination caused by topoisomerase inactivation is not caused by breaks that might occur as a consequence of local conflicts generated between transcription and replication. This may contrast with the hyper-recombination in the rDNA (8).
In non-rDNA regions, evidence has recently shown that in human cells Top1 prevents replication obstacles in transcribed sequences, suggesting that such obstacles are mediated by R-loops (44). Indeed it has recently been shown in yeast that R-loops form in yeast cells deleted for TOP1 (45), consistent with previous work in E. coli showing R-loop accumulation in the rDNA in topA mutants (26). Even though it seems clear that transcribed regions are more prone to obstacles, and recombination in our assays are determined in transcribed genes, our data clearly shows that the topological constrains generated in topoisomerases mutants causes instability in DNA regions that are not transcribed either. In our systems in particular, hyper-recombination is observed in the direct repeat regardless of whether or not is transcribed. Another report in yeast (46) showed that Top2 prevents chromosome fragility in regions that are transcribed during S phase. Again, we are able to observe hyper-recombination even in the absence of transcription. Therefore, there are two possibilities, either the chromosome fragility reported in human and yeast in top1 or top2 cells caused by replication impairment in transcribed DNA regions is not resolved primarily by detectable homologous recombination or if they are, transcription-dependent recombination only reflects a minor fraction of chromosome fragility generated by topoisomerase inactivation. The strong impact of topoisomerase inactivation on replication progression would therefore be sufficient to generate the hyper-recombination phenotype observed in top1 top2 mutants. This would fit the expected role of topoisomerases in regulating DNA supercoiling changes occurring during replication fork progression (4,5). Failure to resolve this supercoiling accumulation may cause replication stalls that can resolve in ssDNA breaks or gaps.
Our results exclude the possibility that a co-transcriptional R-loop that could putatively form at the direct repeats could be responsible for recombinogenic breaks. Global supercoiling of the plasmid may also be affected by transcription of DNA regions away from the direct repeat used for the analysis of recombination, but whether such a transcriptional activity influences at distance the levels of recombination of the direct repeat we cannot be certain. However, given the low levels of transcription of the plasmid marker as compared to the high transcript levels of the direct-repeat recombination system driven from the GAL1 promoter, this is an unlikely possibility. In any case recombination would not initiate at the transcribed plasmid marker but away of it in the non-transcribed direct repeat. For the same reason, an R-loop putatively formed in the plasmid outside the direct-repeat recombination system would not be expected to induce the high levels of recombination determined in this study. In conclusion, the genome instability phenotype of Topo mutants does not require transcription of the sequence involved in the recombination event.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–4.
FUNDING
Spanish Ministry of Science and Innovation (BFU2006-05260, BFU2010-16372 and Consolider Ingenio 2010 CSD2007-0015); the Junta de Andalucía (BIO102 and CVI4567). Funding for open access charge: Spanish Ministry of Science and Innovation BFU2010-16372.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank R. Wellinger for critical reading of the manuscript and D. Haun for style supervision.
REFERENCES
- 1.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.French SL, Sikes ML, Hontz RD, Osheim YN, Lambert TE, El Hage A, Smith MM, Tollervey D, Smith JS, Beyer AL. Distinguishing the Roles of Topoisomerases I and II in Relief of Transcription-Induced Torsional Stress in Yeast rRNA Genes. Mol. Cell. Biol. 2011;31:482–494. doi: 10.1128/MCB.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crooke E, Hwang DS, Skarstad K, Thony B, Kornberg A. E. coli minichromosome replication: regulation of initiation at oriC. Res. Microbiol. 1991;142:127–130. doi: 10.1016/0923-2508(91)90019-7. [DOI] [PubMed] [Google Scholar]
- 4.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 5.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook DN, Ma D, Pon NG, Hearst JE. Dynamics of DNA supercoiling by transcription in Escherichia coli. Proc. Natl Acad. Sci. USA. 1992;89:10603–10607. doi: 10.1073/pnas.89.22.10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- 9.Brill SJ, Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54:403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 10.Osborne BI, Guarente L. Transcription by RNA polymerase II induces changes of DNA topology in yeast. Genes Dev. 1988;2:766–772. doi: 10.1101/gad.2.6.766. [DOI] [PubMed] [Google Scholar]
- 11.Drolet M, Wu HY, Liu LF. Roles of DNA topoisomerases in transcription. Adv. Pharmacol. 1994;29A:135–146. doi: 10.1016/s1054-3589(08)60543-8. [DOI] [PubMed] [Google Scholar]
- 12.Chen D, Bowater R, Lilley DM. Topological promoter coupling in Escherichia coli: delta topA-dependent activation of the leu-500 promoter on a plasmid. J. Bacteriol. 1994;176:3757–3764. doi: 10.1128/jb.176.12.3757-3764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DM, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
- 14.Schultz MC, Brill SJ, Ju Q, Sternglanz R, Reeder RH. Topoisomerases and yeast rRNA transcription: negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev. 1992;6:1332–1341. doi: 10.1101/gad.6.7.1332. [DOI] [PubMed] [Google Scholar]
- 15.Gartenberg MR, Wang JC. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc. Natl Acad. Sci. USA. 1992;89:11461–11465. doi: 10.1073/pnas.89.23.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capranico G, Ferri F, Fogli MV, Russo A, Lotito L, Baranello L. The effects of camptothecin on RNA polymerase II transcription: roles of DNA topoisomerase I. Biochimie. 2007;89:482–489. doi: 10.1016/j.biochi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Christman MF, Dietrich FS, Fink GR. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988;55:413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- 18.Trigueros S, Roca J. Failure to relax negative supercoiling of DNA is a primary cause of mitotic hyper-recombination in topoisomerase-deficient yeast cells. J. Biol. Chem. 2002;277:37207–37211. doi: 10.1074/jbc.M206663200. [DOI] [PubMed] [Google Scholar]
- 19.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 20.Keil RL, Roeder GS. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 21.Voelkel-Meiman K, Keil RL, Roeder GS. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luna R, Gaillard H, Gonzalez-Aguilera C, Aguilera A. Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma. 2008;117:319–331. doi: 10.1007/s00412-008-0158-4. [DOI] [PubMed] [Google Scholar]
- 24.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Wellinger RE, Prado F, Aguilera A. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol. Cell. Biol. 2006;26:3327–3334. doi: 10.1128/MCB.26.8.3327-3334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc. Natl Acad. Sci. USA. 1995;92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguilera A, Klein HL. HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol. Cell. Biol. 1990;10:1439–1451. doi: 10.1128/mcb.10.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hecht A, Strahl-Bolsinger S, Grunstein M. Mapping DNA interaction sites of chromosomal proteins. Crosslinking studies in yeast. Methods Mol. Biol. 1999;119:469–479. doi: 10.1385/1-59259-681-9:469. [DOI] [PubMed] [Google Scholar]
- 29.Huertas P, Garcia-Rubio ML, Wellinger RE, Luna R, Aguilera A. An hpr1 point mutation that impairs transcription and mRNP biogenesis without increasing recombination. Mol. Cell. Biol. 2006;26:7451–7465. doi: 10.1128/MCB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Rubio M, Huertas P, Gonzalez-Barrera S, Aguilera A. Recombinogenic effects of DNA-damaging agents are synergistically increased by transcription in Saccharomyces cerevisiae. New insights into transcription-associated recombination. Genetics. 2003;165:457–466. doi: 10.1093/genetics/165.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez-Barrera S, Garcia-Rubio M, Aguilera A. Transcription and double-strand breaks induce similar mitotic recombination events in Saccharomyces cerevisiae. Genetics. 2002;162:603–614. doi: 10.1093/genetics/162.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piruat JI, Aguilera A. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 1998;17:4859–4872. doi: 10.1093/emboj/17.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prado F, Aguilera A. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gellert M. DNA topoisomerases. Annu. Rev. Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- 35.Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- 36.Pruss GJ, Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989;56:521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- 37.Dunaway M. Inhibition of topoisomerase II does not inhibit transcription of RNA polymerase I and II genes. Mol. Cell. Biol. 1990;10:2893–2900. doi: 10.1128/mcb.10.6.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merino A, Madden KR, Lane WS, Champoux JJ, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 39.Mondal N, Parvin JD. DNA topoisomerase IIalpha is required for RNA polymerase II transcription on chromatin templates. Nature. 2001;413:435–438. doi: 10.1038/35096590. [DOI] [PubMed] [Google Scholar]
- 40.Salceda J, Fernandez X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25:2575–2583. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Proudfoot N, Gullerova M. Gene silencing CUTs both ways. Cell. 2007;131:649–651. doi: 10.1016/j.cell.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 43.Trigueros S, Arsuaga J, Vazquez ME, Sumners DW, Roca J. Novel display of knotted DNA molecules by two-dimensional gel electrophoresis. Nucleic Acids Res. 2001;29:E67–E67. doi: 10.1093/nar/29.13.e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell. Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bermejo R, Capra T, Gonzalez-Huici V, Fachinetti D, Cocito A, Natoli G, Katou Y, Mori H, Kurokawa K, Shirahige K, et al. Genome-organizing factors Top2 and Hmo1 prevent chromosome fragility at sites of S phase transcription. Cell. 2009;138:870–884. doi: 10.1016/j.cell.2009.06.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.