Abstract
Human La protein is an essential factor in the biology of both coding and non-coding RNAs. In the nucleus, La binds primarily to 3′ oligoU containing RNAs, while in the cytoplasm La interacts with an array of different mRNAs lacking a 3′ UUUOH trailer. An example of the latter is the binding of La to the IRES domain IV of the hepatitis C virus (HCV) RNA, which is associated with viral translation stimulation. By systematic biophysical investigations, we have found that La binds to domain IV using an RNA recognition that is quite distinct from its mode of binding to RNAs with a 3′ UUUOH trailer: although the La motif and first RNA recognition motif (RRM1) are sufficient for high-affinity binding to 3′ oligoU, recognition of HCV domain IV requires the La motif and RRM1 to work in concert with the atypical RRM2 which has not previously been shown to have a significant role in RNA binding. This new mode of binding does not appear sequence specific, but recognizes structural features of the RNA, in particular a double-stranded stem flanked by single-stranded extensions. These findings pave the way for a better understanding of the role of La in viral translation initiation.
INTRODUCTION
La is an exceedingly abundant protein functioning in various intracellular processes involving RNA. Originally identified as an autoantigen in patients affected by the rheumatic diseases Sjögren's syndrome and systemic lupus erythematosus, La was found ubiquitously expressed throughout eukaryotes (1). Within the nucleus, La associates with all newly synthesized RNA polymerase (pol) III transcripts, including precursors to 5S rRNA and tRNAs, as well as a subset of pol II small nuclear and nucleolar RNA intermediates, by binding specifically to the common (U)n-OH moiety present at the 3′ termini of these RNAs (1–4). La is a key player in the metabolism, maturation, processing, folding and subcellular localization of these regulatory non-coding RNA precursors, with the protection of the 3′ ends from exonuclease cleavage being the best-characterized role of La from yeasts to humans (1,2,5–8). The ubiquitous nature of this function relates to a highly conserved region that maps to the N-terminal half of the human protein and contains an elaborated winged-helix domain, the La motif (LaM), neighbouring an RNA recognition motif, the RRM1 (Figure 1) (1,2,9–11). The LaM and RRM1 perform as a single RNA-binding unit (recently re-named the ‘La module’) to recognize 3′ UUUOH single-stranded (ss) RNA sequences and structural studies have delineated how these domains are configured to achieve high-specificity binding to 3′ oligoU RNA (Figure 1) (12–14). In particular, both domains make specific contacts with the 3′ oligoU sequence, inducing it to fold into a conformation that stacks the third- or fourth-last U onto the terminal nucleotide base (Figure 1). The primary contacts for specificity are made by the LaM with the 2′- and 3′-hydoxyl groups of the terminal nucleotide and by both domains with the penultimate U, which is splayed out in the conformation adopted by the bound RNA (12–14).
Figure 1.
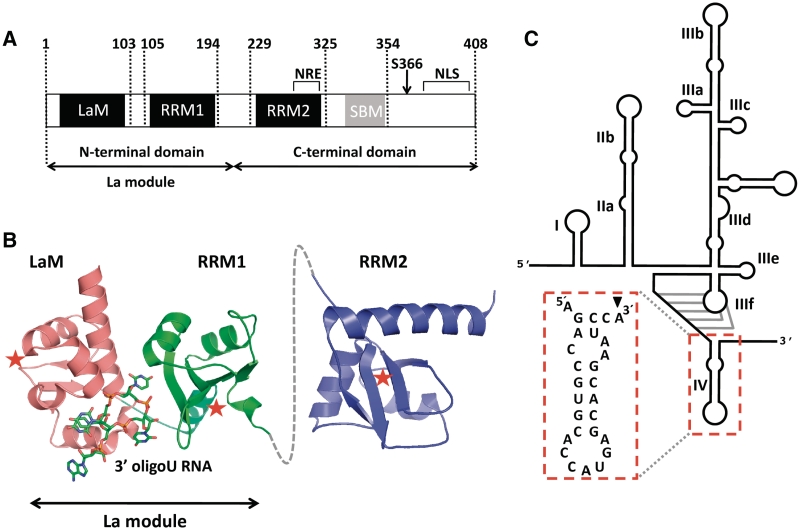
Details of the human La protein and the HCV IRES domain. (A) Domain organization of human La showing the La motif (LaM) and RRM1 (forming the La module), the RRM2, the nuclear retention element (NRE), the Short Basic Motif (SBM) and nuclear localization signal (NLS). (B) Structured domains of human La, depicting the crystal structure of the La module in complex with a 3′ oligoU ssRNA (PDB ID 2VOP) and the solution structure of the isolated RRM2 (PDB ID 1OWX). A dotted grey line denotes the linker between the RRM1 and RRM2. Red stars indicate the canonical RNA-binding surfaces on the three domains. (C) Schematic representation of the secondary structure elements and domains of the HCV IRES. The domain fragment used in biophysical experiments (domain IV) is highlighted. The black arrowhead indicates the base that was mutated from U to A in our experiments.
This mode of RNA binding is unusual and intriguing since it uses neither of the canonical RNA-binding surfaces—the winged-helix of the LaM or the β-sheet surface of RRM1—which may therefore be potentially available to interact with other portions of larger RNA ligands. Indeed, recognition of 3′ oligoU sequences appears to be only one facet of the RNA interactions made by La and recent investigations of the functional interaction with pre-tRNA targets suggest that as well as clamping onto the 3′ oligoU trailer the protein establishes additional points of contact with the RNA (15–17).
However the complexity and diversity of the RNA-binding repertoire of La does not stop here, as in some cases La–RNA interactions occur that appear to be entirely independent of binding to a 3′ oligoU trailer. This is exemplified by the cytoplasmic role of human La protein (hLa) where it can associate with internal ribosome entry sites (IRES) found in a subset of cellular mRNAs or in the positive-sense RNA genomes of viruses such as poliovirus, human immunodeficiency virus (HIV) or hepatitis C virus (HCV) (18–26). Mostly, the hLa–IRES interaction appears to augment translation, although the molecular mechanism of how this is achieved remain obscure, in part because the details of the hLa–RNA binding have not been elucidated.
Most work on hLa–IRES interactions has been done with HCV, where the stimulatory role of hLa in viral mRNA translation has been linked to a direct interaction between hLa and the HCV IRES element (19,21–23), in particular involving the IRES domain IV in the vicinity of the AUG start codon (Figure 1) (21,22). Significantly, this RNA contains neither the free 3′ end nor the terminal oligoU sequences that are invariably found in the nuclear pol III nascent transcripts and would activate the canonical and well-characterized 3′-termini recognition (9,12,13).
The key question of how exactly hLa recognizes ‘internal’ structured RNA sequences lacking a 3′ oligoU trailer has yet to be addressed. It has been suggested that the C-terminal half of the human La protein, which contains a second RRM domain (RRM2) followed by an unstructured stretch of polypeptide (27) punctuated by a short basic motif (SBM) (Figure 1A), may be implicated in interactions with mRNAs rather than pre-tRNAs (2,21,22,28). However, definitive evidence for a contribution of this region to the RNA-binding properties of hLa has yet to be produced. Although some early studies based on deletion mutagenesis experiments suggested such a role for RRM2 (23,29), these were performed used a truncated form of the domain that may have been misfolded and therefore prone to artefacts (27). Notably, the functional significance of the C-terminal half of La proteins has been clouded by the fact that they display a lower degree of conservation varying in both size and sequence between species; neither the second RRM nor the SBM, which could potentially confer additional RNA-binding capability to hLa, are found in the yeast proteins (27,30) (Figure 1).
To shed new light on the versatility of La–RNA interactions and the molecular basis of La's role in mRNA translation, we set out to analyse in detail the ability of hLa to bind to 3′ oligoU-deficient RNA molecules. Steered by the current knowledge (21,22), for these investigations we chose a 27-nt RNA fragment comprising domain IV of HCV IRES (residues 330–356) that includes the AUG start codon and spans part of the core-coding sequence (Figure 1) (31,32). An array of quantitative biophysical and biochemical techniques were employed to examine the interaction between this RNA fragment and hLa; several structure-based variants of HCV IRES domain IV and deletion mutants of hLa were used to fully dissect the interaction. Unexpectedly, these experiments revealed a mode of RNA recognition for hLa that results from the synergistic action of all three structured domains—the LaM, RRM1 and RRM2—and is therefore quite distinct from the interaction hLa makes with 3′ oligoU sequences. By RNA mutagenesis analysis we also demonstrated that this alternative mode of binding by hLa involves a sequence-independent recognition of a short double-stranded stretch flanked by a single-stranded extension, which is in line with a more general chaperone role for La in mRNA-related activities. Our results therefore attest to the remarkable adaptability of the hLa protein and provide the first evidence of RNA contacts by the La RRM2 domain, which was previously shown not to be involved in binding 3′ UUUOH sequences and pre-tRNA ligands.
MATERIALS AND METHODS
Plasmid construction
A variety of plasmids encoding the full-length human La protein and several deletion mutants were used to produce recombinant proteins. La(1–408) and La(1–194) were cloned as reported previously (27). The fragment containing the three structured domains, La(4–325), and the tandem RRM construct, La(105–325), were amplified from pET-La1–408 using PCR and subcloned into a modified pETM-11 vector to include a TEV NIa protease (TEVpro) cleavable N-terminal His6-tag. La(1–354) was cloned into an in-house modified pET30 vector, so as to have a TEVpro following the N-terminal His6-tag. La(1–229) was cloned in a pET-30 vector using a LIC methodology (Novagen) (33).
Protein expression and purification
All the La proteins were expressed in Escherichia coli strain Rosetta II at 37°C in rich media with induction by 1 mM IPTG (isopropyl β-d-thiogalactoside). Expressions were also performed in minimal media containing 0.8 g/l 15-ammonium chloride for La(1–194) and La(4–325) and in minimal media containing 0.8 g/l 15N-ammonium chloride, 2 g/l 13C glucose and 95% D2O for La(4–325). La(1–408) and La(1–194) were expressed and purified as described before (27). Cell pellets containing La(4–325), La(1–354), La(105–325) or La(1–229) were lysed by sonication in 50 mM Tris, 300 mM NaCl, 10 mM imidazole, 5% glycerol, pH 8.0, 2 mM PMSF (phenylmethanesulfonyl fluoride), pH 8.0. Lysates were clarified by centrifugation and applied to 5 ml HisTrap columns (GE Healthcare) with protein elution performed over a gradient of 20–300 mM imidazole. The His6-tag was cleaved from La(4–325) by incubating the protein overnight with TEVpro [at TEVpro:La(4–325) molar ratio of 1:50] at 30°C in 50 mM Tris, 100 mM NaCl, 1 mM DTT (dithiothreitol), pH 8. The cleaved tags, the His-tagged TEVpro and any undigested product from the reaction mixture were removed by a further round of purification on a Ni–NTA column (Qiagen). The La proteins were then loaded on a 5 ml Hi-Trap heparin column, mainly to eliminate nucleic acids contaminants, and eluted with a linear 0–2 M KCl gradient. Following overnight dialysis, final purification was performed by gel filtration on a Superdex™ 75 10/300 GL column run at 0.5 ml/min in 20 mM Tris, 100 mM KCl, 0.2 mM EDTA (ethylenediaminetetraacetic acid), 1 mM DTT, pH 7.25. Protein concentrations were calculated based upon the near-UV absorption (ε280) using theoretical extinction coefficients derived from ExPASY (34).
RNA sample preparation
RNA 4-nt oligoU (5′-UUUU-3′) was synthesized and gel purified by Dharmacon Inc. Domain IV of HCV IRES and the following mutants were synthesized and gel purified by IBA GmbH (Göttingen, Germany): IV, a 27-mer of sequence 5′-AGACCGUGCACCAUGAGCACGAAUCCA-3′, corresponding to the entire domain IV (encompassing residues 330–356) with the 3′ U (U356) mutated to A; IVOMe, bearing a methyl group at the 3′-OH; IV3′ext of sequence 5′-CGUGCACCAUGAGCACGAAUCCA-3′; IV5′ext of sequence 5′-AGACCGUGCACCAUGAGCACG-3′; 27-mer ssRNA of sequence 5′-ACCUAACCACCACUACCACCUCCCACA-3′. The RNA oligo IVUUUU, a mutant version of the domain IV in which the first 3 nt were changed to C and the last three to U (5′-CCCCCGUGCACCAUGAGCACGAAUUUU-3′) was purchased from Primm srl (Milan, Italy).
Domain IV and two mutants thereof were prepared by in vitro T7 polymerase transcription: IVhalves, where the two halves of the domain IV RNA, 5′-AGACCGUGUGCACCA-3′ and 5′-UGAGCACGAAUCCA-3′, were made separately and then annealed together (see below); IVlowerSL, containing only the lower stem-loop of the domain IV, of sequence 5′-CGUGCACCAUGAGCACG-3′. The 22-mer RNA oligo (22-mer structRNA) of sequence 5′-CACCUAUAUAGUUAUAUAAUAA-3′ was a kind gift from Ian Taylor, NIMR.
For in vitro transcription, large-scale homogeneous RNA production was performed as described earlier (35). Briefly, a 5′ hammerhead ribozyme and target RNA sequence(s) were cloned between the T7 promoter and hepatitis δ ribozyme site in the plasmid pUC119δv, using XbaI and PstI restriction sites (36). The ribozyme construct was linearized with HindIII and transcribed in a standard large-scale T7 polymerase reaction at 10–14 ml scale. The reaction mix was then annealed for 10 min at 65°C, slow cooled to 55°C and held at this temperature for 30 min. Product RNAs were purified on an 8 M urea 10% polyacrylamide denaturing gel and then eluted with PAGE elution buffer, 0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA, 0.1% SDS (sodium dodecyl sulphate). Following ethanol precipitation, the RNA was extensively dialysed in 20 mM Tris, 100 mM KCl, 10 mM MgCl2, 1 mM DTT, pH 7.25 and then concentrated using centrifugal concentrators (Vivaspin).
Solutions of IV, IVOMe, IV3′ext, IV5′ext, IVUUUU, 22-mer structRNA, 27-mer ssRNA and 4-nt oligoU were prepared by solubilizing the oligonucleotides in 20 mM Tris, 100 mM KCl, 10 mM MgCl2, 1 mM DTT, pH 7.25; the RNAs were annealed by heating at 95°C for several minutes followed by slow cooling to room temperature (>4 h). The IVhalves RNA was prepared by mixing together equal concentrations of the two complementary RNAs and annealing the resulting solution as described before. The concentration of the dissolved oligonucleotides was evaluated by UV measurement at 95°C, using the molar extinction coefficients at 260 nm calculated by the nearest-neighbour model (37).
Nuclear magnetic resonance spectroscopy
Nuclear Magnetic Resonance (NMR) samples of 15N-labelled La(1–194) and La(4–325) and 2H/15N/13C-labelled La(4–325) were prepared by dialysing the purified protein against 20 mM Tris, 100 mM KCl, 1 mM DTT, pH 7.25. NMR spectra were recorded at 298 K on a Varian Inova spectrometer operating at 18.8 T and on a Bruker Avance spectrometer operating at 16.4 T both equipped with triple resonance cryoprobes. The spectra were processed using NMRPipe/NMRDraw (38) and analysed using NEASY (39).
Two sets of NMR titration experiments were performed by adding increasing amounts of unlabeled IV RNA solution into a sample containing 15N-labelled La(1–194) or 2H/15N/13C-labelled La(4–325). 1H-15N HSQC or 1H-15N TROSY-HSQC spectra were recorded at protein:IV RNA molar ratio of 1:0, 1:0.3, 1:0.6, 1:1.0 1:1.3.
A 2H/15N/13C-labelled La(4–325) sample was used to collect 1H-15N TROSY-HSQC, TROSY-HNCA, TROSY-HNCACB, TROSY-HNCOCACB and TROSY-HNCO, to yield an almost complete backbone assignment of the apo protein (deposited to BMRB, accession number 17 878). To this sample was then added IV RNA to a final protein:RNA molar ratio of 1:1.2 and 1H-15N TROSY-HSQC, TROSY-HNCA, TROSY-HNCOCA and TROSY-HNCO were acquired on the complex.
Chemical shift variations were determined where possible by comparing 3D spectra of the apo La(4–325) protein with those of La(4–325) complexed with HCV RNA. When the poor spectral quality and/or instability of the complex precluded the unambiguous identification of resonance signals in the bound state, the analysis was conducted as follows: (i) in regions of reasonable spectral clarity, signals were transferred to the resonances of the complex that would have the smallest chemical shift variation (ΔδAV); (ii) where the peak unambiguously moved but severe spectral overlap prevented a clear identification of the nearest neighbour, the chemical shift variations were broadly classified as ‘not quantified’; (iii) resonances that clearly disappeared because of line broadening were classified as ‘disappearing signals’; (iv) several resonances were left unassigned or unclear (Supplementary Figure S2C).
The weighted average of 15N and 1HN chemical shift variation (ΔδAV) was calculated as follows (9):
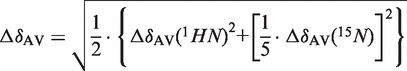 |
Chemical shift variations upon RNA binding were divided in five major categories: weak 0.035 ≤ ΔδAV ≤ 0.06; medium 0.06 < ΔδAV ≤ 0.1; strong ΔδAV > 0.1; not quantified; and signals that disappear upon interaction (Supplementary Figure S2C).
Isothermal titration calorimetry
For most experiments protein and RNA solutions were prepared in 20 mM Tris, 100 mM KCl, 10 mM MgCl2, 1 mM DTT, pH 7.25 (exceptions are noted in the text). Experiments in which the La proteins were titrated with RNA solutions were performed at 298 K using an Isothermal Titration Calorimetry (ITC)-200 microcalorimeter from Microcal (GE Healthcare) following the standard procedure reported previously (40). In each titration twenty injections of 2 µl each of a RNA solution, at a concentration of 120–140 µM, were added into a 15 µM protein solution. A spacing of 180 s between each injection was applied to enable the system to reach the equilibrium. Heat produced by titrant dilution was verified to be negligible by a control experiment, titrating into buffer alone, under the same conditions. Integrated heat data obtained for the titrations corrected for heats of dilution were fitted using a non-linear least-squares minimization algorithm to a theoretical titration curve, using the MicroCal-Origin 7.0 software package. ΔH (reaction enthalpy change in kJ/mol), Kb (equilibrium binding constant in per molar) and n (molar ratio between the protein and the RNA in the complex) were the fitting parameters. The reaction entropy was calculated using the relationships ΔG = −RT·lnKb (R = 8.314 J/(mol·K), T 298 K) and ΔG = ΔH−TΔS.
Where explicitly specified in the text, ITC experiments on La(4–325) were repeated in absence of MgCl2: 20 mM Tris, 100 mM KCl, 1 mM DTT, pH 7.25; or in absence of MgCl2 and varying KCl concentrations (115, 150, 200 mM respectively).
Circular dichroism spectroscopy
Circular Dichroism (CD) spectra of RNA and protein samples were recorded on the Applied Photophysics Ltd Chirascan Plus Spectrometer (Leatherhead, UK). Rectangular Suprasil cells with 1-cm path lengths were employed to record spectra in the regions between 340 and 220 nm. The parameters used to acquire the spectra were: spectral bandwidth of 1 nm, data step-size of 1 nm with a time-per-data-point of 1.5 s. Spectra were baseline corrected by subtracting the spectrum of the buffer alone. In all the experiments the protein concentration was in the range of 0.1–0.2 mg/ml (3.6–9 µM) and the RNA concentration was between 6 and 10 µM. The CD spectra of the protein-containing samples were acquired in 20 mM Tris, 100 mM KCl, 1 mM DTT, pH 7.25. Thermal unfolding curves of domain IV, IVhalves, IVlowerSL, IV3′ext, IV5′ext and 22-mer structRNA were recorded in the temperature mode monitoring the change of the signal at 260 nm from 5 to 95°C with a heating rate of 1.0°C/min. For melting experiments, RNA concentrations of 6 µM were used, in two different buffers, 20 mM Tris, 100 mM KCl pH 7.25; and 20 mM Tris, 100 mM KCl, 10 mM MgCl2 pH 7.25.
Electrophoretic mobility shift assays
La proteins were incubated for 15 min at room temperature with 100 nM γ-32P end-labelled RNA in 20 µl binding reactions in 100 mM NaCl, 10 mM Tris, 5 mM EDTA pH 8 before loading onto a native 12% polyacrylamide gel. Gels were run at 35 mA for 2.5 h in 1× TAE, dried and autoradiographed.
RESULTS
Identification of the regions of hLa involved in domain IV HCV RNA interaction
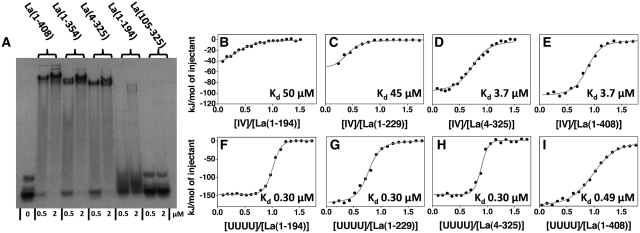
The human La protein has been shown to bind to domain IV of the HCV IRES RNA (nucleotides 330–356, Figure 1, hereafter referred to as IV), yet the domains of La responsible for this interaction have not been thoroughly investigated. A number of deletion mutants of hLa were therefore designed, prepared and tested for domain IV RNA binding: La(1–408) (full-length); La(1–354) (spanning the three structured domains and the SBM); La(4–325) (spanning the La module and the RRM2); La(1–229) (spanning the La module and the interdomain linker between RRM1 and RRM2); La(1–194) (spanning the La module); La(105–325) (spanning the two RRMs) (Figure 1). Electrophoretic mobility shift assays (EMSA) showed that La(1–408) binds to domain IV to form a single, well-defined complex, and that progressively trimming the C-terminus up to residue 325 had no observable impact on the affinity of the protein for domain IV RNA (Figure 2). Further deletions, to remove RRM2 in La(1–229) or La(1–194) or the LaM in La(105–325), resulted in a marked loss of binding affinity (Figure 2 and data not shown). Moreover, none of the individual domains (LaM, RRM1 and RRM2) exhibited binding capability for domain IV (data not shown).
Figure 2.
Interaction of the human La protein with domain IV of the HCV IRES. (A) Representative EMSA experiment of domain IV RNA (20 nM) mixed with two different concentrations (0.5 and 2.0 µM) of the following proteins: full-length La(1–408), La(1–354), La(4–325), La(1–194) and La(105–325). (B, C, D, E) ITC experiments of the interaction of domain IV RNA with La(1–194), La(1–229), La(4–325) and La(1–408) respectively. The normalized heat of interaction for the titrations was obtained by integrating the raw data and subtracting the heat of ligand dilution into the buffer alone. The grey line represents the best fit obtained by a non-linear least-squares procedure based on an independent binding sites model. Dissociation constants (Kd) are shown for each interaction. The binding profile observed for the full-length protein is unchanged in the mutant La(4–325), but a dramatic decrease in binding affinity is observed for mutants lacking RRM2. (F, G, H, I) ITC experiments of the interaction of UUUUOH RNA with La(1–194), La(1–229), La(4–325) and La(1–408). All the proteins show similar affinities and thermodynamic profiles for the interaction with this RNA, confirming that La(1–194) is sufficient for binding to short 3′ U-rich RNA sequences.
Thus, the deletion mutant La(4–325), comprising the three structured domains of hLa, appears to contain the minimum set of domains required to bind to domain IV of HCV RNA. Intriguingly, this differs from the 3′ oligoU ssRNA recognition that is confined to the La module La(1–194), thereby pointing to an alternative mechanism of recognition by hLa for structured RNA sequences that lack a 3′ UUUOH.
Addition of molar excesses of ssRNA oligos (10- to 48-nt long that did not contain 3′ oligoU sequences) used as competitor ligands in EMSA experiments had only a minor effect on binding. In contrast, it is intriguing to note that the formation of La-domain IV RNA complexes was efficiently inhibited by a subset of structured RNA sequences and by 3′ oligoU ssRNAs (see below and data not shown).
hLa-domain IV interaction involves a distinct recognition mechanism
To provide quantitative detail on how the interaction of La with domain IV differs from the 3′ oligoU ssRNA recognition, we performed ITC measurements. This technique has the advantage of affording more precise measurements of the affinity constants while simultaneously revealing the stoichiometry and the thermodynamic signature of binding interactions, which can provide useful mechanistic clues. Two sets of experiments were carried out, titrating La(1–408) and deletion mutants La(1–194), La(1–229) and La(4–325) with 4-nt oligoU and HCV IRES domain IV, respectively (Figure 2 and Table 1).
Table 1.
Thermodynamic parameters of the association of the human La protein and mutants thereof with different RNA targets in standard conditions (20 mM Tris, 100 mM KCl, 10 mM MgCl2, 1 mM DTT, pH 7.25)
| n | Kd (µM) | ΔH (kJ/mol) | –TΔS (kJ/mol) | ΔG (kJ/mol) | |
|---|---|---|---|---|---|
| La(1–194)/UUUU | 1.0 | 0.30 | −149 | 112 | −37 |
| La(1–229)/UUUU | 0.9 | 0.30 | −170 | 133 | −37 |
| La(4–325)/UUUU | 0.9 | 0.30 | −150 | 113 | −37 |
| La(1–408)/UUUU | 1.0 | 0.49 | −179 | 143 | −36 |
| La(1–194)/IV | 0.4 | 50 | −42 | 18 | −24 |
| La(1–229)/IV | 0.4 | 45 | −45 | 20 | −25 |
| La(4–325)/IV | 0.9 | 3.7 | −93 | 62 | −31 |
| La(1–408)/IV | 0.9 | 3.7 | −102 | 71 | −31 |
| La(4–325)/IVMet | 0.9 | 5.5 | −101 | 71 | −30 |
| La(4–325)/IVhalves | 0.9 | 3.7 | −68 | 37 | −31 |
| La(4–325)/IV3′ext | 0.9 | 1.4 | −68 | 35 | −33 |
| La(4–325)/IV5′ext | 0.9 | 1.0 | −64 | 30 | −34 |
| La(4–325)/IVlowerSL | 0.4 | 43 | −50 | 25 | −25 |
| La(4–325)/22-mer structRNA | 0.9 | 1.6 | −72 | 39 | −33 |
| La(1–194)/IVUUUU | 0.9 | 0.73 | −209 | 174 | −35 |
| La(4–325)/IVUUUU | 0.9 | 0.73 | −251 | 216 | −35 |
| 27-mer ssRNA/ La(4–325) | – | – | – | – | – |
| In absence of MgCl2 (20 mM Tris, 100 mM KCl, 1 mM DTT, pH 7.25) | |||||
| La(4–325)/UUUU | 0.9 | 0.49 | −185 | 149 | −36 |
| La(4–325)/IV | 0.9 | 0.48 | −121 | 85 | −36 |
| In absence of MgCl2 and varying KCl concentrations | |||||
| La(4–325)/UUUU (115 mM KCl) | 0.9 | 0.52 | −179 | 143 | −36 |
| La(4–325)/UUUU (150 mM KCl) | 0.8 | 0.55 | −175 | 139 | −36 |
| La(4–325)/UUUU (200 mM KCl) | 0.9 | 0.59 | −167 | 131 | −36 |
| La(4–325)/IV (115 mM KCl) | 0.9 | 0.50 | −100 | 64 | −36 |
| La(4–325)/IV (150 mM KCl) | 0.8 | 0.56 | −92 | 57 | −35 |
| La(4–325)/IV (200 mM KCl) | 0.8 | 0.62 | −71 | 36 | −35 |
The values reported are averaged over three different experiments, and the errors in the thermodynamic parameters are <5%
The binding of hLa mutants to UUUUOH RNA adheres closely to the interaction profile expected from previous work (9,13,27), with the La module La(1–194) solely in charge of the recognition (Table 1 and Figure 2). Consistent with this idea, the enthalpic and entropic contributions to binding are broadly similar for La(1–194) and the full-length La(1–408).
In agreement with the EMSA results, ITC experiments confirmed specific binding of full-length hLa to domain IV of HCV IRES, in that this association generated a well interpolated sigmoid-shaped curve based on an independent and equivalent binding sites model centred on a 1:1 stoichiometry, indicative of the formation of a unique complex with definite intermolecular interactions and defined energetics of association (41,42). The binding affinity is ∼7-fold lower than that for UUUUOH RNA (Kd = 3.7 µM), under the experimental conditions used (Figure 2 and Table 1). Comparison of the thermodynamic binding parameters revealed that La(4–325) behaved essentially as the full-length protein with respect to domain IV recognition, whereas removal of RRM2 [in the deletion mutants La(1–229) and La(1–194)] was accompanied by a substantial decrease in binding affinity and a markedly different thermodynamic signature of the molecular association, in which the magnitudes of the enthalpic and entropic contributions were significantly reduced, as well as the binding stoichiometry (Figure 2 and Table 1).
The association of hLa with oligoU and domain IV at 25°C is enthalpically driven with an unfavourable entropic contribution in both cases, although the thermodynamic signatures of binding are not identical: binding of hLa to HCV IRES domain IV involves smaller changes in enthalpy and entropy (Table 1). ITC was also used to examine the perturbations of the binding energetics of La(4–325) for both RNA ligands by varying concentrations of MgCl2 (0–10 mM) or KCl (100–200 mM). In agreement with previous reports (16), the interaction of La with 3′ UUUOH RNA was found to be Mg2+-independent while reduction of Mg2+ concentration from 10 to 0 mM produced a 7-fold enhancement of binding affinity for La(4–325)-domain IV association (Table 1). Furthermore, a very different dependence on KCl concentration was observed for the two interactions (Table 1).
These results show two distinct and diverse modes of RNA binding by the La protein: (i) the well-known sequence-specific 3′ oligoU recognition supported exclusively by the La module and relatively unresponsive to changes in buffer ionic strength; and (ii) the newly characterized interaction with domain IV of HCV IRES (which lacks a 3′ oligoU element) that requires the La module and the RRM2 and has exposed, for the first time, a requirement for RRM2 domain to RNA binding. Data also suggest that the electrostatic contribution is likely to play a more substantial role in binding of hLa to domain IV than in its interaction with 3′ oligoU RNA, underscoring the differences in the mechanisms of recognition by hLa protein.
Domain IV adopts a stem–loop conformation
Which particular features of domain IV RNA are being recognized by the La protein in this novel mode of binding? As depicted in Figure 1C, the 27-nt fragment containing the HCV IRES domain IV is predicted by mfold (43) to form a stem–loop structure (with a 5-bp lower stem and a 7-nt loop), topped by a small bulge, a short 2-bp upper stem and short 5′ and 3′ extensions. As no detailed structural information is available on this IRES domain to date, the mfold prediction was scrutinized by CD and NMR spectroscopy.
The CD spectrum of domain IV at 25°C indicated that the RNA duplexes adopt an A-conformation, with a large positive band ∼260 nm and a large negative signal at 210 nm (44,45) (Supplementary Figure S1). The CD magnitude at 260 nm was followed in CD melting experiments, performed by heating the RNA samples from 5°C to 95°C and intended to inform on the veracity of the predicted secondary structure in conjunction with deletion mutagenesis. Under these conditions and in standard buffer, domain IV unfolds with two major apparent transitions with melting temperatures (Tm) of 40°C and 78°C (Supplementary Figure S1). Removal of the 2-bp upper stem in IV3′ext, IV5′ext and IVlowerSL variants affected the transition at 40°C but left the higher temperature transition largely unchanged (Supplementary Figure S1), thereby tentatively assigning the opening of the upper stem to the first and the unfolding of the lower stem to the second apparent transition. Elimination of MgCl2 from the buffer did not alter the shape of the melting curve for domain IV RNA but resulted in a shift of Tm for both transitions (to 30°C and 70°C respectively) (Supplementary Figure S1).
NMR experiments conducted on domain IV were also consistent with the mfold predicted secondary structure and CD melting profiles. In particular, the 1H NMR spectrum confirmed the presence of 6 or 7 resonances corresponding to base-paired imino protons, two of which at 25°C were only visible (albeit as broad signals) in the presence of 10 mM MgCl2 (data not shown).
Although in absence of structural data alternative interpretations of the CD melting curves are still possible, these analyses appear to be consistent with the mfold predicted secondary structure arrangement of domain IV. This provided a framework for designing mutants to be tested for protein recognition.
Identification of RNA elements within HCV IRES domain IV recognized by hLa
In order to identify the elements within domain IV that are involved in its interaction with the hLa protein, a number of RNA variants (Figure 3) were constructed and their ability to bind La(4–325) determined by ITC. The mutants included:
1. Modification to block 3′-OH group
2. Mutations to destroy duplex formation
3. Mutation to interfere with lower loop
4. Mutations at the bulge and upper stem.
Figure 3.
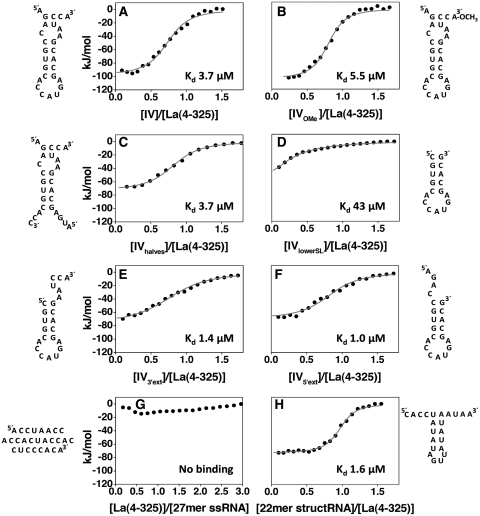
Interaction of La(4–325) with RNA targets. (A–H) ITC experiments showing the thermal effect of mixing La(4–325) with different RNA targets as follows: (A) IV; (B) IVOMe; (C) IVhalves; (D) IVlowerSL; (E) IV3′ext; (F) IV5′ext; (G) 27-mer ssRNA; (H) 22-mer structRNA. The grey lines represent the best fit derived by a non-linear least-squares procedure based on an independent binding sites model. The expected secondary structures of the RNA molecules obtained with the software mfold, which matched well with CD thermal unfolding data (Supplementary Figure S1), are shown beside each graph. Taken together, these data show that La(4–325) recognizes a double-stranded stem flanked by extensions at the 5′ or 3′, independently of nucleotide sequence.
In each case, correct folding of the variant RNAs was verified by CD spectroscopy (see above and Supplementary Figure S1).
1. The 3′-OH of the HCV domain IV is not involved in binding hLa
Since domain IV of the HCV IRES domain is an ‘internal’ sequence embedded within the 5′-UTR of the viral genome, it was imperative to verify that its association with La would not hinge on the free 3′-OH artificially created in the truncated RNAs used in our biophysical measurements. To this end, a 3′ O-methylated version of domain IV (IVOMe) was synthesized and its association with La(4–325) examined, confirming a binding profile analogous to the 3′-OH counterpart (Figure 3B). In marked contrast, the same chemical modification of a 3′ UUUOH ligand resulted in a 38-fold reduction of binding affinity (12). The loss of discrimination for the 3′-OH denotes that the interaction between La and domain IV is a genuine ‘internal’ mode of binding which, contrary to oligoU sequences, is independent from 3′ termini recognition.
2. The RNA duplex is necessary for binding
Next, we asked whether the double-strand structure in domain IV is essential for interaction with La(4–325). To address this, we introduced a number of G or A to C substitutions within the RNA that would disrupt existing base pairings and prevent formation of any other stable duplex, thereby generating a predominantly ss 27-nt RNA (27-mer ssRNA, Figure 3G and Supplementary Figure S1). This resulted in complete abrogation of binding, suggesting that a double-stranded structure, either in isolation or within a stem–loop moiety, may be a feature recognized by the hLa protein.
3. The lower loop of the HCV RNA plays no role in hLa recognition
Loops of 7–15 nt located in the context of RNA hairpin structures have been shown to be interacting partners of single or tandem RRM containing proteins (46–49). To gauge the role of domain IV lower loop in La recognition, a cleavage in the phosphate backbone was effectively introduced between the third and fourth loop nucleotides by re-constructing domain IV from two partially complementary fragments (in the mutant hereafter referred to as IVhalves). As this modification had virtually no effect on the binding affinity for La(4–325) (Figure 3C), we conclude that an intact lower loop is not implicated in the interaction with hLa. This was further confirmed by the experiments with the 22-mer structRNA (see below).
4. 3′ or 5′ ss extensions are key determinants for hLa recognition
The mutants IV5′ext, IV3′ext and IVlowerSL (Figure 3D–F) were designed to dissect the contributions to La binding of the bulge, upper stem and 5′/3′ extensions of the RNA. Deletion of all these elements in IVlowerSL generated an unadorned hairpin-loop structure which bound an order of magnitude more weakly to La(4–325) than the intact domain IV (Figure 3D). Interestingly, the addition of a ss extension at the 5′ or 3′ end of the lower stem–loop (in the IV5′ext and IV3′ext mutants respectively) completely restored high-affinity binding to La(4–325). In fact, these RNAs even displayed a slightly enhanced affinity compared to domain IV (Figure 3A, E, F and Table 1).
Taken together, these experiments suggest that the minimal element of domain IV required for La interaction encompasses the lower stem flanked by a ss extension on either the 5′ or 3′ end.
hLa–domain IV RNA interaction is a shape-directed recognition
The analysis of the binding requirements acquired thus far points towards shape-dependent rather than a sequence-specific type of recognition. To further support this hypothesis, we repeated ITC and EMSA experiments with an RNA molecule that would retain the key structural features of the domain IV—a duplex stem with ss 5′ and 3′ extensions—but with a completely different nucleotide sequence as well as a shorter lower loop (22-mer structRNA, Figure 3H and Supplementary Figure S1).
ITC experiments confirmed that La(4–325) binds to 22-mer structRNA with an affinity and overall thermodynamic parameters that are closely comparable to domain IV, indicative that in both cases the interaction follows a similar mechanism of recognition (Figure 3).
These data therefore endorse a shape-dependent mechanism of recognition, in contrast to the sequence-specific 3′ oligoU interaction. While hLa may make a number of hydrogen bonding and stacking ‘cation-π’ interactions with domain IV RNA, shape-selective recognition probably mainly occurs through contacts with the sugar–phosphate backbone, since, as shown by ITC measurements, the interaction is salt sensitive and therefore involves a significant electrostatic component (Table 1).
Domain IV undergoes minor conformational changes upon La binding
Next we examined the influence of La binding on the conformation of domain IV, as other IRES-binding proteins are known to induce conformational changes in their RNA targets with consequent enhancement of IRES function (50). Moreover, hLa itself has recently been shown to harbour both strand-annealing and strand-dissociation RNA chaperone capabilities (A.R. Naeeni, M.R.C. and M.A. Bayfield, unpublished data).
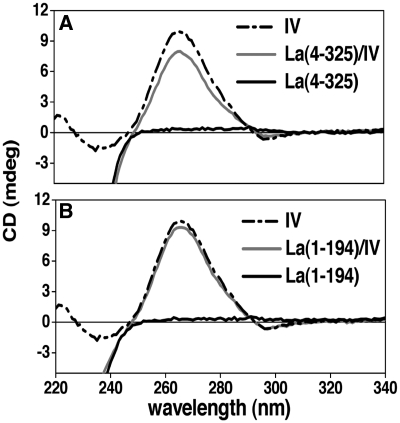
Comparison of the near-UV CD spectra arising from the RNA in the free and La(4–325)-bound state reveals a modest reduction in the intensity of the RNA CD signal (at 260 nm) in the complex (Figure 4). Such a profile, often brought about by double-stranded nucleic acid binding proteins, is indicative of a somewhat decreased stacking of the DNA/RNA bases, and it may denote localized melting and/or distortion of the double helix of domain IV following La(4–325) binding (51). Although the CD data do not allow us to elaborate on the exact nature of this conformational rearrangement, the observed effect is small, implying that a large portion of the double-stranded region of domain IV would still be retained in the complex with La(4–325). Attempts to analyse further the conformation of domain IV bound to La using NMR spectroscopy failed because of the poor linewidth of the RNA resonances in the complex.
Figure 4.
CD analysis of domain IV conformation upon interaction with La. (A) Near-UV CD spectra of domain IV RNA alone (dotted line) and in complex with the La(4–325) (grey line) show a modest reduction in the intensity of the CD signal at 263 nm following protein binding. Since the signal from the RNA dominates the CD spectrum at wavelengths >250 nm (near UV), with the signal of the protein being negligible in this range (black line), any spectral difference here could therefore be attributed solely to changes in the polynucleotide conformation. (B) Near-UV CD spectra of domain IV RNA alone (dotted line) and in complex with the La(1–194) (grey line). In this case a much smaller change in the RNA CD signal was detected.
The three structured domains and both inter-domain linkers are affected by La(4–325) binding to domain IV
The surfaces of the hLa protein involved in shape-dependent recognition of domain IV were delineated by NMR chemical shift perturbation analysis. To begin with, backbone assignment for apo La(4–325) was obtained by analysis of TROSY-based experiments on a 70% non-exchangeable deuterated sample. For structured portions (LaM, RRM1 and RRM2), most of the chemical shifts were directly transferred from previously obtained assignments of the individual domains (33,52–54) indicating that these maintain essentially the same structure in the context of the longer protein and that in solution they largely tumble independently of one another. Backbone NMR assignments of the linker regions connecting the structured domains proved more challenging. Although the linker stretch between the LaM and RRM1 had already been assigned in the apo La(1–194) protein (54), weak, missing and/or severely overlapped resonances prevented their unambiguous identification in free La(4–325). No prior information was available for the 30 residue inter-RRM segment and several residues remain unassigned (Supplementary Figure S2C).
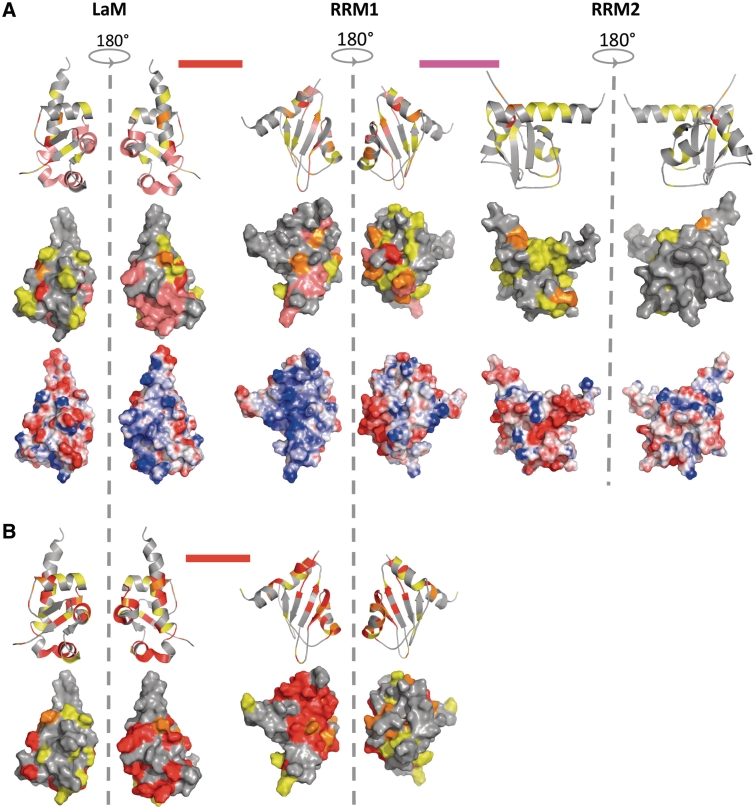
To identify regions of the protein affected by complex formation, chemical shift variations experienced by protein residues upon addition of domain IV RNA were mapped onto the structures of the isolated domains or annotated to inter-domain linker regions (Figure 5). This analysis shows variations across the entire molecule, i.e. in residues belonging to LaM, RRM1 and RRM2, as well as the inter-RRM linker (Figure 5 and Supplementary Figure S2C); despite the lack of resonance assignment for the linker between the LaM and RRM1 in La(4–325) spectra, the experiments performed on La(1–194) described below suggest that this region is also affected by domain IV complex formation.
Figure 5.
NMR analysis of La-domain IV interaction. (A) Structure mapping of the chemical shift variations obtained in La(4–325) on domain IV binding for the LaM, RRM1, RRM2 and inter-domain linkers and comparison with the electrostatic surface potential. The LaM, RRM1 and RRM2 are shown as structures of individual domains (PDB ID: 1S7A, 1S79 and 1OWX respectively), whereas the inter-domain linkers are shown as bars between domains. The positions of the residues that show substantial shifts are indicated on the protein secondary structures (top) and surfaces (middle). The chemical shift perturbations (ΔδAV) are represented with the following colour code: yellow (0.035 ≤ ΔδAV ≤ 0.06), orange (0.06 < ΔδAV ≤ 0.1), red (ΔδAV > 0.1), pink (disappearing signals) and magenta (not quantified) (Supplementary Figure S2C). (B) Structure mapping of the chemical shift variations obtained in La(1–194) on 3′ oligoU binding for the LaM, RRM1 and inter-domain linker.
The majority of the strong chemical shift changes are spread over the LaM and RRM1, and the patches thereby delineated overlap to some degree with the surfaces involved in 3′ oligoU recognition, especially for the former domain (Figure 5). Nonetheless, intriguing differences emerge with respect to the RRM1: in particular, while the titration with UUUUOH leaves the α1/α2 face of this domain completely devoid of perturbations, binding of IRES domain IV is sensed by several residues clustered on this ‘hind’ region (Figure 5). However it remains unclear whether this identifies a potentially new RNA-binding surface on RRM1 or reflects conformational changes on binding that are propagated to the helical face of RRM1.
In accord with our EMSA and ITC results, NMR chemical shift variations were also detected in regions of the protein beyond the La module, in particular affecting the RRM2 and the inter-RRM linker, although the associated shifts appear to be less pronounced. Intriguingly, the observed perturbations on the RRM2 do not map to the canonical β-platform RNA-binding surface, but rather interest part of the atypical C-terminal α-helix that in this domain lies across the upper part of the β-sheet (27). Assuming that domain IV binding elicits conformational rearrangements of the modular structured domains of the protein, it is likely that the chemical shift variations experienced by the inter-RRM linker are coupled with induced conformational changes, although the possibility of a direct contact with RNA cannot be ruled out.
HCV IRES domain IV binding by hLa involves a bipartite mode of recognition
Given that the La module is a ssRNA-binding unit (13) and strengthened by our observations that many of the surfaces mediating 3′ oligoU interaction also appear to be involved in the interaction with domain IV, we speculated that this portion of the protein would be implicated mainly in contacting the ss stretches of domain IV, with the rest of the molecule engaging the double-stranded element. To test this hypothesis, we analysed the NMR chemical shift variations generated by titrating domain IV into a sample containing 15N-labelled La(1–194). Although ITC and EMSA experiments showed that La(1–194) did not possess all the determinants to behave as the full-length protein with respect to domain IV binding, NMR experiments carried at millimolar concentrations could be used to investigate lower affinity interactions. Intriguingly, the chemical shift variations experienced by La(1–194) upon addition of domain IV were almost completely superposable on the shifts observed in this part of the protein in the context of the longer La(4–325) (Supplementary Figure S2B). Furthermore, a similar result (albeit with some variations in the shift profile) was obtained by titrating a single strand of IVhalves (data not shown), endorsing a model in which the La module is responsible for making contacts with a ss portion of IV RNA, whereas the inter-RRM linker and RRM2 are important for dsRNA interaction.
Finally here, the higher quality NMR spectra obtained with the smaller La(1–194) protein indicated that the linker between the LaM and RRM1 experienced chemical shift variations upon domain IV titration, probably more as a result of conformational changes than direct contacts with RNA, as observed before for 3′ oligoU RNAs (12,13). Since most of the other resonances in La(1–194) complexed with domain IV could be directly transferred onto the spectra of La(4–325)–domain IV, we assume that this linker also changes in the context of the longer protein.
Comparison between domain IV binding and pre-tRNA type of interaction
On the basis of the proposed bipartite model of interaction we were intrigued to know whether the mode of interaction revealed for the La-domain IV complex—recognition of a stem bordered by a ss extension—might also apply for pol III nascent transcripts, where the ss tail would terminate with a 3′ oligoU sequence. To address this, we constructed an RNA mutant that bears the lower stem of domain IV appended to a 3′ UUUOH trailer, termed IVUUUU, and monitored its interaction with La(1–194) and La(4–325) by ITC. Figure 6 shows that La(1–194) and La(4–325) bind IVUUUU with indistinguishable thermodynamic signatures of association which are somewhat reminiscent of the ss oligoU interaction profile (Figure 6). In other words, for this small structured RNA mutant that harbours a 3′ oligoU end, all the determinants for interaction seemingly reside within the La module La(1–194), and no clear evidence for an additional contribution to binding by the RRM2 and inter-RRM linker (perhaps to the stem region) could be seen in the longer La(4–325). We deduce that the interactions of La with domain IV (short double-stranded stem flanked by a short single-strand extension) and structured 3′ oligoU containing RNA targets appear to be mutually exclusive (see ‘Discussion’ section).
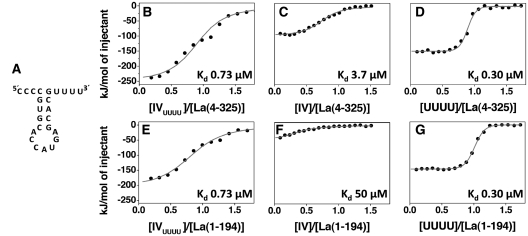
Figure 6.
Comparative analysis of the interaction between La and IVUUUU RNA. (A) Sequence and secondary structure for IVUUUU, which retains the lower stem–loop portion of domain IV while the 5′ and 3′ extensions are mutated to oligoC and oligoU, respectively. (B and E) ITC experiments examining the interaction of IVUUUU with La(4–325) and La(1–194) respectively. The grey line represents the best fit obtained by a non-linear least-squares procedure based on an independent binding sites model. For comparison, the interactions of domain IV (C and F) and UUUUOH (D and G) with La(4–325) and La(1–194) respectively are reported. For the interactions of La(4–325) and La(1–194) with IVUUUU, the thermodynamic signature of binding, evaluated as the relative ΔH/–TΔS value, is identical in both cases and is closer to the UUUUOH associations (Table 1).
DISCUSSION
While a large number of reports designate hLa as an established IRES trans-acting factor (18–26,28,55,56), a rigorous analysis of the mechanism underpinning the recognition of these internal RNA sequences by La has been lacking. The study presented here detailing the interaction of hLa with domain IV of the HCV IRES provides important new insights into this mode of binding and so into the adaptability of the La protein in recognizing structured RNA targets that lack a 3′ oligoU element.
We performed a systematic biophysical investigation closely informed by the structure of human La to provide evidence that the protein binds HCV IRES domain IV using an alternative mode of RNA recognition that differs markedly from its well-characterized interaction with 3′ UUUOH ssRNAs. Several attributes distinguish the two mechanisms: whereas recognition of 3′ oligoU sequences is directed by an exposed 3′ end, a specific nucleotide sequence and a ssRNA conformation, the interaction with domain IV is truly ‘internal’, selecting for particular RNA structural motifs (short duplexes with single-stranded trailers) but independent of nucleotide sequence composition.
Our results provide a molecular explanation of how the human La protein can effectively handle such profoundly different modes of RNA recognition, in that it relies exclusively on the La module for the 3′-termini binding but summons the three modular domains for the internal type of recognition. Remarkably, while the La module appears precisely tailored to recognize a free 3′-OH and the penultimate uridine base (U-2) in 3′ oligoU RNA sequences (12,13), in conjunction with the inter-RRM linker and RRM2 it can bind with comparable affinity to radically different RNA targets, using a mode of binding that alleviates the dependence of 3′ termini recognition. These findings significantly extend the known plasticity of La–RNA interactions (13,14).
Interestingly, our experiments with the variant IVUUUU RNA suggest a mutually exclusive behaviour for the 3′ UUUOH versus the internal type of RNA recognition. In particular, since no difference could be detected in the binding of IVUUUU to La(4–325) and La(1–194), it appears that the clamping onto the intervening 3′ ss oligoU stretch within IVUUUU by the La module renders it incapable of working together with the RRM2 and inter-RRM linker to generate the composite RNA-binding surface required for contacting the double-stranded portion of IVUUUU. Given that binding of the La module to UUUOH dictates a precise orientation of both protein and RNA through formation of the specific RNA-binding cleft and coordination of the 2′ and 3′ hydroxyls by D33 (13), it may be conceivable to invoke incompatibility between this imposed geometry and the simultaneous rearrangement of the three modular domains underlying the alternative mode of RNA binding. Whatever the precise molecular basis of the competition between the two modes of interaction, the dominance of 3′ oligoU binding, which is mediated only by the La module, provides a ready explanation for the fact that the possible contribution of RRM2 binding to structured RNAs containing a 3′ oligoU tail, such as pre-tRNAs, has not been detected in previous work (16,17). Nevertheless, although our results suggest that RRM2 is not involved in binding IVUUUU, it is possible that for larger RNA targets binding to a 3′ oligoU tail by the La module would not preclude involvement of RRM2 in contacting other, perhaps more distal, parts of the RNA.
Indeed, the interest in the alternative mechanism of binding by La is heightened by the fact that for the first time a clear contribution of RRM2 to RNA binding has been revealed by a systematic study based on the known structure of the domain (27). Intriguingly, chemical shift analysis suggests that the RRM2 does not behave as an ordinary RRM: first, the canonical β-sheet RNA-binding surface appears unaffected by the presence of domain IV; second, NMR data indicate that the RRM2 largely retains its free form conformation in the complex, implying that the β-sheet platform will remain in part blocked by the long C-terminal helix (27); third, such an atypical helix may itself be a feature for domain IV recognition. NMR analysis also highlights the unexpected involvement of the α1/α2 helical face of the RRM1 in complex formation. In this scenario, our proposed bipartite model posits that the La module interacts with the single-stranded extension of domain IV, with the rest of the protein involved in contacting the double-stranded portion of the RNA (Supplementary Figure S3). Intriguingly, the 5′/3′ direction of the ss extension appears not to be critical for La binding, perhaps suggesting a degree of flexibility for the contacts with the backbone of this region.
Verification of this model and elucidation of the arrangement of the RNA-binding domains within the La–domain IV complex awaits a full structure determination.
Nevertheless, our work assigns a clear molecular role to the C-terminal region of hLa in HCV IRES domain IV recognition, and builds on past functional studies (2,19,21,22) to provide a solid framework to investigate how hLa contributes to IRES-directed translation initiation. Moreover, the essential features of the alternative mode of RNA recognition unveiled here may hold true for other La proteins given their resemblance across higher eukaryotes, though will not be shared by yeast homologues that do not harbour an RRM2. It appears therefore that some of the key RNA-binding activities of La are a prerogative of higher eukaryotes and for the first time a molecular explanation for the different RNA recognition properties of the human and yeast La proteins is unambiguously provided. Our observations not only suggest plausible solutions for some of the conflicting results observed in the past decade regarding additional roles of La (e.g. modulation of removal of pre-tRNA 5′ trailers, nuclear retention of certain transcripts, stabilization of viral mRNA) (1), but also offers new insights for a sounder appreciation of the functional difference between the La proteins across species.
ACCESSION NUMBER
Nuclear Magnetic Resonance (NMR) assignment submitted to the Biological Magnetic Resonance Bank (BMRB), accession number 17878.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures (1–3).
FUNDING
The Wellcome Trust through a project grant to M.R.C. and S.C. (075295/A/04/Z) and a Capital Award for the Centre for Biomolecular Spectroscopy to M.R.C. and A.F.D. (085944/Z/08/Z); Long term EMBO Fellowship to L.M. OKK was supported by BBSRC funding BB/E02209X/1 (to S.C.); Medical Research Council, UK (U117584228) to S.J.S. Funding for open access charge: Wellcome Trust/University funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Caterina Alfano, Elizabeth Valentine, Cyril Gaudin and Jingjie Yang for help in the initial stages of this project and to Dr Ian Taylor (NIMR) for the kind gift of RNA reagents. The authors thank Drs Richard Maraia and Mark Bayfield for helpful discussions.
REFERENCES
- 1.Wolin SL, Cedervall T. The La protein. Annu. Rev. Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 2.Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs) Biochim. Biophys. Acta. 2010;1799:365–378. doi: 10.1016/j.bbagrm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue D, Rubinson DA, Pannone BK, Yoo CJ, Wolin SL. U snRNP assembly in yeast involves the La protein. EMBO J. 2000;19:1650–1660. doi: 10.1093/emboj/19.7.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kufel J, Allmang C, Chanfreau G, Petfalski E, Lafontaine DL, Tollervey D. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol. 2000;20:5415–5424. doi: 10.1128/mcb.20.15.5415-5424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoo CJ, Wolin SL. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]
- 6.Pannone BK, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Intine RV, Dundr M, Misteli T, Maraia RJ. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Mol. Cell. 2002;9:1113–1123. doi: 10.1016/s1097-2765(02)00533-6. [DOI] [PubMed] [Google Scholar]
- 8.Chakshusmathi G, Kim SD, Rubinson DA, Wolin SL. A La protein requirement for efficient pre-tRNA folding. EMBO J. 2003;22:6562–6572. doi: 10.1093/emboj/cdg625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, Conte MR. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 2004;11:323–329. doi: 10.1038/nsmb747. [DOI] [PubMed] [Google Scholar]
- 10.Dong G, Chakshusmathi G, Wolin SL, Reinisch KM. Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J. 2004;23:1000–1007. doi: 10.1038/sj.emboj.7600115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curry S, Conte MR. A terminal affair: 3′-end recognition by the human La protein. Trends Biochem. Sci. 2006;31:303–305. doi: 10.1016/j.tibs.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Teplova M, Yuan YR, Phan AT, Malinina L, Ilin S, Teplov A, Patel DJ. Structural basis for recognition and sequestration of UUU(OH) 3′ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol. Cell. 2006;21:75–85. doi: 10.1016/j.molcel.2005.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotik-Kogan O, Valentine ER, Sanfelice D, Conte MR, Curry S. Structural analysis reveals conformational plasticity in the recognition of RNA 3′ ends by the human La protein. Structure. 2008;16:852–862. doi: 10.1016/j.str.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curry S, Kotik-Kogan O, Conte MR, Brick P. Getting to the end of RNA: structural analysis of protein recognition of 5′ and 3′ termini. Biochim. Biophys. Acta. 2009;1789:653–666. doi: 10.1016/j.bbagrm.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Kucera NJ, Hodsdon ME, Wolin SL. An intrinsically disordered C terminus allows the La protein to assist the biogenesis of diverse noncoding RNA precursors. Proc. Natl Acad. Sci. USA. 2011;108:1308–1313. doi: 10.1073/pnas.1017085108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayfield MA, Maraia RJ. Precursor-product discrimination by La protein during tRNA metabolism. Nat. Struct. Mol. Biol. 2009;16:430–437. doi: 10.1038/nsmb.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Bayfield MA, Intine RV, Maraia RJ. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat. Struct. Mol. Biol. 2006;13:611–618. doi: 10.1038/nsmb1110. [DOI] [PubMed] [Google Scholar]
- 18.Meerovitch K, Svitkin YV, Lee HS, Lejbkowicz F, Kenan DJ, Chan EK, Agol VI, Keene JD, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa-Mattioli M, Svitkin Y, Sonenberg N. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 2004;24:6861–6870. doi: 10.1128/MCB.24.15.6861-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svitkin YV, Pause A, Sonenberg N. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol. 1994;68:7001–7007. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ali N, Pruijn GJ, Kenan DJ, Keene JD, Siddiqui A. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J. Biol. Chem. 2000;275:27531–27540. doi: 10.1074/jbc.M001487200. [DOI] [PubMed] [Google Scholar]
- 22.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl Acad. Sci. USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pudi R, Abhiman S, Srinivasan N, Das S. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by specific interaction of independent regions of human La autoantigen. J. Biol. Chem. 2003;278:12231–12240. doi: 10.1074/jbc.M210287200. [DOI] [PubMed] [Google Scholar]
- 24.Holcik M, Korneluk RG. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svitkin YV, Meerovitch K, Lee HS, Dholakia JN, Kenan DJ, Agol VI, Sonenberg N. Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J. Virol. 1994;68:1544–1550. doi: 10.1128/jvi.68.3.1544-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sommer G, Dittmann J, Kuehnert J, Reumann K, Schwartz PE, Will H, Coulter BL, Smith MT, Heise T. The RNA-binding protein La contributes to cell proliferation and CCND1 expression. Oncogene. 2011;30:434–444. doi: 10.1038/onc.2010.425. [DOI] [PubMed] [Google Scholar]
- 27.Jacks A, Babon J, Kelly G, Manolaridis I, Cary PD, Curry S, Conte MR. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure. 2003;11:833–843. doi: 10.1016/s0969-2126(03)00121-7. [DOI] [PubMed] [Google Scholar]
- 28.Kenan DJ, Keene JD. La gets its wings. Nat. Struct. Mol. Biol. 2004;11:303–305. doi: 10.1038/nsmb0404-303. [DOI] [PubMed] [Google Scholar]
- 29.Pudi R, Srinivasan P, Das S. La protein binding at the GCAC site near the initiator AUG facilitates the ribosomal assembly on the hepatitis C virus RNA to influence internal ribosome entry site-mediated translation. J. Biol. Chem. 2004;279:29879–29888. doi: 10.1074/jbc.M403417200. [DOI] [PubMed] [Google Scholar]
- 30.Fan H, Goodier JL, Chamberlain JR, Engelke DR, Maraia RJ. 5′ processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol. Cell. Biol. 1998;18:3201–3211. doi: 10.1128/mcb.18.6.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honda M, Brown EA, Lemon SM. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 33.Sanfelice D, Babon J, Kelly G, Curry S, Conte MR. Resonance assignment and secondary structure of the La motif. J. Biomol. NMR. 2004;29:449–450. doi: 10.1023/B:JNMR.0000032556.32985.89. [DOI] [PubMed] [Google Scholar]
- 34.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnvig KB, Pennell S, Gopal B, Colston MJ. A high-affinity interaction between NusA and the rrn nut site in Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA. 2004;101:8325–8330. doi: 10.1073/pnas.0401287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jovine L, Hainz T, Oubridge C, Nagai K. Crystallization and preliminary X-ray analysis of the conserved domain IV of Escherichia coli 4.5S RNA. Acta Crystallogr. D. Biol. Crystallogr. 2000;56:1033–1037. doi: 10.1107/s0907444900006910. [DOI] [PubMed] [Google Scholar]
- 37.SantaLucia J, Jr, Allawi HT, Seneviratne PA. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 38.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 39.Bartels C, Xia T, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer supported NMR spectral-analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 40.Hands-Taylor KL, Martino L, Tata R, Babon JJ, Bui TT, Drake AF, Beavil RL, Pruijn GJ, Brown PR, Conte MR. Heterodimerization of the human RNase P/MRP subunits Rpp20 and Rpp25 is a prerequisite for interaction with the P3 arm of RNase MRP RNA. Nucleic Acids Res. 2010;38:4052–4066. doi: 10.1093/nar/gkq141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holbrook JA, Tsodikov OV, Saecker RM, Record MT., Jr Specific and non-specific interactions of integration host factor with DNA: thermodynamic evidence for disruption of multiple IHF surface salt-bridges coupled to DNA binding. J. Mol. Biol. 2001;310:379–401. doi: 10.1006/jmbi.2001.4768. [DOI] [PubMed] [Google Scholar]
- 42.Oda M, Furukawa K, Ogata K, Sarai A, Nakamura H. Thermodynamics of specific and non-specific DNA binding by the c-Myb DNA-binding domain. J. Mol. Biol. 1998;276:571–590. doi: 10.1006/jmbi.1997.1564. [DOI] [PubMed] [Google Scholar]
- 43.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wells BD, Yang JT. A computer probe of the circular dichroic bands of nucleic acids in the ultraviolet region. II. Double-stranded ribonucleic acid and deoxyribonucleic acid. Biochemistry. 1974;13:1317–1321. doi: 10.1021/bi00704a002. [DOI] [PubMed] [Google Scholar]
- 45.Gratzer WB, Richards EG. Evaluation of RNA conformation from circular dichroism and optical rotatory dispersion data. Biopolymers. 1971;10:2607–2614. doi: 10.1002/bip.360101220. [DOI] [PubMed] [Google Scholar]
- 46.Hall KB. Interaction of RNA hairpins with the human U1A N-terminal RNA binding domain. Biochemistry. 1994;33:10076–10088. doi: 10.1021/bi00199a035. [DOI] [PubMed] [Google Scholar]
- 47.Finger LD, Johansson C, Rinaldi B, Bouvet P, Feigon J. Contributions of the RNA-binding and linker domains and RNA structure to the specificity and affinity of the nucleolin RBD12/NRE interaction. Biochemistry. 2004;43:6937–6947. doi: 10.1021/bi049904d. [DOI] [PubMed] [Google Scholar]
- 48.Skrisovska L, Bourgeois CF, Stefl R, Grellscheid SN, Kister L, Wenter P, Elliott DJ, Stevenin J, Allain FH. The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction. EMBO Rep. 2007;8:372–379. doi: 10.1038/sj.embor.7400910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B”-U2A' protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 50.Yu Y, Abaeva IS, Marintchev A, Pestova TV, Hellen CU. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 2011;39:4851–4865. doi: 10.1093/nar/gkr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan R, Frankel AD. Circular dichroism studies suggest that TAR RNA changes conformation upon specific binding of arginine or guanidine. Biochemistry. 1992;31:10288–10294. doi: 10.1021/bi00157a016. [DOI] [PubMed] [Google Scholar]
- 52.Alfano C, Babon J, Kelly G, Curry S, Conte MR. Resonance assignment and secondary structure of an N-terminal fragment of the human La protein. J. Biomol. NMR. 2003;27:93–94. doi: 10.1023/a:1024741228802. [DOI] [PubMed] [Google Scholar]
- 53.Jacks A, Kelly G, Curry S, Conte MR. Resonance assignment and secondary structure determination of a C-terminal fragment of the lupus autoantigen (La) protein containing a putative RNA recognition motif (RRM) J. Biomol. NMR. 2002;22:387–388. doi: 10.1023/a:1014928117895. [DOI] [PubMed] [Google Scholar]
- 54.Sanfelice D, Kelly G, Curry S, Conte MR. NMR assignment of the N-terminal region of human La free and in complex with RNA. Biomol. NMR Assign. 2008;2:107–109. doi: 10.1007/s12104-008-9097-5. [DOI] [PubMed] [Google Scholar]
- 55.King HA, Cobbold LC, Willis AE. The role of IRES trans-acting factors in regulating translation initiation. Biochem. Soc. Trans. 2010;38:1581–1586. doi: 10.1042/BST0381581. [DOI] [PubMed] [Google Scholar]
- 56.Buchkovich NJ, Yu Y, Pierciey FJ, Jr, Alwine JC. Human cytomegalovirus induces the endoplasmic reticulum chaperone BiP through increased transcription and activation of translation by using the BiP internal ribosome entry site. J. Virol. 2010;84:11479–11486. doi: 10.1128/JVI.01330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.