Abstract
Besides the fact that miR-96 and miR-182 belong to the miR-182/183 cluster, their seed region (UUGGCA, nucleotides 2–7) is identical suggesting potential common properties in mRNA target recognition and cellular functions. Here, we used the mRNA encoding Glypican-3, a heparan-sulfate proteoglycan, as a model target as its short 3′ untranslated region is predicted to contain one miR-96/182 site, and assessed whether it is post-transcriptionally regulated by these two microRNAs. We found that miR-96 downregulated GPC3 expression by targeting its mRNA 3′-untranslated region and interacting with the predicted site. This downregulatory effect was due to an increased mRNA degradation and depended on Argonaute-2. Despite its seed similarity with miR-96, miR-182 was unable to regulate GPC3. This differential regulation was confirmed on two other targets, FOXO1 and FN1. By site-directed mutagenesis, we demonstrated that the miRNA nucleotide 8, immediately downstream the UUGGCA seed, plays a critical role in target recognition by miR-96 and miR-182. Our data suggest that because of a base difference at miRNA position 8, these two microRNAs control a completely different set of genes and therefore are functionally independent.
INTRODUCTION
Post-transcriptional regulations are complex cellular mechanisms involving cis-acting RNA sequences located throughout the messenger RNA, and their associated trans-regulatory factors (1,2). Typical cis-acting RNA sequences are microRNA sites, the AU-rich element (ARE) or the major protein-coding region determinant of instability (1,3–5). AREs are mainly located in 3′-untranslated regions (UTR) and predicted to control 5–8% of cellular mRNAs (6). By comparison microRNA sites are particular in the sense that they are found in both untranslated and coding regions, and in many organisms, microRNAs (miRNAs) are predicted to exert their regulatory effects on at least 50% of cellular mRNAs (5,7).
MiRNAs are ∼22-nt-long non-coding RNAs, originally discovered in worms and plants, which control expression of a plethora of genes involved in transient and adaptable cellular processes, such as induced proliferation, metabolism or stress. Mainly described as translation regulators in earlier studies (8), more recent reports show that some miRNAs downregulate protein output at the mRNA level by inducing mRNA decay (7,9–12). Many mammalian miRNAs are organized in miRNA families (13) or in gene clusters transcribed as a long polycistronic primary miRNA (e.g. miR182-183 cluster or miR-17-92 cluster) (14). Clustered miRNAs are under intense post-transcriptional controls which fine tune their cellular abundance and biological functions (15). Finally, although double-stranded once matured, miRNAs are loaded into an Argonaute (AGO) family protein as single-guide strand, this association forming the active miRNA-induced silencing complex (miRISC) (7,12).
At a molecular level, miRNAs control gene expression by annealing to their mRNA targets through perfect or imperfect matching following base-pairing rules (5,12). However this interaction depends on many other molecular features amongst which the miRNA-site neighborhood, proximity of poly(A) tail or of termination codon, proximal AU-richness, the number of miRNA sites, as well as protein trans-regulatory factors [Argonautes, GW182/Trinucleotide repeat-containing gene 6A protein (TNRC6) and their accessory proteins] (5,7,12,13). Target recognition is mainly based on conserved and continuous Watson–Crick pairing centered on miRNA positions 2–7, the so-called seed sequence (5,7). Sometimes seed pairing can tolerate wobble (i.e. Let-7a:MYC recognition) or requires an extra match at position 8 (5,9). In other cases, miRNAs can recognize their targets through contiguous base pairing to their central region (16). A match at miRNA position 1 seems not necessary for miRNA:target recognition, nor miRNA function. In fact the miRNA 5′-end nucleotide is hidden in AGO and not accessible for target recognition (5,7). Involvement of 3′-compensatory pairing in miRNA:target recognition is still a matter of debate as functional data failed to demonstrate its general requirement for miRNA function (5). Intriguingly some mammalian miRNAs have multiple isoforms (paralogues) with the same seed region but a variable remaining sequence. Based on this feature, those were classified in families and are predicted to target the same genes (5,13). However questions about the role of such a functional redundancy at the level of a cell or a whole organism mostly remain unanswered.
Here, we evaluated the regulatory potential of specific miRNAs selected by in silico approaches on the Glypican-3 (GPC3), a gene encoding a cell-membrane-embedded glycosyl-phosphatidylinositol-anchored extracellular glycoprotein which belongs to the heparan-sulfate proteoglycan family. GPC3 was chosen as model because of its involvement in various human pathologies (17,18). We found that miR-96 post-transcriptionally controls GPC3 whereas miR-182, a miRNA bearing a 2–7 seed region identical to that of miR-96, had no effect on GPC3 expression. Using the lentiviral- and fluorescent reporter-based method named FunREG method (9,10,19) and molecular approaches, we deciphered the differential mechanism governing the regulation of GPC3 by miR-96 and miR-182.
MATERIAL AND METHODS
Plasmid constructs
The pTRIP-eGFP plasmid has been described previously (9). pTRIP-eGFP-GPC3 was constructed as follows. The GPC3 3′-UTR was amplified with the primers 5′-CAGACTCGAGCTGCCTGGTGCCCAGC-3′ and 5′-GAGAGGTACCCAAAGAAATCCATGCAAAGAG-3′ using normal liver cDNA. The PCR product was cloned into the pGEM-T plasmid (Promega) creating the pGEM-GPC3 plasmid and integrity of the insert was controlled by DNA sequencing. The pGEM-GPC3 plasmid was digested by XhoI and KpnI, and the resulting insert was gel purified and cloned into the pTRIP-eGFP. The plasmids pGEM-GPC3ΔA was constructed by PCR-site directed deletion using the pGEM-GPC3 plasmid as template, two primers spanning the sequence to delete (5′-CATATAGATTGTCCCCATCAAGTTGTGCC-3′ and 5′-GGCACAACTTGATGGGGACAATCTATATGC-3′) as well as the primers spanning the GPC3 3′-UTR (see above). The final PCR product was cloned into the pTRIP-eGFP plasmid as described above. Similarly the pTRIP-eGFP-mutGPC3 and pTRIP-eGFP-G>U GPC3 plasmids were constructed by PCR-site directed mutagenesis using the pTRIP-eGFP-GPC3 plasmid as template. The miRNA site was mutated using either the primers 5′-CCATCAAGTTGTCCGATATTATTCTCCTATG-3′ and 5′-CATAGGAGAATAATATCGGACAACTTGATGG-3′ or the primers 5′-CCATCAAGTTTTGCCAAATTAT-3′ and 5′–ATAATTTGGCAAAACTTGATGG-3′. Each PCR product was digested by XhoI and KpnI, cloned into pTRIP-eGFP and DNA sequenced as described above.
Cultures of cell lines and primary hepatocytes, small RNA synthesis and transfection
The hepatocellular carcinoma (HCC)-derived HuH7 and SNU398 cell lines were grown in D-MEM medium (Invitrogen) containing 10% FCS and penicillin/streptomycin antibiotics. The Luciferase small interfering RNA (siLuc, sense 5′-CGUACGCGGAAUACUUCGA-3′) was from Eurofin MWG Operon. The miRNA mimics and hairpin inhibitors, as well as Argonaute protein siRNAs were from Thermo Scientific Dharmacon Products. When mentioned, artificial double-strand wild type or mutated miRNAs (bearing a RNA backbone) were chemically synthesized and purified (Supplementary Table S2). Then the corresponding strands were annealed. Small RNAs were transferred into the target cells by reverse-transfection using Lipofectamine RNAi Max (Invitrogen) following manufacturer's instructions at a final concentration of 12 nM.
Lentiviral production, titration and cell transduction
Production and titration of infectious lentiviral particles, as well as biosafety considerations, procedures and policies have been described previously (9). Lentiviral particles were added to the target cells and incubated for 24 h. Then the cells were washed twice in PBS and grown in the presence of medium for 6 days before experimental use.
FunREG analysis
Flow cytometry
One week after transduction, cells were washed in PBS, detached with trypsin/EDTA, collected and analyzed by FACS using a BD FACSCanto II (BD Biosciences, San Jose, CA, USA) and the BD FACSDiva software as described previously (9).
Real-time quantitative PCR and RT–PCR
Genomic DNA and total RNA were respectively isolated with the Nucleospin Tissue kit (Macherey-Nagel) and the TRI Reagent (Sigma) following manufacturer's instructions. Complementary DNA was synthesized with the AMV Reverse Transcriptase (Promega). Real-time quantitative PCR (QPCR) amplifications were performed in 12-µl multiplex PCR reactions containing 1× SYBR® Green Supermix (Quanta Biosciences). The primers used were as described previously (9). TaqMan microRNA assays (Applied Biosystems) were used to quantify the expression levels of mature miRNAs. The Albumin gene and 18s ribosomal RNA served as internal controls for normalization when using, respectively, genomic DNA or cDNA as template (9). Subsequent data analyses were performed using the Step One Plus Quantitative PCR System (Applied Biosystems).
Antibodies and western blot analyses
Whole cell extracts were prepared by treating cells with RIPA buffer (Sigma). Proteins were separated by 10% SDS–PAGE and blotted onto nitrocellulose membrane (Protran, Whatman). After blotting, total loaded proteins were quantified with SYPRO Ruby following manufacturer's instructions (Invitrogen). Stained membranes were imaged with the Molecular Imager PharosFX Plus System (Biorad) and proteins were analyzed with the Quantity One (Biorad) basic software. Then membranes were saturated in Odyssey Blocker and successively incubated with the indicated primary antibodies and adequate InfraRed-labeled secondary antibody (either IRDye-680 or -800 conjugated secondary antibodies) following manufacturer's instructions. Fluorescence signals were detected and quantified using the Odyssey infrared imaging system. Blocker and Odyssey infrared imaging system were from LI-COR Biosciences (ScienceTec, Les Ulis, France). Specific protein staining was normalized to the quantity of total proteins. Anti-GPC3 was from Biomosaics. Anti-FOXO1 (C-20) was from Santa Cruz Biotechnology, anti-FN1 was from BD Biosciences and anti-ADCY6 (SAB2100054) was from Sigma.
Statistical analyses
All analyses were done using GraphPad Prism 5.0 software. Data are represented as mean with standard deviation (SD) from the indicated number of independent experiments. When experiment contained three groups of values or more, regular one-way analysis of variance (ANOVA) was used for the comparison of multiple means. Means were considered significantly different if the P < 0.05. NS means ‘not significant’. The ANOVA test was followed by a Bonferroni's multiple-comparison post-test and selected pairs of data were compared. Significant variations were represented by asterisks above the corresponding bar when comparing the test with the control condition or above the line when comparing the two indicated conditions.
RESULTS
miR-96, but not miR-182, controls Glypican-3 expression
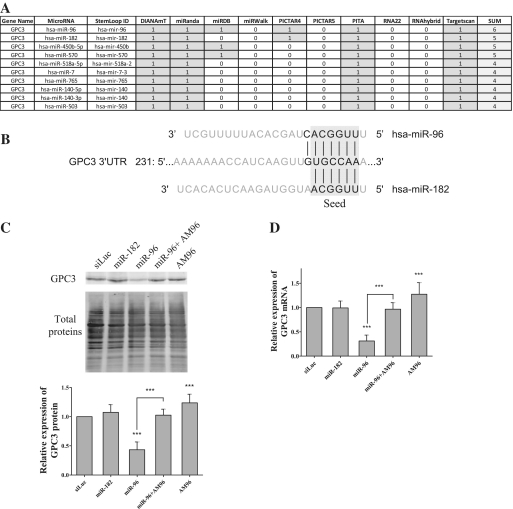
To find miRNAs involved in GPC3 post-transcriptional regulation, we submitted the GPC3 mRNA to 10 established prediction programs gathered on miRWalk [miRWalk—A Database on Predicted and Validated microRNA Targets, (http://mirwalk.uni-hd.de)] and selected those targeting the 3′-UTR. As shown in Figure 1A, miR-96 appeared in top position with 6 positive predictions over 10. MiR-182, which displays the same 5′-UUGGCA-3′ seed sequence to positions 2–7 (Figure 1B) than miR-96, appeared just below miR-96 among the four top-ranked miRNAs with 5 positive predictions over 10 (Figure 1A). By individually testing different programs, miR-96 and miR-182 were predicted to target GPC3 using Targetscan (Figure 1B and Supplementary Figure S1A) (13), Diana microT v.4 (Supplementary Figure S1B) (20), miRanda/mirSVR (Supplementary Figure S1C) (21) and Pictar (Supplementary Figure S1E) (22), whereas only miR-96 was found amongst the miRNAs predicted using PITA (23) and miRDB (24,25) (Supplementary Figure S1D and F, respectively). The unique 5′-UGCCAA-3′ miRNA site located in the GPC3 3′-UTR and predicted to pair with the seed of miR-96 or miR-182 is very conserved amongst species (Supplementary Figure S2A) suggesting an apparent evolutionary-conserved partnership between GPC3 and these two miRNAs. In silico, miR-96:GPC3 3′-UTR pairing is defined as an 8-mer site by Targetscan, whereas miR-182:GPC3 3′-UTR interaction is defined as 7-mer-1A (13). The only difference between these two types of sites resides in the fact that in miR-96, the nucleotide at position 8 immediately downstream the seed can pair the GPC3 3′-UTR (G–C pair), whereas that of miR-182 cannot (Figure 1B). Finally miR-96 and miR-182, together with miR-183, belong to the same intergenic miRNA cluster (miR-182-183, miRBase (26)).
Figure 1.
GPC3 is a target of miR-96, but not of miR-182. (A) The 10 highest-ranking miRNA:GPC3-3′-UTR pairings as predicted by miRWalk using the indicated algorithms (1/grey: pairing predicted; 0/empty: pairing not predicted). (B) Schematic representation of the pairing of GPC3 3′-UTR with miR-96 or miR-182 as predicted by TargetScan. In this figure and the following ones, base pairings between indicated mRNAs and miRNAs are represented by vertical lines. (C and D): HuH7 cells were transfected with small RNAs as indicated. Three days later, the amounts of GPC3 protein (C) and mRNA (D) were quantified (ANOVA: P < 0.0001; n = 6). In Panel C, a representative western blot experiment is shown on top and a bar graph recapitulating means with standard deviation (SD) of six experiments is shown on bottom. In this figure and the following ones, bars represent means, error bars indicate SD and the ANOVA test was followed by a Bonferroni's multiple comparison post-test. ***P < 0.001.
Based on these observations, experimental analyses were undertaken. Expression of miR-96 and miR-182 in HuH7 and SNU398 cells, two human HCC-derived GPC3-expressing cell lines, was first confirmed by RT-qPCR (Supplementary Figure S2B). Results showed that miR-182 was apparently more abundant in both HCC cell lines than miR-96 (compare ΔCt values in Supplementary Figure S2B) suggesting the existence of post-transcriptional regulations in the course of miR-182-183 cluster biogenesis. Then we tested the ability of the two miRNAs to control GPC3 expression in HCC-derived HuH7 cells by over-expressing each miRNA using cell transfection. As shown in Figure 1C, miR-96 significantly decreased GPC3-protein amount by more than half. As expected, a specific hairpin inhibitor, AM96, counterbalanced the negative effect induced by miR-96. However when used alone, AM96 slightly, but significantly, increased GPC3-protein expression suggesting that it efficiently interacted with the endogenous miR-96 and inhibited its function. Comparable results were obtained when monitoring GPC3-transcript amounts (Figure 1D) suggesting that miR-96 downregulated GPC3 expression at the mRNA level. Surprisingly, although miR-96 and miR-182 carry the same seed (Figure 1B), miR-182 had no effect on GPC3 protein expression, nor mRNA level (Figure 1C and D). This absence of effect could not originate from a poor cell-transfection efficiency as miR-182 overexpression was observed in the corresponding transfected HuH7 cells (Supplementary Figure S2C). To assess miR-182 functioning, we studied its regulatory effect on Adenylate cyclase type 6 (ADCY6), one of its validated target (27). Noticeably ADCY6 3′-UTR is predicted to contain one 8-mer site for miR-182 and three 8-mer sites for miR-96 (Figure 4A). As shown in Supplementary Figure S2D, both miR-96 and miR-182 downregulated expression of ADCY6 mRNA and protein, despite the fact positions of their predicted sites into the ADCY6 3′-UTR remain to be confirmed. Together these results showed that (i) miR-96 specifically controls GPC3 expression at the mRNA level, and (ii) miR-182 is functionally inefficient on GPC3.
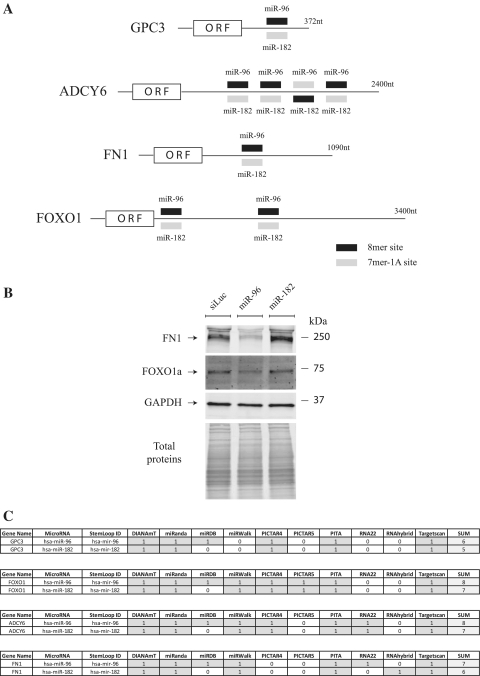
Figure 4.
Target preference of miR-96 and miR-182 depends on presence of a 8-mer site. (A) Schematic representation of the indicated mRNAs with their miR-96/182 sites. The type of site as defined by Targetscan is as indicated (see the legend at the bottom-right). (B) HuH7 cells were transfected with small RNAs as indicated. Three days later, the amounts of FN1, FOXO1 and GAPDH proteins were measured. Representative of three independent experiments. (C) miRNA:target 3′-UTR pairings as predicted by miRWalk using the indicated programs.
miR-96 post-transcriptionally controls GPC3 expression through the 3′-UTR of its transcript
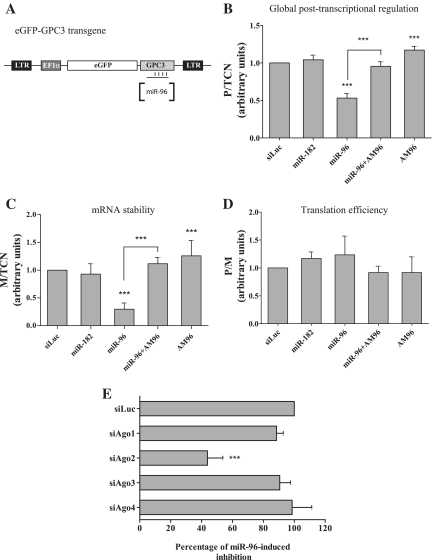
To assess the regulatory role of miR-96 on GPC3 and identify the molecular process, we followed the FunREG experimental pipeline (9,19). Infectious lentiviral particles were used to deliver an eGFP-reporter transgene bearing the GPC3 3′-UTR (Figure 2A) into HuH7 cells (and SNU398 cells, Supplementary Figure S3A). A multiplicity of infection (moi) of 0.5 was used to generate a cell population with no more than one transgene copy per cell (9). One week later the average number of lentiviral transgene copies per cell (‘transgene copy number’, TCN) was assessed by quantitative PCR. Then cells expressing the eGFP-GPC3 transgene were transfected with the above-mentioned small RNAs and 3 days later, the eGFP protein (P) and mRNA (M) expressions were determined by FACS and RT-qPCR, respectively. Finally the three ratios (P/TCN, M/TCN and P/M) were calculated (9). With a fluorescent reporter transgene bearing the GPC3 3′-UTR, results obtained with the FunREG method fully recapitulated the ones obtained with endogenous GPC3. Indeed expression of the eGFP-GPC3 transgene significantly decreased by 47% in HuH7 cells transfected with miR-96 (Figure 2B). A similar but less profound effect was observed with miR-96 when using another HCC cell line (SNU398 cells, Supplementary Figure S3A). This difference could be due to the fact that SNU398 cells are generally more refractory to cell transfection than HuH7 cells. Whereas the specific AM96 neutralized miR-96 effect, it led to a slight but significant increase in eGFP expression when used alone (Figure 2B). All these results demonstrated that in HuH7 cells, the GPC3 3′-UTR is targeted by miR-96 in either its mimic or endogenous form. Moreover M/TCN (Figure 2C) and P/M (Figure 2D) ratios, which are respectively indicative of mRNA stability and translation efficiency (9), showed that miR-96 induced destabilisation of the chimeric eGFP-GPC3 mRNA with minor apparent effect on its translation. Finally as AM96 suppressed miR-96-induced mRNA decay, it led to a significant increase in eGFP-GPC3 mRNA stability. In the same conditions, miR-182 had no effect on eGFP-GPC3 expression in either HuH7 or SNU398 cells (Figure 2B–D, Supplementary Figure S3A). To further substantiate our findings, eGFP-GPC3-expressing HuH7 cells were transfected with increasing amounts of miR-96 (0–16 nM). As shown in the Supplementary Figure S3B, the downregulation mediated by miR-96 on eGFP-GPC3 expression was dose-dependent and reached its maximal effect when using miR-96 at a concentration of 12 nM or more.
Figure 2.
Molecular basis of the post-transcriptional regulation mediated by miR-96 on GPC3. (A) Schematic representation of the eGFP-GPC3 transgene with the 3′-UTR being targeted by miR-96. (B–D) HuH7 cells were transduced once with lentiviral particles expressing the transgene. After one week, the TCN was calculated using genomic DNA extracted from the eGFP-GPC3-expressing HuH7 cell population. Then cells were transfected with the indicated small RNAs. Three days later, the eGFP protein expression and mRNA amount were measured and data were analyzed following the FunREG experimental pipeline (9). (B) Global post-transcriptional regulation (ANOVA: P < 0.0001; n = 6). (C) mRNA stability (ANOVA: P < 0.0001; n = 6). (D) Translation efficiency (ANOVA: P = NS; n = 6). Panel (E): EGFP-GPC3-expressing HuH7 cells were first transfected with the indicated siRNA. Twenty-four hours later, cells were transfected with miR-96. Forty-eight hours later, eGFP protein expression was analyzed by FACS and data normalized to that of siLuc (ANOVA: P < 0.0001; n = 3). ***P < 0.001.
As miRNA-induced effect depends on miRISC and its core protein Argonaute (Ago), we looked for the isoform(s) of this protein that was involved in the regulation of GPC3 by miR-96. We reasoned that the negative effect of miR-96 could not take place in absence of the corresponding Ago protein. We therefore pre-depleted HuH7 cells of each Argonaute isoform with specific siRNAs (Supplementary Figure S3C–F). Then depleted cells were transfected with miR-96. Results in Figure 2E clearly showed that miR-96 mainly required Argonaute 2, as its inhibitory effect (shown at its maximum in the siLuc control condition) was decreased by 60% in absence of Ago2. It should be noted that depletion of Ago2 in HuH7 cells was compensated by an increase of Ago1 and Ago3 mRNAs (Supplementary Figure S3D). Altogether these results showed that miR-96 negatively controls GPC3 expression at a post-transcriptional level by targeting the 3′-UTR of its transcript and by inducing its degradation by an Ago-2-dependent mechanism.
Molecular basis of miR-96-target recognition
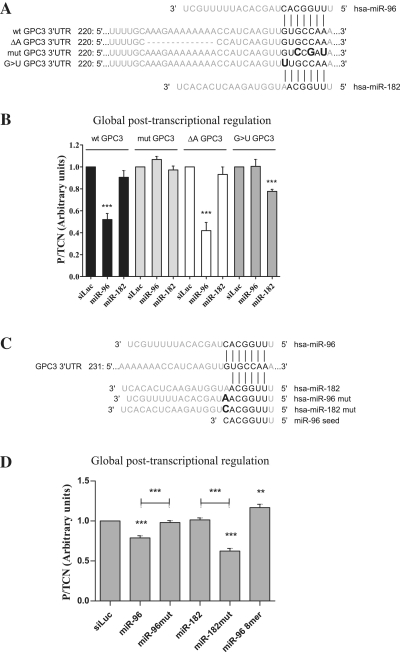
A set of experiments was performed to decipher the mechanism of miR-96-target recognition. First we confirmed the predicted miR-96 site using eGFP transgenes bearing mutated GPC3 3′-UTRs (Figure 3A). The functional consequences of these mutations were studied by FunREG, as described above, using HuH7 cells expressing the corresponding transgene (Figure 3A) and transfected with siLuc, miR-96 or miR-182 (Figure 3B). Results in terms of global post-transcriptional regulation were compared with those obtained with the wild-type 3′-UTR (Figure 3B, ‘wt GPC3’ transgene). As shown, three-point mutations (r.250G>C, r.252C>G and r.254A>U) inserted at the positions predicted to pair with miR-96 seed (‘mut GPC3’ transgene, Figure 3B) fully abrogated miR-96 effect validating the bioinformatic predictions. As miR-96 displays a uridine (U)-rich stretch in its 3′-end (5 consecutive Us at positions 16–20) and the GPC3 3′-UTR contains an adenosine (A)-rich sequence just upstream miR-96 site (Figure 3A), we assessed whether this potential A/U pairing could play a role in miR-96:GPC3 mRNA interaction. As shown in Figure 3B, absence of the A-rich sequence (‘ΔA GPC3’ transgene) did not hamper the targeting of GPC3 3′-UTR by miR-96. Similar results were obtained with miR-1271, the paralog of miR-96, and the different transgenes presented in Figure 3A (data not shown). Together these results suggested that the seed is important in GPC3 mRNA recognition by miR-96 and that a 3′-end A/U pairing mechanism does not seem necessary to make this miRNA functional (5). Based on these evidences, we tried to explain why miR-182 was unable to control GPC3 expression. Contrarily to miR-182, miR-96 pairs the GPC3 3′-UTR through its seed incremented of nucleotide 8 (a cytidine, C; Figure 1B). Therefore we hypothesized that pairing of the target with the seed + nucleotide 8 might be a prerequisite to yield a stable miRNA:mRNA complex. Consequently the guanosine (G) at position 248 of GPC3 3′-UTR, which normally matches a C at position 8 of miR-96, was mutated in U (‘G>U GPC3’ transgene, Figure 3A). Results in Figure 3B (‘G>U GPC3’ transgene) showed that as expected, the r.248G>U mutation abrogated the regulation of eGFP-GPC3 by miR-96. In addition it allowed miR-182 to target the GPC3 3′-UTR and control eGFP-GPC3 expression. To confirm these findings, we synthesised wild type and mutated double-stranded miRNAs (Figure 3C and Supplementary Table S2). miRNAs were mutated as follows: a C>A mutation was created at position 8 of miR-96 to specifically mimic the seed sequence plus nucleotide 8 of miR-182. Conversely, a A>C mutation was created at position 8 of miR-182 to mimic the seed + position 8 of miR-96 (Figure 3C). We then tested the efficiency of these miRNAs to control expression of the eGFP-GPC3 transgene in HuH7 cells. As expected, miR-96 downregulated the expression of the eGFP-GPC3 transgene in HuH7 cells (Figure 3D) whereas its mutated counterpart (miR-96mut) was inefficient. It should be noted however that the synthetic miR-96 that we produced was less efficient than its commercial mimic form. Although miR-182 had still no effect, its mutated counterpart (miR-182mut) was fully functional and decreased eGFP expression by 47%. Because of the central role of the first 8 nt of miR-96 in its functioning, we also tested the impact of transfecting cells with the 8-mer oligonucleotide (‘miR-96 8-mer’, Figure 3C) on eGFP-GPC3 expression. Unexpectedly miR-96 8-mer did not reduce, but rather slightly increased eGFP-GPC3 expression (Figure 3D). This increase was similar to that obtained with AM96 in Figure 2B suggesting that the 8-mer nucleotide cannot recapitulate miR-96 activity, but rather acts as a specific competitive inhibitor.
Figure 3.
Base requirements for miR-96:GPC3 3′-UTR recognition and miR-96 function. (A) Schematic representation of miR-96 or miR-182 pairing with the GPC3 3′-UTR sequence in its wild-type, deleted or mutated versions. (B) HuH7 cells expressing the indicated transgene (shown above the bars; TCN value measured) were transfected with the indicated small RNAs. Three days later, eGFP protein expression was analyzed by FACS as described in Figure 2B (ANOVA: P < 0.0001; n = 3). (C) Schematic representation of the wild type GPC3 3′-UTR sequence and the various synthetic double-strand small RNAs used (only the guide strand is shown). (D) eGFP-GPC3-expressing HuH7 cells were transfected with the indicated small RNAs. Three days later, eGFP protein expression was analyzed as described in panel B (ANOVA: P < 0.0001; n = 5). **P < 0.01; ***P < 0.001.
Finally we evaluated whether the differential regulation of GPC3 expression observed with miR-96 and miR-182 was transposable to other genes. Forkhead box protein O1 (FOXO1) was first chosen as a model, as its 3′-UTR was predicted to contain two miR-96/182 sites (two 8-mer sites for miR-96 and consequently, two 7-mer-1A sites for miR-182, see Figure 4A) (13). Moreover FOXO1 was described as a target of miR-96 and miR-182 (28–30). As expected from our data using GPC3 as target, transfection of HuH7 cells with miR-96 led to a slight decrease of FOXO1 protein, whereas transfection with miR-182 had no effect (Figure 4B). The differential effect of these two miRNAs on FOXO1 expression was confirmed at the mRNA level as only miR-96 significantly decreased FOXO1 mRNA expression (Supplementary Figure S4). To further support these results, we assessed our findings using a second predicted miR-96/182 target. Fibronectin-1 (FN1) was chosen as it is predicted to contain one 8-mer site for miR-96 and consequently one 7-mer-1A sites for miR-182 (Figure 4A) (13). As shown in Figure 4B, miR-96 very efficiently down-regulated FN1 expression whereas miR-182 was still non-functional. Altogether these results demonstrated that despite sharing similarities in their seeding region, miR-96 and miR-182 do not control the same genes. The differential regulation depicted by these two miRNAs depends on the presence of a site containing a seed match augmented by a match at position 8 in targeted 3′-UTRs.
DISCUSSION
Here, we reported that GPC3 is post-transcriptionally regulated by miR-96. We also confirmed the regulation of ADCY6 and FOXO1 by this microRNA and reported for the first time the downregulation of FN1 by miR-96. Using FunREG, we showed that miR-96 induces GPC3 mRNA degradation by targeting its 3′-UTR through a mechanism requiring the RISC-core protein Ago2 (Figure 2). Similar results were obtained with miR-1271 (31), a miR-96-paralog which bears the same seed + 1 region but a different base at position 1 (data not shown). The mRNA destabilizing effect mediated by miR-96 is in agreement with current findings showing that some miRNAs induce mRNA decay rather than repressing translation (7,9–11,31,32). Using silencing of individual Ago proteins, we found that miR-96 effect depended exclusively on Ago2 but not on other Ago proteins. As all four Ago proteins can mediate miRNA repression and bind to a nearly similar set of mRNAs (33,34), it is not clear why the down-regulation of GPC3 by miR-96 specifically requires Ago2. However, a specific requirement for Ago2 in miRNA-mediated gene repression has already been reported for Let7-a (35). It is possible that during miRNA biogenesis, factors associating with the pri- and/or the pre-RNAs direct the mature miR-96 to the Ago2-associated RISC. Alternatively, the cellular concentration of each individual Ago protein, their competition for the mRNAs or their post-translational status might determine the sorting of miR-96 to Ago2 (12,32).
As predicted by several bioinformatic programs, miR-96 recognized a single 8-mer site located at positions 248–254 downstream the stop codon of the GPC3 3′-UTR (370 nt long) (Figure 3). Intriguingly we found that miR-182 was inefficient on GPC3, despite the presence of one 7-mer-1A site in its 3′-UTR (Figure 4A) and the fact the seed borne by miR-182 was identical to that of miR-96 (Figure 1B). However miR-182 functioned on its validated target ADCY6 (Supplementary Figure S2D), the 3′-UTR of which contains an 8-mer site for miR-182 (Figure 4A) (27). It should be reminded that an 8-mer site for miR-182 constitutes a 7-mer-1A site for miR-96 and reciprocally. By studying in more details these paradoxical findings, we demonstrated that miR-182 functioning on GPC3 depended on an extra nucleotide match associating its nucleotide 8 and the mRNA target. Indeed by either introducing the mutation r.248G>U in GPC3 3′-UTR (Figure 3A) or changing the A>C at miRNA position 8 (Figure 3C), miR-182 became functional on GPC3 (Figure 3C and D). Conversely the mutation r.248G>U in GPC3 3′-UTR or r.8C>A in miR-96 abrogated the regulatory effect of this miRNA on GPC3. We therefore concluded that the ‘seed-dependent target recognition’ hypothesis per se is not applicable to miR-96 and miR-182 sites (5). Indeed target recognition by miR-96/182 5′-UUGGCA-3′ seed is not sufficient to make the miRNA functional in cellulo and this required an additional nucleotide match at position 8 in order to generate a stable miRNA:target complex. This assumption was supported by (i) the differential effects of miR-96 and miR-182 on FOXO1, FN1 and GPC3, and (ii) the regulatory effect of miR-96 and miR-182 on ADCY6 (27). Indeed both miRNAs recognized their targets only when they contained at least one 8-mer site in their 3′-UTR (seed + 1 match with the target, Figure 4A). Our findings are also in agreement with the regulatory role of miR-182 on Cortactin and on Regulator of G-protein signaling 17 (RGS17), two genes which contain two miR-182 8-mer sites in their 3′-UTR (13) and which are regulated by miR-182 at the mRNA level (36,37). However the work reported by Moskwa et al. (38) suggests that the ‘seed+1-matched site recognition’ model is not a general rule for miR-182. Moreover our data apparently disagree with other reports that suggested a regulation of FOXO1 by miR-182 (28–30). But the demonstration was partly based on the mutation of the miR-182 site in the region matching the seed (and not the nucleotide matching the miRNA position 8) (29,30), therefore leading to a loss of target recognition not only by miR-182, but also by miR-96 and its paralog, miR-1271. In addition, the use of different experimental conditions or cell types may also affect the results. Indeed, miR-182 site accessibility in FOXO1 mRNA may be different from a cell type to another depending on specific 3′-UTR secondary structures, 3′-UTR contexts, associated RNA-binding proteins or site multiplicity which are all determinants influencing miRNA activity (13,39). Interestingly a small RNA corresponding to the first 8 nt of miR-96 functionally acted as a competitive inhibitor of the endogenous miR-96/RISC complex (Figure 3D). We therefore concluded that an 8-mer RNA is sufficient to recognize its intracellular target but is unable to support miRISC loading, probably because the miRNA 3′-end region is lacking. These data are consistent with a recent report showing that small locked nucleic acids (LNA) targeting miRNA seed can abrogate miRNA function (40). Therefore any interference in miRNA:target recognition by using either seed complementary or 8-mer mimic (comprising the seed flanked by a base at positions 1 and 8) LNAs should specifically block miRNA action, a property which could lead to therapeutic effects (40). At last we showed that miR-96:target pairing does not require a 3′-compensatory mechanism. Indeed both miR-96 and miR-182mut (as well as miR-1271, data not shown) controlled GPC3 expression, although the remaining sequences downstream nucleotide 8 are completely different (Figure 3). Together these observations are in accordance with the proposed ‘seed nucleation’ model where the miRNA 3′-end is necessary for the biological function, but does not participate in target recognition (5). Therefore such a 3′-compensatory mechanism seems not to be required in target recognition by miR-96 nor miR-182. Because of the central role played by the miRNA seed+1 region in our model, we used the RNAhybrid software to estimate the strength of these miRNA-seed:target hybridisations (41). The analyses showed that the free energy hybridisations of miR-96:GPC3 (nucleotides 2–8) and miR-182:GPC3 (nucleotides 2–7) complexes were −14.8 and −12.8 kCal/mol, respectively. In the same condition, the free energy hybridisation of the miR-182mut:GPC3 (nucleotides 2–8) complex was −13.7 kCal/mol. Therefore in our experimental conditions, a minimum free energy of at least −13.7 kCal/mol is apparently required to make these miRNAs functional. Together our data demonstrated that the nucleotide 8 of miR-96 and miR-182 is as critical as nucleotides 2–7 for target recognition and miRNA:mRNA complex stabilization. Our results are in accordance with other reports showing that introducing single nucleotide changes in the target site complementary to nucleotides 2–8 of Human miR-96/1271 or Drosophila miR-7 abrogates gene regulation (31,42). Therefore although dissimilar (Figure 1B), the seeds of miR-96 and miR-182 comprise nucleotides 2–8 (31,32,43) rather than 2–7 (5) and their matching site can be classified as 5′ dominant seed site (42) or 8-mer site (5). However we cannot exclude the possibility that these miRNA could act differently under specific circumstances as previously reported for miR-182 (38).
By taking into account all these findings, we can speculate that the sets of cellular genes regulated by miR-96 and by miR-182 are profoundly different with the exception of those carrying at least one 8-mer site for both miRNAs (e.g. ADCY6, Figure 4A). Moreover it can be proposed that any C>A variations at position 8 of miR-96 (or conversely A>C change in miR-182) or any G>U changes in the target at the corresponding matching position might lead to a gene reprogramming with deep cellular consequences. Indeed such miRNA modifications might take place in physiological conditions (i.e. edition (12)) and play a role in cell plasticity especially during organism development or cell differentiation. They may also be linked to evolutionary mechanisms such as single nucleotide polymorphisms or to post-transcriptional RNA modifications. Finally they may also be linked to human illness since several single nucleotide mutations involving miRNA:target recognition have been reported lately, some involving miR-96 or miR-433 (10,44,45). With the advent of genomic deep sequencing programs and the recent discovery of gene deregulations associated with 3′-UTR mutations, maybe some of these questions will find answers.
Our data also point to the lack of accuracy of the bioinformatic prediction tools gathered on miRWalk (http://mirwalk.uni-hd.de). Indeed none of the algorithms in Figure 4C were in complete accordance with our functional data: the regulatory effect of miR-96 on GPC3, ADCY6, FOXO1 and FN1, and that of miR-182 on ADCY6. It should be specified however that miRDB (24,25) gave the best prognostication as it efficiently predicted a regulation of GPC3, FN1 and FOXO1 by miR-96, and not by miR-182. However it missed the regulation of ADCY6 by miR-182. Because the number of cell types used in our work was limited, we cannot pretend that our results reflect a general mechanism. However it is clear that room remains for bioinformatic tool improvement and that functional studies, associated with new valuable experimental and screening tools, should greatly contribute to their enhancement.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–4, Supplementary References (13,20–25).
FUNDING
This work was supported by the Agence Nationale pour la Recherche (ANR)—Programme Jeunes Chercheurs (Paris, France) (Grant no: JC07_184264 to C.G.) and by La Ligue Nationale Contre le Cancer. M.M. and V.M. were recipients of fellowships from the Ministère de l'Enseignement Supérieur et de la Recherche (MESR). Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Hélène Jacquemin-Sablon, Violaine Moreau, Frédéric Saltel, Patrick Lestienne and Eric Chevet are warmly acknowledged for their critical comments of the manuscript.
REFERENCES
- 1.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 3.Grosset C, Chen CY, Xu N, Sonenberg N, Jacquemin-Sablon H, Shyu AB. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 4.Grosset C, Boniface R, Duchez P, Solanilla A, Cosson B, Ripoche J. In vivo studies of translational repression mediated by the granulocyte-macrophage colony-stimulating factor AU-rich element. J. Biol. Chem. 2004;279:13354–13362. doi: 10.1074/jbc.M308003200. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Laloo B, Simon D, Veillat V, Lauzel D, Guyonnet-Duperat V, Moreau-Gaudry F, Sagliocco F, Grosset C. Analysis of post-transcriptional regulations by a functional, integrated, and quantitative method. Mol. Cell Proteomics. 2009;8:1777–1788. doi: 10.1074/mcp.M800503-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon D, Laloo B, Barillot M, Barnetche T, Blanchard C, Rooryck C, Marche M, Burgelin I, Coupry I, Chassaing N, et al. A mutation in the 3′-UTR of the HDAC6 gene abolishing the post-transcriptional regulation mediated by hsa-miR-433 is linked to a new form of dominant X-linked chondrodysplasia. Hum Mol Genet. 2010;19:2015–2027. doi: 10.1093/hmg/ddq083. [DOI] [PubMed] [Google Scholar]
- 11.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 13.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 15.Slezak-Prochazka I, Durmus S, Kroesen BJ, van den Berg A. MicroRNAs, macrocontrol: regulation of miRNA processing. RNA. 2010;16:1087–1095. doi: 10.1261/rna.1804410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filmus J, Capurro M. The role of glypican-3 in the regulation of body size and cancer. Cell Cycle. 2008;7:2787–2790. doi: 10.4161/cc.7.18.6672. [DOI] [PubMed] [Google Scholar]
- 18.Jakubovic BD, Jothy S. Glypican-3: From the mutations of Simpson-Golabi-Behmel genetic syndrome to a tumor marker for hepatocellular carcinoma. Exp. Mol. Pathol. 2007;82:184–189. doi: 10.1016/j.yexmp.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Laloo B, Maurel M, Jalvy-Delvaille S, Sagliocco F, Grosset CF. Analysis of post-transcriptional regulation using the FunREG method. Biochem. Soc. Trans. 2010;38:1608–1614. doi: 10.1042/BST0381608. [DOI] [PubMed] [Google Scholar]
- 20.Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 23.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 24.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 28.Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S, Lam EW. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;70:367–377. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J. Biol. Chem. 2009;284:23204–23216. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, Panse I, Riedel R, Flossdorf M, Dong J, Fuhrmann F, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat. Immunol. 2010;11:1057–1062. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 31.Jensen KP, Covault J. Human miR-1271 is a miR-96 paralog with distinct non-conserved brain expression pattern. Nucleic Acids Res. 2011;39:701–711. doi: 10.1093/nar/gkq798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 33.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34:4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Liu T, Huang Y, Liu J. microRNA-182 inhibits the proliferation and invasion of human lung adenocarcinoma cells through its effect on human cortical actin-associated protein. Int. J. Mol. Med. 2011;28:381–388. doi: 10.3892/ijmm.2011.679. [DOI] [PubMed] [Google Scholar]
- 37.Sun Y, Fang R, Li C, Li L, Li F, Ye X, Chen H. Hsa-mir-182 suppresses lung tumorigenesis through down regulation of RGS17 expression in vitro. Biochem. Biophys. Res. Commun. 2010;396:501–507. doi: 10.1016/j.bbrc.2010.04.127. [DOI] [PubMed] [Google Scholar]
- 38.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell. 2011;41:210–220. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 40.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 44.Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 45.Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M, Redshaw N, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat. Genet. 2009;41:614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.