Abstract
The development of economical and high-throughput gene synthesis technology has been hampered by the high occurrence of errors in the synthesized products, which requires expensive labor and time to correct. Here, we describe an error correction reaction (ECR), which employs Surveyor, a mismatch-specific DNA endonuclease, to remove errors from synthetic genes. In ECR reactions, errors are revealed as mismatches by re-annealing of the synthetic gene products. Mismatches are recognized and excised by a combination of mismatch-specific endonuclease and 3′→5′ exonuclease activities in the reaction mixture. Finally, overlap extension polymerase chain reaction (OE-PCR) re-assembles the resulting fragments into intact genes. The process can be iterated for increased fidelity. With two iterations, we were able to reduce errors in synthetic genes by >16-fold, yielding a final error rate of ∼1 in 8700 bp.
INTRODUCTION
Gene and genome syntheses are playing an increasingly important role in synthetic biology and biotechnology (1–5). To increase throughput and reduce cost, new gene synthesis methods that take advantage of DNA microarrays (6–8) and microfluidic devices (9–11) have recently been demonstrated. However, removing errors that arise from oligonucleotide (oligo) synthesis and gene assembly remains a significant challenge, especially for gene synthesis using microarray-produced oligos, where error rates tend to be higher (6,7). Cloning and sequencing large numbers of synthetic constructs in order to identify correct clones has become a bottle neck for gene and genome syntheses.
A number of methods have been used to reduce synthesis errors. To improve the quality of gene-construction oligos, size exclusion purification using polyacrylamide gel electrophoresis (PAGE) (12) or high performance liquid chromatography (HPLC) (13) can be used to remove large insertions and deletions. An array hybridization method has also been developed to reduce errors in chip-generated oligo pools, which requires special microarrays of complementary oligos (6). Using next-generation sequencing technology, it may also be feasible to sequence and select correct oligo sequences for gene construction, as a recent proof-of-concept experiment has demonstrated (14).
To eliminate errors in longer synthetic gene constructs, slow and labor-intensive cloning and sequencing methods are traditionally used. If the error rate is high or the sequence is long, large numbers of clones need to be sequenced in order to identify a correct sequence (15). If a perfect clone cannot be isolated, site-directed mutagenesis needs to be used to fix errors identified by sequencing (5,16–19). Multiple rounds of cloning, sequencing and site-directed mutagenesis can significantly increase the cost and turn around time for gene synthesis.
In order to increase the chance of finding a correct clone, the overall error frequency in the synthetic gene pool needs to be significantly reduced. Methods of using mismatch-binding proteins (e.g. MutS) to remove error-containing DNA heteroduplexes have been developed (15,20,21). However, MutS-based methods theoretically do not work well for error-rich sequences, because the correct sequences have to outnumber the erroneous sequences in order to avoid being depleted from the synthetic pool.
In comparison, methods using mismatch-cleaving enzymes show an advantage as these enzymes can cleave the heteroduplexes at the vicinity of the mismatch sites, which allows the mutant bases to be subsequently removed by exonuclease activity present in the reaction mixture. A number of enzymes have been tested, including T7 endonuclease I, T4 endonuclease VII and Escherichia coli endonuclease V, which showed various effectiveness due to various specificities of the enzymes (22–24).
CEL endonuclease is a new member of the S1 nucleases isolated from celery and prefers double-stranded mismatched DNA substrates (25,26). It is not inhibited by high GC content, and can cut mismatch-containing heteroduplexes efficiently at neutral pH whether the mismatches are base substitutions, insertions or deletions anywhere from 1 to 12 nt. CEL nuclease is able to act efficiently on molecules with multiple mismatches, even with only 5 nt between mismatches. Additionally, it can handle substrates anywhere from 40 bp to ∼30 kb. Its broad substrate specificity and low non-specific activity has made CEL nuclease one of the best tools for mismatch detection (25,27–30). In a previous study, we first reported that Surveyor nuclease, a commercialized form of the CEL endonuclease, was effective in removing errors during chip-based gene synthesis (31). Here, we describe detailed characterization of the molecular mechanism of the Surveyor-based error correction reaction (ECR) and the development of an optimized ECR protocol, which further reduced the error rate down to 1 error in ∼8700 bp.
MATERIALS AND METHODS
Reagents
Chemicals were purchased either from Sigma-Aldrich or VWR. Enzymes were from New England Biolabs. The Surveyor nuclease was purchased from Transgenomic as part of the Surveyor Mutation Detection Kit. GC5 chemical competent cells were purchased from Invitrogen.
Oligonucleotide synthesis and on-chip gene assembly
Oligonucleotides were synthesized on a plastic chip using a custom-made inkjet DNA microarray synthesizer (32). Gene-construction oligos were designed to be 60-nt long with overlapping regions of similar melting temperatures (Tm = 65 ± 2°C). The exact oligos synthesized are listed in Supplementary sequences. On-chip oligo amplification and gene assembly using combined nicking strand displacement and polymerase cycle assembly (nSDA–PCA) reaction was performed as described with minor modifications (31). Briefly, an 8-well incubation adapter (Sigma-Aldrich) was fitted onto the COC slide so that each well contained a synthesized oligo array. The wells were filled with the nSDA–PCA reaction cocktail composed of 0.4 mMdNTP, 0.2 mg/ml BSA, Nt. BstNBI, Bst large fragment and Phusion polymerase in an optimized Thermopol II buffer. The slides with sealed chambers were placed on the in situ slide-adapter of a Mastercycler Gradient thermocycler (Eppendorf) to perform combined nSDA–PCA reactions. nSDA involved incubation at 50°C for 2 h followed by 80°C for 20 min; the PCA reaction involved an initial denaturation at 98°C for 30 s, followed by 40 cycles of denaturation at 98°C for 7 s, annealing at 60°C for 60 s and elongation at 72°C for 15 s/kb, and finished with an extended elongation step at 72°C for 5 min.
After the nSDA–PCA reaction, 1–2 μl of the reaction from each chamber was used for PCR amplification with Phusion polymerase and end primers (Supplementary sequences). End primers were employed at a concentration of 0.5 µM. The PCR reaction involved an initial denaturation at 98°C for 30 s, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 60°C for 60 s and elongation at 72°C for 30 s/kb, and finished with a final elongation at 72°C for 5 min.
ECR of assembled genes
Once PCR amplification of the on-chip assembled gene was completed, the gene products were purified by agarose gel electrophoresis and extracted to yield a concentration of >100 ng/μl (measured using a Nanodrop analyzer). These PCR products were then diluted with either 1X Taq buffer or 1X Phusion HF buffer to yield a final concentration of 50 ng/μl. This was then melted by heating at 95°C for 10 min, cooled to 85°C at 2°C/s and held for 1 min. It was then cooled down to 25°C at a rate of 0.3°C, holding for 1 min at every 10°C interval.
For ECR using a 20 min Surveyor cleavage incubation, 4 μl (200 ng) of the re-annealed gene product was mixed with 0.5 μl of Surveyor nuclease and 0.5 μl enhancer [which is known to be DNA ligase in nature and enhances the reaction (29–31)] and incubated at 42°C for 20 min. Two microliter of the reaction mixture was used for subsequent overlap extension–PCR (OE-PCR) using the same reaction conditions as the PCR above. The OE-PCR product was cloned and sequenced to serve as the result from the first iteration of error correction. For the second iteration of error correction, the OE-PCR product band was diluted to 50 ng/μl using 1X Taq buffer and re-annealed as before. Similar to the first iteration, a 5 μl reaction consisting of 4 μl re-annealed product, 0.5 μl of Surveyor nuclease and 0.5 μl enhancer was incubated at 42°C for 20 min. Two microliter of the product was subjected to OE-PCR, cloned and sequenced to serve as the result from the second iteration of error correction.
For ECR using a 60 min Surveyor cleavage incubation, 8 μl of the re-annealed gene product in 1X Phusion buffer (final DNA concentration of 50 ng/μl) was added to 2 μl of Surveyor nuclease and 1 μl enhancer to yield a total of 11 μl that was then incubated at 42°C for 60 min. Two microliter of the reaction mixture was then subjected to OE-PCR, and the resulting PCR product was cloned and sequenced to serve as the result from the first iteration of error correction. For the second iteration, the product from the first iteration was diluted to 50 ng/μl using 1X Phusion buffer and re-annealed as before. Similar to the first iteration, an 11-μl reaction consisting of 8 μl of re-annealed product, 2 μl of Surveyor nuclease and 1 μl of enhancer was incubated at 42°C for 60 min. Two microliter of the product was used for OE-PCR and the PCR product was cloned and sequenced to serve as the result from the second iteration of error correction.
Cloning, sequencing and functional analysis of synthetic genes
Synthetic gene products, before or after ECR, were cloned into pAcGFP1 vector using circular polymerase extension method (CPEC) (33,34). Briefly, 250 ng of the linear vector was mixed with the synthetic gene products at 1:2 molar ratios in a 25-μl CPEC reaction using Phusion polymerase. The reaction involved 10 cycles of denaturation at 98°C for 10 s, annealing at 55–60°C for 30 s and extension at 72°C for 15 s, and finished with an extended elongation step at 72°C for 5 min.
Two microliter of the cloning product was transformed into GC5 chemically competent cells (Invitrogen) according to the manufacturer's instructions. Cells were grown on agar plates with 100 μg/ml carbenicillin for ∼16 h and then kept at room temperature for 48 h before been imaged in an AlphaImage gel documentation system. The percentage of fluorescent colonies was automatically determined using CellC program (http://sites.google.com/site/cellcsoftware/download). The results were verified by thresholding the UV images using Adobe Photoshop and counting using ImageJ. Sequence analysis was done by extracting plasmids from randomly selected colonies using a miniprep kit (Qiagen) and sequencing at the Duke University Sequencing Facility.
RESULTS AND DISCUSSION
General design of the ECR using Surveyor nuclease
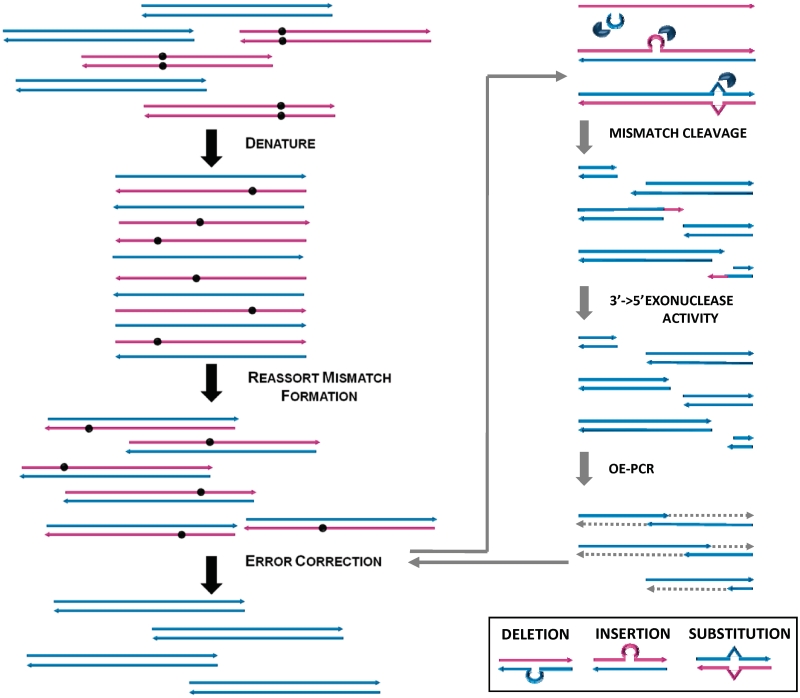
In this study, we aimed to develop a simple and convenient method to effectively remove errors from synthetic genes. The general strategy of using the Surveyor endonuclease to correct errors in synthetic genes is illustrated in Figure 1. After gene synthesis, the products are denatured and re-annealed to form mismatch-containing heteroduplexes (left panel). The subsequent ECR, right panel involves incubation of the re-annealed product with the Surveyor nuclease, followed by OE-PCR using a proofreading DNA polymerase. The 3′→5′ exonuclease activity of the DNA polymerase removes 3′ overhangs that contain the mismatch base(s) and allows OE to proceed efficiently.
Figure 1.
Outline of steps involved in error correction of synthetic DNA constructs. Gene synthesis products are heat denatured and then slowly cooled down to form heteroduplexes containing mismatches at the error sites (left panel). Heteroduplexes are cleaved by the Surveyor nuclease at the sites flanking the mismatch bulges. The resulting single-stranded overhangs, where mismatch bases are located, are removed by the proofreading exonuclease activity of Phusion polymerase used in the OE-PCR. The resulting fragments with mismatch bases removed are efficiently assembled back into full-length gene constructs during OE-PCR (right panel).
Mismatch structures formed at the deletion, insertion and substitution sites in the heteroduplexes are recognized by the Surveyor mismatch-specific endonuclease, which cuts each strand at the phosphodiester bond at the 3′ side of the mismatch site (29). During the subsequent OE-PCR reaction, the 3′→5′ exonuclease activity of the proof-reading DNA polymerase chews away any 3′ overhangs that contain the mismatch base(s) (substitutions and insertions). Finally, the error-free fragments are extended and amplified into full-length gene constructs by the DNA polymerase. One round of ECR may not completely remove all errors and we are interested to determine whether more iterations of the ECR can be used to further remove any remaining errors.
Determine error frequency of on-chip gene synthesis
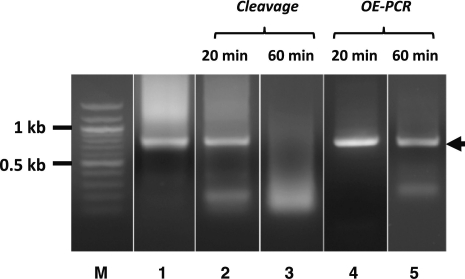
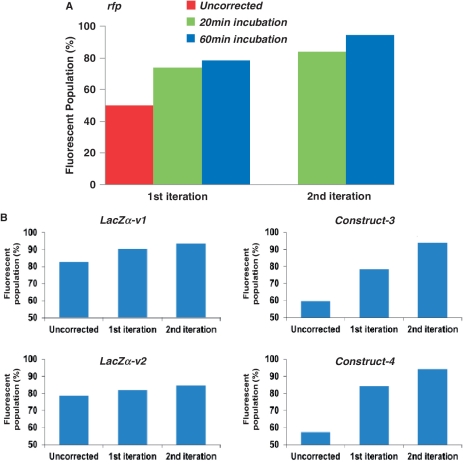
Integrating oligo synthesis with gene assembly on a microchip can significantly reduce synthesis cost and increase throughput. As described in the ‘Materials and Methods’ section, we synthesized DNA microarrays using a custom inkjet DNA synthesizer and used a combined nSDA–PCA reaction for on-chip oligo amplification and gene assembly. To determine error frequency of on-chip gene synthesis without error correction, we chose red fluorescent protein (rfp) as a test gene for convenient screen of functionally correct genes, which served as a good approximation of sequence correct genes. After the nSDA–PCA reaction, the 723-bp rfp construct was amplified by PCR (Figure 2, lane 1) and inserted into a modified pAcGFP1 expression vector using the CPEC cloning method as described in the ‘Materials and Methods’ section. After transformation into bacteria, the colonies produced were either non-fluorescent, dimly or brightly fluorescent. A rough approximation of synthesis quality without error correction could be made using colony counts on agar plates. Using automated colony counting, it was found that 50.2% of the rfp colonies formed from uncorrected product fluoresced brightly (Figure 3A).
Figure 2.
Cleavage and reassembly of synthetic gene product during ECR. Synthetic rfp gene (lane 1) was incubated with Surveyor nuclease for 20 min (lane 2) and 60 min (lane 3) at 42°C. The cleavage reaction products (lanes 2 and 3) were then re-assembled by OE-PCR into full-length gene products (lanes 4 and 5, respectively, marked by arrow). The reaction products were analyzed by agarose gel electrophoresis with DNA molecular weight marker (lane M).
Figure 3.
ECR results as measured by gene function or reporter assays. Percentage of functional or fluorescent clones was measured before and after one or two iterations of ECR for five different gene constructs. (A) The effects of Surveyor incubation time (20 and 60 min) and number of ECR iterations on the synthesis of rfp gene by counting fluorescent colonies. (B) The percentage of blue (lacZα-v1&2) or fluorescent colonies (constructs 3 and 4) after 1 or 2 ECR iterations.
DNA sequencing was performed on 42 randomly picked rfp colonies from both directions. The sequencing results indicate an error rate of ∼1.9/kb (Table 1). Deletions were found to be the dominant form of errors (75.4%), which was similar to column DNA synthesis where monomers are not successfully added to all of the growing polymer chains.
Table 1.
Error analysis of synthetic gene sequences before and after ECR with Surveyor nuclease
| Error type | Without ECR | ECR1 (20 min) | ECR1 (60 min) | ECR2 (20 min) | ECR2 (60 min) |
|---|---|---|---|---|---|
| Deletion (total) | 43 | 3 | 0 | 0 | 0 |
| Single-base deletion | 30 | 2 | 0 | 0 | 0 |
| Multi-base deletion | 13 | 1 | 0 | 0 | 0 |
| Insertion (total) | 4 | 0 | 0 | 0 | 0 |
| Single-base insertion | 3 | 0 | 0 | 0 | 0 |
| Multi-base insertion | 1 | 0 | 0 | 0 | 0 |
| Substitution (total) | 10 | 7 | 11 | 5 | 6 |
| Transition | |||||
| G/C to A/T | 3 | 2 | 3 | 1 | 2 |
| A/T to G/C | 3 | 4 | 0 | 2 | 1 |
| Transversion | |||||
| G/C to C/G | 0 | 0 | 2 | 0 | 0 |
| G/C to T/A | 1 | 1 | 4 | 1 | 1 |
| A/T to C/G | 2 | 0 | 1 | 0 | 2 |
| A/T to T/A | 1 | 0 | 1 | 1 | 0 |
| Total errors | 57 | 10 | 11 | 5 | 6 |
| Bases sequenced | 29 958 | 31 866 | 42 714 | 27 798 | 52 206 |
| Error frequency (errors per kb) | 1.9 | 0.31 | 0.26 | 0.18 | 0.11 |
| Error frequency (bases per error) | 526 | 3187 | 3883 | 5560 | 8701 |
Random clones of synthetic genes before (without ECR) or after one or two ECR iterations (ECR1, ECR2) were sequenced in both directions. Surveyor incubation time (20 min or 60 min) was indicated. The occurrence of different type of errors was counted.
ECR with Surveyor nuclease
Surveyor nuclease has typically been used for mutation detection. We devised a strategy of using it for eliminating errors in synthetic genes, as shown in Figure 1. To determine the optimal reaction conditions of using Surveyor nuclease for error correction, we systematically varied reaction parameters, such as reagent amount, buffer composition, incubation time, temperature and number of iterations.
In the first set of experiments, varying amounts of the Surveyor nuclease reagents, including the enzyme and the enhancer were tested. 0.5, 1 and 2 μl of Surveyor nuclease reagents were mixed with 200 ng of re-annealed synthetic rfp product. Incubations were performed either at 42°C for 20 min or 25°C for 60 min. After OE-PCR amplification, products from all variations were run on an agarose gel (Supplementary Figure S1). All bands on the gel appeared to be similar, indicating little difference with increased enzyme concentration.
Depending on the length and sequence quality of the synthetic gene products, after re-annealing and incubation with the Surveyor nuclease, the amount of intact full-length product that can survive the cleavage may be very limited. To assess the extent of cleavage of our on-chip synthesized rfp genes, we incubated the re-annealed product with Surveyor for either 20 or 60 min at 42°C. Figure 2 shows that after 20 min of Surveyor treatment, a fraction of the synthetic genes was cleaved into smaller fragments (lane 2); after 60 min, the majority of the genes were cleaved (lane 3). The results suggested that the cleavage by Surveyor nuclease was relatively efficient. It also suggests that the Surveyor cleavage assay can be used as a quick assessment of the sequence quality of the synthetic products. Following cleavage, OE-PCR was able to assemble and extend the fragments back to full-length genes, as shown in Figure 2 (lanes 4 and 5).
Reduction of error frequencies after ECR
Both functional colony counting and DNA sequencing were performed to estimate error frequencies of chip-synthesized genes after ECR with 20-min or 60-min Surveyor treatment. It was reasoned that in one round of ECR, error sequences could form homodimers by chance during annealing and thus escape detection and cleavage. We therefore tested whether an additional round of ECR could eliminate more errors. Two iterations of ECR were performed with both 20 min and 60 min incubations as outlined in the ‘Materials and Methods’ section. Full-length gene products were cloned and used for functional assays and Sanger sequencing in order to estimate error frequencies.
As shown in Figure 3A, increasing Surveyor cleavage time and number of iterations led to increases in the number of brightly fluorescent colonies. Using 20-min Surveyor treatment, the fluorescent population increased from 50.2% (untreated) to 74% and 84% in the first and second iteration, respectively. Using 60-min Surveyor treatment resulted in 78.4% and 94% fluorescent colonies after the first and second iteration. Example images showing the fluorescent colonies can be found in Supplementary Figure S2.
To investigate the repeatability and robustness of the method, we applied it to synthesis of four additional gene constructs and measured its effectiveness using functional or reporter assays. Of the four constructs, two were codon variants of the lacZα gene, the expression of which cause the colony to turn blue in the presence of X-gal. The other two constructs could not be screened by their own functions and therefore were fused to the N-terminus of the green fluorescent protein (GFP) (Figure 3B). Blue or fluorescent colonies indicated that there were no frame shifts or mutations in the gene constructs that could abolish the function or expression of the genes. Therefore, the percentage of positive colonies could be used as an approximate indicator of the quality of the sequences. The results from the four additional constructs showed iterative increase in positive populations after each round of ECR (Figure 3B). As expected from the model predictions shown in Figure 4A, the small lacZα genes had a large fluorescent population even before error correction (∼80% positive) as it had fewer errors to begin with due to their short length (174 bp). In comparison, the longer constructs (#3 and 4) had lower percentages of correct colonies to begin with (∼500 bp, ∼55–60% positive), but the effect of ECR was more obvious, reaching >90% positive after two iterations (Figure 3B).
Figure 4.
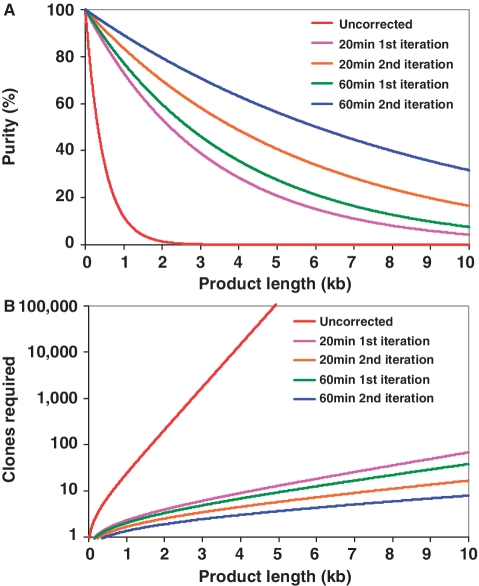
Predicted effects of ECR as a function of sequence length. (A) Purity of gene synthesis products (percentage of error-free clones) decreases exponentially with the length of the product synthesized. Employing ECR (1 error in 8701 bp, blue line) dramatically increases the probability of locating an error-free clone than the uncorrected population (1 error in 526 bp, red line). (B) Employing ECR significantly reduces the number of colonies that need to be screened to have a high (95%) probability of obtaining at least one error-free clone. Two iterations of 60 min cleavage incubations with Surveyor (blue line) could yield a correct 10 kb product by sequencing eight random clones. Plots are derived from the result of model calculations as described in the text.
Results from DNA sequencing analysis of randomly selected colonies correlated with the observations made with the colony counting experiments and revealed more details on the correction efficiency of different types of errors. The results in Table 1 showed that ECR with Surveyor was very efficient in reducing errors arising from deletion and insertion events. Most deletion and insertion type of errors could be eliminated after one round of 60-min treatment or two rounds of 20-min treatment. Surveyor treatment was also effective toward substitutions albeit with reduced efficiency. Substitution types of errors were still present after two rounds of 60-min incubations.
For the purpose of developing the most efficient ECR procedure, data in Table 1 indicated that increasing incubation time from 20 to 60 min reduced error frequency from 0.31 to 0.26 error/kb (∼16% reduction); while adding another round reduced error frequency from 0.31 to 0.18 error/kb for 20-min incubations (∼42% reduction) and from 0.26 to 0.11 error/kb for 60-min incubations (∼58% reduction). It appeared that adding a second round of ECR was more effective than increasing the Surveyor incubation time with only one round of ECR, although the accumulative effects of more iterations and longer Surveyor incubation was most dramatic.
Following the model predictions of Carr et al. (15) and Furhmann et al. (23), we performed statistical analysis to better understand the implication of our results. As can be seen in Figure 4A, the percentage of gene synthesis products that yield error-free clones decreases exponentially with the length of the product synthesized. Employing ECR for error correction (1 error in 8701 bp for two iterations of 60-min ECR, blue line) significantly increases the probability of locating an error-free clone than without error correction (1 error in 526 bp, red line). From the practitioner's perspective, this means that dramatically fewer clones need to be sequenced (Figure 4B). For example, as predicated in Figure 4B with ECR, one will have to screen, on average, only 8–10 clones of a 10 kb treated or a single 1 kb clone in order to locate a correct one. The model prediction correlated well with our sequencing analysis results. Analyzing sequencing data of 77 random colonies from the second iteration of the 60-min ECR, we found 72 of the colonies contained the correct rfp gene. The determined error rate of 0.11/kb meant a >16-fold reduction of errors present in the synthetic pool. With such an improvement, larger DNA targets can be conveniently synthesized and corrected within 2–3 h without resorting to additional cloning or excessive sequencing.
CONCLUSIONS
The method presented here performs enzymatic error correction on synthetic genes using Surveyor nuclease, which has the broadest substrate specificity toward all types of mismatches as compared to other known mismatch-specific binding proteins or endonucleases. The method utilizes the mismatch-specific endonuclease activity of the Surveyor enzyme to cut heteroduplex sequences at the mismatch sites and uses the exonuclease activity of the proof-reading DNA polymerase to remove the mismatch bases, followed by an OE-PCR reaction to re-assemble the cleaved fragments into full-length gene constructs. The results from the current study demonstrate that the optimized ECR procedure is robust and effective for all error types, especially insertions and deletions, yielding superior results than previous methods. The ECR method is probably more suitable for long and error-rich synthetic products and can be performed in less time than MutS-based procedures, which require gel-shift assay and DNA extraction from PAGE. Additionally, in comparison to the commercial ErrASE kit (8), the ECR reaction mitigates the need for tittering and excessive enzyme usage. Using the protocol developed in the current study, two ECR iterations could be completed in <5 h and reduces the error frequency by >16-fold. Future research to improve ECR may involve increasing its efficiency toward substitution types of errors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2, Supplementary sequences.
FUNDING
Funding for open access charge: National Institutes of Health (grant R01HG005862).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
JT was a Beckman Young Investigator and a recipient of The Hartwell Foundation Individual Biomedical Research Award.
REFERENCES
- 1.Tian J, Ma K, Saaem I. Advancing high-throughput gene synthesis technology. Mol. Biosys. 2009;5:714–722. doi: 10.1039/b822268c. [DOI] [PubMed] [Google Scholar]
- 2.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al. Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 3.Carr PA, Church GM. Genome engineering. Nat. Biotechnol. 2009;27:1151–1162. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- 4.Wimmer E, Mueller S, Tumpey TM, Taubenberger JK. Synthetic viruses: a new opportunity to understand and prevent viral disease. Nat. Biotechnol. 2009;27:1163–1172. doi: 10.1038/nbt.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 6.Tian J, Gong H, Sheng N, Zhou X, Gulari E, Gao X, Church G. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- 7.Borovkov AY, Loskutov AV, Robida MD, Day KM, Cano JA, Le Olson T, Patel H, Brown K, Hunter PD, Sykes KF. High-quality gene assembly directly from unpurified mixtures of microarray-synthesized oligonucleotides. Nucleic Acids Res. 2010;38:e180. doi: 10.1093/nar/gkq677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kosuri S, Eroshenko N, LeProust EM, Super M, Way J, Li JB, Church GM. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat. Biotech. 2010;28:1295–1299. doi: 10.1038/nbt.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong DS, Carr PA, Chen L, Zhang S, Jacobson JM. Parallel gene synthesis in a microfluidic device. Nucleic Acids Res. 2007;35:e61. doi: 10.1093/nar/gkm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C-C, Snyder TM, Quake SR. A microfluidic oligonucleotide synthesizer. Nucleic Acids Res. 2010;38:2514–2521. doi: 10.1093/nar/gkq092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang MC, Ye H, Kuan YK, Li M-H, Ying JY. Integrated two-step gene synthesis in a microfluidic device. Lab. Chip. 2009;9:276–285. doi: 10.1039/b807688j. [DOI] [PubMed] [Google Scholar]
- 12.Ellington A, Pollard JD., Jr Introduction to the synthesis and purification of oligonucleotides. Curr. Protoc. Nucleic Acid Chem. 2001 doi: 10.1002/0471142700.nca03cs00. Appendix 3, A.3C. [DOI] [PubMed] [Google Scholar]
- 13.Andrus A, Kuimelis RG. Analysis and purification of synthetic nucleic acids using HPLC. Curr. Protoc. Nucleic Acid Chem. 2001 doi: 10.1002/0471142700.nc1005s01. Chapter 10, Unit 10 15. [DOI] [PubMed] [Google Scholar]
- 14.Matzas M, Stahler PF, Kefer N, Siebelt N, Boisguerin V, Leonard JT, Keller A, Stahler CF, Haberle P, Gharizadeh B, et al. High-fidelity gene synthesis by retrieval of sequence-verified DNA identified using high-throughput pyrosequencing. Nat. Biotechnol. 2010;28:1291–1294. doi: 10.1038/nbt.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carr PA, Park JS, Lee YJ, Yu T, Zhang S, Jacobson JM. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res. 2004;32:e162. doi: 10.1093/nar/gnh160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabhi I, Guedel N, Chouk I, Zerria K, Barbouche MR, Dellagi K, Fathallah DM. A novel simple and rapid PCR-based site-directed mutagenesis method. Mol. Biotechnol. 2004;26:27–34. doi: 10.1385/mb:26:1:27. [DOI] [PubMed] [Google Scholar]
- 17.Xiong AS, Yao QH, Peng RH, Duan H, Li X, Fan HQ, Cheng ZM, Li Y. PCR-based accurate synthesis of long DNA sequences. Nat. Protoc. 2006;1:791–797. doi: 10.1038/nprot.2006.103. [DOI] [PubMed] [Google Scholar]
- 18.Linshiz G, Ben Yehezkel T, Kaplan S, Gronau I, Ravid S, Adar R, Shapiro E. Recursive construction of perfect DNA molecules from imperfect oligonucleotides. Mol. Syst. Biol. 2008;4:191. doi: 10.1038/msb.2008.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsic D, Hughes R, Byrne-Steele M, Ng J. PCR-based gene synthesis to produce recombinant proteins for crystallization. BMC Biotechnol. 2008;8:44. doi: 10.1186/1472-6750-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith J, Modrich P. Removal of polymerase-produced mutant sequences from PCR products. Proc. Natl Acad. Sci. USA. 1997;94:6847–6850. doi: 10.1073/pnas.94.13.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Binkowski BF, Richmond KE, Kaysen J, Sussman MR, Belshaw PJ. Correcting errors in synthetic DNA through consensus shuffling. Nucleic Acids Res. 2005;33:e55. doi: 10.1093/nar/gni053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young L, Dong Q. Two-step total gene synthesis method. Nucleic Acids Res. 2004;32:e59. doi: 10.1093/nar/gnh058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuhrmann M, Oertel W, Berthold P, Hegemann P. Removal of mismatched bases from synthetic genes by enzymatic mismatch cleavage. Nucleic Acids Res. 2005;33:e58. doi: 10.1093/nar/gni058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bang D, Church GM. Gene synthesis by circular assembly amplification. Nat. Methods. 2008;5:37–39. doi: 10.1038/nmeth1136. [DOI] [PubMed] [Google Scholar]
- 25.Yang B, Wen X, Kodali NS, Oleykowski CA, Miller CG, Kulinski J, Besack D, Yeung JA, Kowalski D, Yeung AT. Purification, cloning, and characterization of the CEL I nuclease. Biochemistry. 2000;39:3533–3541. doi: 10.1021/bi992376z. [DOI] [PubMed] [Google Scholar]
- 26.Oleykowski CA, Bronson Mullins CR, Godwin AK, Yeung AT. Mutation detection using a novel plant endonuclease. Nucleic Acids Res. 1998;26:4597–4602. doi: 10.1093/nar/26.20.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oleykowski CA, Mullins CRB, Godwin AK, Yeung AT. Mutation detection using a novel plant endonuclease. Nucleic Acids Res. 1998;26:4597–4602. doi: 10.1093/nar/26.20.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulinski J, Besack D, Oleykowski CA, Godwin AK, Yeung AT. CEL I enzymatic mutation detection assay. Biotechniques. 2000;29:44. doi: 10.2144/00291bm07. [DOI] [PubMed] [Google Scholar]
- 29.Yeung A, Hattangadi D, Blakesley L, Nicolas E. Enzymatic mutation detection technologies. Biotechniques. 2005;38:749–758. doi: 10.2144/05385RV01. [DOI] [PubMed] [Google Scholar]
- 30.Qiu P, Shandilya H, D'Alessio JM, O'Connor K, Durocher J, Gerard GF. Mutation detection using Surveyor nuclease. Biotechniques. 2004;36:702–707. doi: 10.2144/04364PF01. [DOI] [PubMed] [Google Scholar]
- 31.Quan J, Saaem I, Tang N, Ma S, Negre N, Gong H, White KP, Tian J. Parallel on-chip gene synthesis and application to optimization of protein expression. Nat. Biotechnol. 2011;29:449–452. doi: 10.1038/nbt.1847. [DOI] [PubMed] [Google Scholar]
- 32.Saaem I, Ma K, Marchi A, LaBean T, Tian J. In situ synthesis of DNA microarray on functionalized cyclic olefin copolymer substrate. ACS Appl. Mater. Interfaces. 2010;2:491–497. doi: 10.1021/am900884b. [DOI] [PubMed] [Google Scholar]
- 33.Quan J, Tian J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc. 2011;6:242–251. doi: 10.1038/nprot.2010.181. [DOI] [PubMed] [Google Scholar]
- 34.Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.