Abstract
Obtaining random homozygous mutants in mammalian cells for forward genetic studies has always been problematic due to the diploid genome. With one mutation per cell, only one allele of an autosomal gene can be disrupted, and the resulting heterozygous mutant is unlikely to display a phenotype. In cells with a genetic background deficient for the Bloom's syndrome helicase, such heterozygous mutants segregate homozygous daughter cells at a low frequency due to an elevated rate of crossover following mitotic recombination between homologous chromosomes. We constructed DNA vectors that are selectable based on their copy number and used these to isolate these rare homozygous mutant cells independent of their phenotype. We use the piggyBac transposon to limit the initial mutagenesis to one copy per cell, and select for cells that have increased the transposon copy number to two or more. This yields homozygous mutants with two allelic mutations, but also cells that have duplicated the mutant chromosome and become aneuploid during culture. On average, 26% of the copy number gain events occur by the mitotic recombination pathway. We obtained homozygous cells from 40% of the heterozygous mutants tested. This method can provide homozygous mammalian loss-of-function mutants for forward genetic applications.
INTRODUCTION
Mammalian cell lines provide a convenient model for mammalian cell biology, particularly in high-throughput applications where using mice is not feasible. Forward genetic screens using various cell lines have identified genes required for many cellular processes (1). Gain-of-function screens, where genes are overexpressed or ectopically expressed, have been successful for many phenotypes (2,3). However, loss-of-function screens using mutant cells that lack expression of a particular gene are harder to conduct in diploid mammalian cells, as in most cases both alleles of a gene must be knocked out to see a phenotype. This difficulty has meant that loss-of-function screens have not been applied as widely as in yeast, Drosophila or Caenorhabditis elegans, where homozygous mutants can easily be obtained on a genome-wide scale (4,5).
This problem of obtaining functional null mutants can be solved using a number of technologies, each with limitations. First, a cell line that is functionally hemizygous can be used. Chinese hamster ovary cells are one such cell type in which many loci are functionally hemizygous and these cells have been used to isolate, for example, X-ray sensitive mutants, with a single random hit (6). However, the extent of hemizygosity in the genome of these cells is unknown and it is likely that not all genes are accessible. A near-haploid human leukemia cell line has also been described, which can be used to isolate loss-of-function mutants in the haploid portion of the genome with a single hit (7,8). Further derivatives of this cell line (9), and the recent report of a haploid ES cell line (10), show promise for screens in the future.
RNA interference (RNAi) screens have also been used in mammalian cells (11). These act at the mRNA level and thus the zygosity of the gene is irrelevant. Many RNAi screens have been performed in mammalian cell lines, and the reagents are applicable to a variety of cell types enabling many phenotypes to be studied. Common problems encountered are incomplete knockdown at the protein level, and off-target effects where transcripts other than the one predicted are affected, which can lead to variable results (12,13).
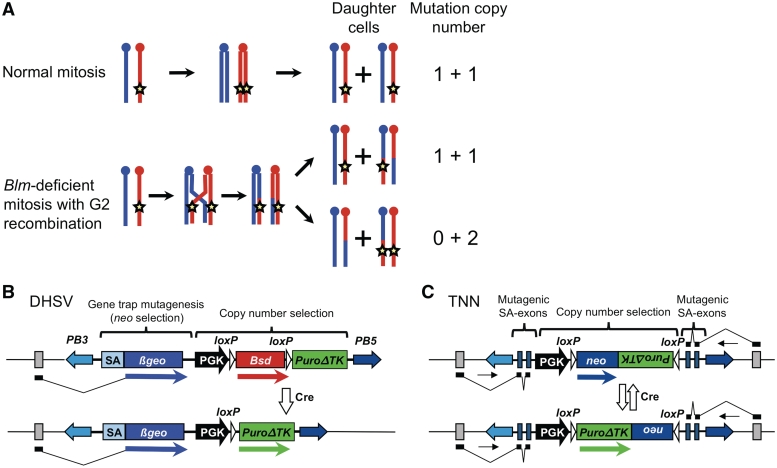
The most robust way to make loss-of-function mutations in a relatively normal cell line would be serially target both alleles in mouse embryonic stem (ES) cells. Although resources of targeting vectors and targeted heterozygous mutant ES cells are increasing in size [www.knockoutmouse.org and ref. (14)], this is still a very time-consuming task on the scale required for genome-wide genetic screens of homozygous mutations. We have previously used ES cells deficient for the Bloom syndrome gene [Blm, ref. (15)] to obtain homozygous mutants. Blm-deficient cells (referred to here as Blm cells for simplicity) have an increased frequency of crossing over following mitotic recombination relative to wild-type cells (Figure 1A). In practice, this means that cells carrying a heterozygous mutation segregate homozygous mutations at division at a low rate of the order of 10−4 events/locus/cell/division.
Figure 1.
Copy number selection for recovery of homozygous mutants. (A) Mechanism of LOH in Blm-deficient cells. Blue and red chromosomes represent homologs. Crossing over between homologous chromosomes is normally suppressed in mitosis (top). In the absence of Blm, crossovers can occur in G2 phase, forming recombinant chromatids. If these segregate to different daughter cells, LOH occurs distal to the point of crossover. If the cells carry a heterozygous mutation (represented by the star), this becomes homozygous after LOH, and the copy number of the mutation increases from one in the starting cell to two in the homozygous daughter. (B and C) Design of vectors for copy number-based selection. The vectors DHSV (B) and TNN (C) are shown integrated into an intron, with grey boxes representing exons of the disrupted gene; note that TNN can trap genes transcribed in either direction. SA, splice acceptor; PB5 and PB3, PB repeats.
Screens of mutant libraries made in Blm cells have been successful for phenotypes where null mutants are selectable, for example resistance to 6-thioguanine (mismatch repair mutants), aerolysin (glycosylphosphatidylinositol anchor synthesis mutants) or retroviral infection (16–18). Reporter systems can also be used to make the phenotype artificially selectable (19,20). The requirement for a selectable phenotype is due to the fact that each potentially interesting homozygous cell in the library is outnumbered by the order of one thousand cells heterozygous for the insertion, which are unlikely to display a loss-of-function phenotype. We were therefore interested in extending this method to other non-selectable phenotypes by increasing the proportion of homozygous cells in the library.
We present here a method to isolate homozygous cells from these libraries independent of their phenotype. We use the piggyBac (PB) transposon (21) to cause loss-of-function insertion mutations and also to deliver a selection construct that carries two drug resistance genes but can express only one at a time. The expressed gene can be switched using Cre recombinase, and only cells with two copies of this construct can acquire resistance to both corresponding drugs simultaneously. This allows selection for the increase in copy number of the mutation that occurs after loss of heterozygosity (LOH), and thus for homozygous mutants. We have isolated homozygous mutants in 48 genes using this method, which should be applicable on a larger scale to generate clonal libraries of ES cells with null mutations for genetic screens.
MATERIALS AND METHODS
Construction of the gene trap vectors
The PB transposon vector, which contains 313 bp of the 5′ inverted terminal DNA repeat (TR) and 235 bp of the 3′ inverted TR, has been described previously (19). For the deletion homozygosity selection vector (DHSV), the PB 5′ and 3′ TRs were polymerase chain reaction (PCR) amplified and cloned upstream and downstream of the selection cassettes. The SA-βgeo-bpA gene trap cassette was derived from RGTV1 (18). The Bsd-bpA fragment was derived from pYTC77 (22). The PGK-loxP-Bsd-bpA-loxP-PuroΔtk-bpA fragment was derived from pYTC86 (22).
The TNN vector was assembled using a neo gene and SV40 polyA from pcDNA3 (Invitrogen), and puΔTK-bpA from pYTC86. The mutagens are pairs of terminal exons with their preceding introns from the genes Ccdc107 and Dom3z, and were amplified by PCR and cloned in opposite orientations. These exon pairs were chosen for small size, constitutive splicing, lack of protein domains and potential to cause frameshift and premature termination. The QuikChange site-directed mutagenesis kit (Stratagene) was used to introduce premature stop codons in the native reading frame. This is predicted to cause non-sense-mediated decay of any fusion transcripts produced, thus increasing the chance that insertions toward the 3′-end of genes, which may not disrupt functional domains, will be mutagenic. The final vectors were confirmed by sequencing and full sequence of these vectors is available from GenBank under accession numbers HQ683721 (DHSV1) and HQ683722 (TNN).
ES cell culture and generation of gene trap mutations
Blm-deficient (m3/m4) ES cells (NN5), originally derived from the AB2.2 ES cell line, were cultured on a layer of γ-irradiated SNL76/7 feeder cells as described previously (18). Five million cells expanded from a single-cell clone of NN5 were co-electroporated with 0.5 or 1 µg of each gene trap vector and 10 µg of a PBase expression plasmid (pCAG-PBase). After electroporation, the cells were cultured on 90 mm feeder plates for 3 days without selection, then trypsinized and replated onto new feeder plates to reduce mosaicism in individual clones, before applying G418 selection (180 µg/ml, Invitrogen).
Transient expression of Cre plasmid and Cre protein transduction
Cre was introduced into ES cells by various methods such as electroporation, lipofection and transduction with cell-permeable Cre protein (23,24). For electroporation, 20 µg of pCAGGS-Cre plasmid was electroporated into 1.0 × 107 ES cells, one-tenth of the electroporated cells were plated on one 90 mm feeder cell plate. Puromycin (3 mg/l, Sigma) and blasticidin S (10 mg/l, Invitrogen) selection was initiated 48 h later and continued for 7 days until individual ES cell clones were visible. For Lipofection, 1 µg of pCAGGS-Cre plasmid and 3 µl Lipofectamine 2000 (Invitrogen) were applied to ES cells cultured in one well of 24-well feeder plate (50% confluent) in accordance with instructions from the manufacturer. Two days later, the cells were replated on six-well plates and drug selection was conducted as above. Cre transduction was performed as described (23,24). In brief, one well of 50% confluent ES cells from a 24-well feeder plate were cultured in 200 µl ES cell medium (without serum and antibiotics) containing 1 mM HTNC (His6-Tat-NLS-Cre) protein produced as previously described (24) for 8 h. After treatment, 1.5 ml fresh ES cell medium (without antibiotics) was added to each well. Forty-eight hours later, cells were replated on a six-well plate and drug selection was applied.
For some experiments, an inducible Cre-ERT2 protein stably expressed from the Rosa26 locus was used. The Rosa26 locus was targeted as previously described (25) using a vector provided by Adams,D. and Jonkers,J. (unpublished data). To induce recombination, cells were treated with 1 mM 4-hydroxytamoxifen overnight.
Splinkerette-PCR to identify transposon insertion sites
Isolation of the transposon–chromosome junction was performed using the Splinkerette-PCR method as described (26). Briefly, genomic DNA was isolated from ES cell colonies on 96- or 24-well plates. Two to three micrograms of DNA was digested with Sau3AI and ligated with the corresponding Splinkerette adaptors HMSp-Sau3AI (generated by annealing Splinkerette oligos HMSpBb-Sau3AI with HMSpAa). A first-round of PCR was carried out with Splinkerette primer HMSp1 and PB transposon primers PB5′-1 or PB3′-1. Then 1% volume of the PCR product was directly used for second-round nested PCR that was carried out with Splinkerette primer HMSp2 and the PB transposon primers PB5′-2 or PB3′-2. The nested PCR products were purified by High Pure 96 UF cleanup Plate (Roche) and were used for sequencing with the primer PB5′-seq or PB3′-seq separately. All nucleotide positions are from National Center for Biotechnology Information (NCBI) mouse build 37.
Genomic PCR and reverse transcription PCR
All primers for genomic PCR and reverse transcription–PCR (RT–PCR) are described in Supplementary Table S1.
Karyotyping and Fluorescence in situ hybridisation of metaphase chromosome spreads of ES cells
Metaphase chromosome spreads were prepared from the ES cells cultured on 6-well gelatin-treated plates according to a standard protocol (27). Hybridization was performed by using appropriate labeled mouse whole chromosome paints or a specific DHSV vector probe (27,28).
Adriamycin treatment
Atm+/+ (NN5), Atm+/− and Atm−/− ES cells were plated on γ-irradiated SNL76/7 feeder layer in a series of six-well plates at a density of 600–800 cells/well. Twenty-four hours later, cells were treated with graded concentration of adriamycin (Sigma) for 7 h and then replaced with fresh ES cell medium. The surviving ES cell colonies in each well were counted after 7–9 days of culture.
RESULTS
Copy number-based selection vectors
We considered that the copy number of the mutagenesis vector could be used to discriminate homozygous and heterozygous cells, and thus to select for homozygous mutants independently of phenotype. If the initial mutagenesis is limited to one mutation per cell, then homozygous mutant cells will have two mutations (at allelic positions). We therefore designed reporter constructs that are selectable based on the number of expressed copies in the cell. These constructs contain only one promoter and coding sequence for two different resistance genes. The two genes are expressed mutually exclusively; the expressed gene can be switched by the action of Cre recombinase. This is achieved either by deletion of one of the genes (vector DHSV, Figure 1B) or inversion when the genes are arranged tail-to-tail (vector TNN; Figure 1C). DHSV contains tandemly arranged bsd and puroΔTK genes (29) that confer blasticidin and puromycin resistance, respectively. TNN contains puroΔTK and a neo gene, which confers resistance to the drug G418, in the opposite orientation.
Cells bearing a single copy of such a construct can only be resistant to one drug at a time. However, in the case where cells have two or more copies of the construct, transient treatment with Cre will result in some cells with both forms of the construct, expressing both resistance genes. These cells will be resistant to both drugs. Thus, these constructs are predicted to allow stringent selection against cells with only one copy (i.e. heterozygous mutants) and to recover a subset of cells with two or more copies (homozygous mutants).
Steps to increase the number of mutable loci
As well as the copy number selection cassette, each vector contains mutagenic elements designed to disrupt splicing. The constructs are flanked by TR sequences of the PB transposon for efficient delivery into the genome of mammalian cells. PB is highly active in ES cells and exhibits wide genome coverage (30,31). Moreover, ∼50% of PB insertions occur in genes, making it an ideal mutagenesis vector (32). The DHSV vector contains a conventional selectable gene trap based on the ß-geo gene, which combines G418 resistance with a lacZ reporter (33). Selectable gene trap vectors ensure that each insertion is in a gene, although there is a requirement for this gene to be expressed at the time of mutagenesis. PB appears to have wider genome coverage than the retroviruses used for gene trap mutagenesis—a previous study isolating components of the mismatch repair pathway showed that PB mutagenesis found more known components of the pathway than retroviral mutagenesis (34). As an option to further increase coverage, the TNN vector includes two non-selectable gene trap cassettes. These consist of pairs of exons and splice acceptors, containing multiple premature stop codons in all reading frames that are predicted to cause non-sense-mediated decay of fusion transcripts (35). These were designed to maximize the chance of causing a mutation, given that PB insertions frequently occur in genes, without requiring sustained expression of the gene trap coding sequence.
Homozygous mutants recovered using selection for copy number increase
We applied these constructs to the selection of rare homozygous mutants that arise in cultures of heterozygously mutated Blm-deficient cells. Using transfection conditions in which a PB transposase expression plasmid is cotransfected with a limiting amount of transposon donor plasmid, we obtained hundreds of clonal Blm-deficient mouse ES cell lines. The majority of these contain a single-copy integration of the transposon (Figure 2A). Selection with both drugs at this point did not yield any double-resistant subclones (data not shown). For each cell line, we mapped the transposon integration site by Splinkerette-PCR. After culturing these cell lines for several passages, we could isolate double drug-resistant subclones after providing Cre by transfection of an expression plasmid, transduction with cell-permeable Cre protein or activation of an inducible ERT2-Cre transgene (Figure 2B). Some cell lines produced very large numbers of double-resistant subclones, comparable to the number of cells plated; these lines contained two or more non-allelic transposon insertions (Figure 2C, well 5) and were not analyzed further. Some lines did not yield double-resistant cells. However, for most lines we obtained varying numbers of double-resistant subclones, of the order of one per 102–104 cells plated (Figure 2C). We genotyped these double-resistant subclones to determine whether these represented homozygous mutants. We assessed presence of the wild-type and mutant alleles by PCR using primers designed to flank the transposon integration site, along with a common primer that hybridizes to one transposon terminus and extends outwards (Figure 2D). For some cell lines, all double-resistant subclones showed the expected result for a homozygous mutant—amplification of only the PCR product from the mutant allele, and loss of the wild-type allele (Figure 2E). However, for most cell lines we observed some double-resistant subclones from which a wild-type PCR product could be amplified (Figure 2F), and in some cases all double-resistant subclones retained a wild-type locus as assessed in this PCR assay (Figure 2G).
Figure 2.
Isolation of homozygous mutants by selection for copy number increase. (A) Transposition using limiting amounts of transposon donor plasmid results in most G418 resistant subclones having one transposon per cell. (B) Selection scheme, showing predicted number and type of transposon present at each stage. (C) Typical result of double-selection stage. Well 5 has two non-allelic transposon integrations to begin with and thus all cells can become double resistant. (D) Genotyping scheme—primer positions (1–3) are shown relative to the transposon integration site. (E) All double-resistant subclones (derived from an integration in Dhx35) fail to amplify the wild-type PCR product. (F and G) Examples of double-resistant populations in which some (F) and all (G) subclones have a wild-type allele.
Double-resistant clones that retain a wild-type locus are aneuploid
Further investigation of these subclones that retained the wild-type locus by Southern blot revealed that although there were two allelic copies of the transposon present, these were accompanied by one or more extra wild-type chromosomes. In some cases, the cells had a tetraploid karyotype (Figure 3A–D), while in others only the chromosome with the insertion site was in excess (Figure 3E). These data show that LOH is not the only pathway for copy number increase in Blm-deficient cells, and that acquisition of aneuploidy is the major alternative pathway. Across all cell lines that we investigated, the average proportion of genuine homozygous mutant subclones isolated was 26% (Table 1). As the rate of LOH is low even in Blm-deficient ES cells (4.2 × 10−4 events/locus/cell/division), this represents a 25- to 250-fold enrichment for homozygous mutants, which would be expected to form only 0.1–1% of the culture prior to double selection
Figure 3.
Abnormal karyotype of clones that retain a wild-type allele. (A) Structure of Myo10 locus analyzed in (B–D). (B) PCR genotyping as in Figure 2D. All double-resistant subclones retain the wild-type allele. (C) All these subclones have a single-transposon integration site (top) but contain both pre- (In: insertion) and post-Cre (Del: deleted) forms of the transposon (bottom). (D) Spectral karyotype showing tetraploidy in a double-resistant subclone. (E) An example of a less severe aberration—only chromosome 6, which bears the insertion at the Grid2 locus, is present in excess. Left, FISH showing three copies of chromosome 6 with (white arrows) and without (yellow arrow) transposon insertions (red dots). Right, spectral karyotype.
Table 1.
Number and proportion of homozygous mutant clones isolated using each vector
| Vector | Clones analyzed | Clones with homozygotes | Clones with homozygotes (%) | Mean hom : het ratio in subclones (%)a |
|---|---|---|---|---|
| DHSV | 89 | 36 | 40 | 23 |
| TNN | 19 | 12 | 63 | 34 |
| Combined | 108 | 48 | 44 | 26 |
aIncludes clones for which no homozygous subclones were isolated.
LOH rate at different loci
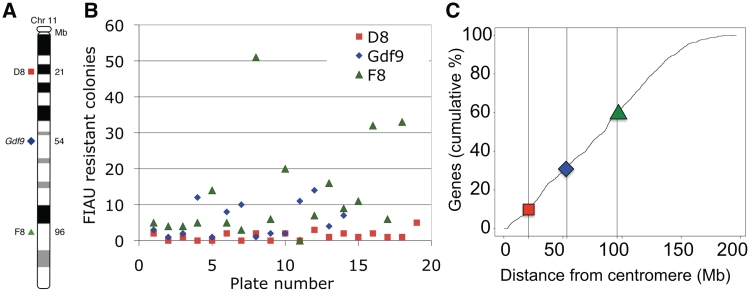
As the process of LOH depends on a recombination event between the locus in question and the centromere, it is possible that it would be difficult to obtain homozygous mutants for loci close to the centromere. To address this, we measured the rate of LOH at three different loci on chromosome 11, one of which was a targeted insertion of a PGK-puroΔTK cassette at the Gdf9 locus, where previous measurements of the LOH rate in Blm-deficient ES cells have been made (15). We isolated other chromosome 11 insertions of TNN in the puro orientation (Figure 4A). In all of these cases, the wild-type cell that results from crossover and loss of the puroΔTK cassette can be isolated by selection in 1-(2-deoxy-2-fluoro-1-d-arabinofuranosyl)-5-iodouracil; 200 nM (FIAU). For each locus we set up replicate cultures starting from a single cell, allowed them to expand to 7.5 × 105 cells and selected cells with LOH events in FIAU (Figure 4B). The rate of LOH was calculated by the fluctuation analysis using the method of medians (36), and did show an increase with distance from the centromere (Table 2). The rates were lower than previously measured using HPRT for negative selection, even for Gdf9 (previously measured as 4.2 × 10−4). This may be related to the use of ΔTK for selection in this case. We found that the LOH rate varied by a factor of four over the 20–96 Mb range tested. As 92% of genes lie >20 Mb from the centromere [Figure 4C, ref. (37)], the probability of crossover is not likely to limit the recovery of homozygous mutants for the vast majority of loci.
Figure 4.
Measurement of LOH rate at different loci. (A) Position of loci investigated on chromosome 11. (B) Number of FIAU resistant colonies obtained (one-tenth of each culture was plated). Data are shown in the original random order, and the rate calculation shown in Table 2. (C) Cumulative frequency plot of all Vega-curated gene start sites (37). The positions of the loci investigated here are marked.
Table 2.
Calculation of the rate of LOH at three loci on chromosome 11 by fluctuation analysis
| Locus | Mb | Cultures | Median FIAUR/culture | Plating efficiency | Mean cells/culture | LOH events/culture (mh)a | LOH rate (10−5 events/cell/division) |
|---|---|---|---|---|---|---|---|
| D8 | 21 | 19 | 1 | 0.35 | 753 000 | 7.53 | 0.99 |
| Gdf9 | 54 | 14 | 3.5 | 0.38 | 762 000 | 18.56 | 2.5 |
| F8 | 96 | 18 | 6.5 | 0.27 | 739 000 | 40.95 | 5.5 |
aSee ref. (34) for details of calculation. FIAUR, FIAU-resistant colonies.
The mutagenesis vectors cause null mutations
Homozygotes were readily isolated using both the deletion (DHSV) and inversion (TNN) vectors. For one insertion of DHSV in the Atm gene (Figure 5A), which encodes the ataxia telangiectasia-mutated kinase involved in the DNA damage response (38,39), we obtained three homozygous subclones after double selection (Figure 5B). Although, the vector was oriented with the splice acceptor in the opposing direction, and is therefore trapping another promoter in this instance, Atm transcription was effectively disrupted (Figure 5C). Mutant subclones showed sensitivity to the DNA damaging drug adriamycin, as expected for an Atm null mutant. In all other cases that we tested, both the DHSV and TNN vectors effectively disrupted transcription of genes that they were inserted into as assessed by RT–PCR (Figure 6). These data show that mutations generated with DHSV or TNN cause null mutations and therefore can be used to screen non-selectable phenotypes.
Figure 5.
Insertion of DHSV at Atm causes DNA damage sensitivity. (A) Structure of mutant Atm locus. (B) Triple primer PCR genotyping using primers a, b and c shown in (A). Clones 2, 3 and 5 lack the wild-type band, consistent with being homozygous mutants. (C) RT–PCR showing loss of Atm transcript. Internal primers to LacZ were included as a positive control. (D) Atm mutants are hypersensitive to adriamycin. Mean surviving fraction (colonies) is plotted. NN5, n =2; Atm−/−, n = 3. Error bars show SEM.
Figure 6.
Robust disruption of transcription in homozygous mutants. (A) PCR genotyping of DHSV insertion in Ddef2 using primers for mutant (Junction, left) and wild-type alleles. All double-resistant subclones are homozygous mutants. (B) RT–PCR showing amplification of fusion transcript (left) and loss of wild-type transcript (right) in double-resistant subclones. (C and D) Insertions of the TNN vector also disrupt transcription. PCR and RT–PCR genotyping is shown for insertions in Dym (C) and Arrb2 (D). Asterisks indicate homozygous mutant subclones, all of which have lost wild-type transcript expression. Minus indicates no template control, Actb is an internal control.
DISCUSSION
We have developed a method for enriching libraries of Blm-deficient ES cells carrying single-copy transposon mutations for homozygous mutants. The method is based on selection for two copies of the transposon construct in the rare homozygous mutants, which is achieved by a switchable drug selection cassette. Using our method of single-copy mutagenesis, clonal expansion, cassette switching by Cre recombinase and selection for two copies, we obtained double-drug resistant populations enriched in homozygous mutants. The proportion of homozygous mutants increased from a predicted 0.1% in the expanded unselected population to a measured average of 26%, depending on the vector used, in the double-resistant population. This increased proportion means that homozygous mutants can be obtained by picking relatively small numbers of subclones. It is possible that this number is an underestimate where clones were genotyped by absence of wild-type locus by PCR, as this may be very sensitive to contamination either between clones or from wild-type feeder cells.
Genotyping of subclones from the double-drug resistant populations revealed two classes of mutants. We found that while some subclones had lost the wild-type locus, consistent with a homozygous mutant, others retained a wild-type locus that could be amplified by PCR. Further analysis of some of these clones revealed numerical chromosome abnormalities, either genome wide (tetraploidy) or affecting the chromosome bearing the transposon insertion (trisomy). We conclude that our method faithfully selects for two copies of the construct, and there are two routes by which Blm cells can increase mutation copy number—the predicted LOH pathway and by numerical chromosome instability (Figure 7). Often we isolated similar numbers of these two classes of double-resistant subclone, suggesting that each of these events occurs at a similar low rate. In some cases, this may be exacerbated by a growth advantage linked to trisomy, particularly for chromosomes 8, 11 and 16 (40–42). However, we did successfully isolate homozygous mutants from insertions on these chromosomes.
Figure 7.
Pathways for copy number gain in Blm-deficient cells. Most cell divisions do not result in copy number increase (left). The two pathways of mitotic recombination (middle) and aneuploidy acquisition (right) result in copy number gain and contribute to the double-resistant population.
The system of selection for copy number increase in Blm-deficient ES cells could be applied more broadly to obtain large collections of mutants for study, for example in genetic screens. Using the procedure described above, we isolated homozygous mutants for 48 out of 108 cell lines tested (Table 1 and Supplementary Tables S1 and S2). We screened relatively small numbers of subclones (usually six or fewer); therefore it is possible that homozygous mutants could be isolated for some of the remaining 60 cell lines by screening further subclones.
We were unable to isolate homozygous mutants for some clones, either because no double-resistant cells were obtained or because all double-resistant subclones analyzed retained the wild-type locus. This could be simply because the events leading to double resistance are rare and may not occur in every expanded clone, or because only a chromosome instability event and not an LOH event occurs. Another explanation could be that the gene mutated is essential for ES cell survival or growth, and therefore homozygous cells do not survive or are at a growth disadvantage compared with wild-type retaining cells. This is likely to be the case for some clones that we tested, for example Faf1, a mutant of which has been reported to cause early embryonic lethality and for which no homozygotes were isolated using our system (43). It should also be noted that when the DHSV vector is used, double-resistant cells may not be recovered if Cre is very efficient and causes deletion in both copies of the resistant cassette. In the case of TNN, the theoretical chance of isolating a cell with two copies is 50% if the Cre reaction reaches an effective equilibrium—this represents the probability that both copies are in different orientations. However, we did not notice an overall difference in the number of double-resistant clones isolated with each vector in practice.
The Blm mutant alleles used here are constitutive, rather than inducible, mutations. Since Blm is a key regulator of homologous recombination (44), there is cause for concern that a constitutive mutation may result in ongoing genome instability. We have not observed high levels of gross chromosomal instability, for example genuine homozygotes isolated from cell lines that also gave aneuploid subclones usually have a normal diploid karyotype (at the microscopic level). An inducible Blm allele (17) is an obvious potential improvement to the system, and a similar system has been developed using such an allele (45). Even in this case, Blm function must be still disrupted to allow LOH, hence an inducible allele will not improve genome stability during the homozygote isolation process, although it would allow for formal separation of the effects of the transposon-induced mutations from the Blm-deficient genetic background.
Even better validation of mutations could come from resources of targeted mouse ES cell knockouts and targeting vectors, which are increasing in size to the point where obtaining a heterozygous mutant in a gene of interest will become trivial. These ES cell lines can be used to generate homozygous mutants after disruption of the remaining wild-type allele using the vectors provided (14,46). Given the ease of obtaining mutants from the public resources in a low-passage cell line with a ‘clean’ genetic background, this represents the best option for further functional study. Mammalian screening methods such as the one we describe here are an excellent complement to these resources, as they provide rapid functional information to prioritize study of the available mutants.
In conclusion, we describe a system using PB transposon constructs that cause null mutations with wide genome coverage in mouse ES cells that are a versatile model mammalian cell type. The mutations generated are stable, but reversion can be induced and selected for by a second round of transposition to allow phenotype rescue experiments (26). This gives a robust link between genotype and phenotype, representing a key advantage compared to RNAi-based screens where causally linking the phenotype to the targeted gene can be problematic. Moreover, other mutations or reporter constructs can easily be introduced into the Blm-deficient cells to create customized libraries for more sophisticated screens. By providing easy access to elusive homozygous mammalian mutant cells, our method significantly expands the scope of genetic screens and future functional studies in mammalian cells.
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3.
FUNDING
Wellcome Trust (79643 to A.B.); Royal Society (to Y.H.); National Natural Science Foundation of China (30971670 to Y.H. and G.L.); National Basic Research Program of China (2011CB965203 to Y.H. and G.L.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Xiaozhong Wang (Northwestern University) for providing the PB transposon vector and PB transposase expression plasmid (pCAG-PBase); Pentao Liu and Leopold Parts for comments on this manuscript; Qi Liang, Frances Law and Hiroko Yusa for help with tissue culture.
REFERENCES
- 1.Grimm S. The art and design of genetic screens: mammalian culture cells. Nat. Rev. Genet. 2004;5:179–189. doi: 10.1038/nrg1291. [DOI] [PubMed] [Google Scholar]
- 2.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 3.Goldfarb M, Shimizu K, Perucho M, Wigler M. Isolation and preliminary characterization of a human transforming gene from T24 bladder carcinoma cells. Nature. 1982;296:404–409. doi: 10.1038/296404a0. [DOI] [PubMed] [Google Scholar]
- 4.St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 2002;3:176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- 5.Jorgensen EM, Mango SE. The art and design of genetic screens: Caenorhabditis elegans. Nat. Rev. Genet. 2002;3:356–369. doi: 10.1038/nrg794. [DOI] [PubMed] [Google Scholar]
- 6.Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, Smithies O. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 7.Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 8.Kotecki M, Reddy PS, Cochran BH. Isolation and characterization of a near-haploid human cell line. Exp. Cell Res. 1999;252:273–280. doi: 10.1006/excr.1999.4656. [DOI] [PubMed] [Google Scholar]
- 9.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leeb M, Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature. 2011;479:131–134. doi: 10.1038/nature10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- 12.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 13.Bushman FD, Malani N, Fernandes J, D'Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, König R, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathogens. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 16.Wang W, Bradley A. A recessive genetic screen for host factors required for retroviral infection in a library of insertionally mutated Blm-deficient embryonic stem cells. Genome Biol. 2007;8:R48. doi: 10.1186/gb-2007-8-4-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yusa K, Horie K, Kondoh G, Kouno M, Maeda Y, Kinoshita T, Takeda J. Genome-wide phenotype analysis in ES cells by regulated disruption of Bloom's syndrome gene. Nature. 2004;429:896–899. doi: 10.1038/nature02646. [DOI] [PubMed] [Google Scholar]
- 18.Guo G, Wang W, Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–895. doi: 10.1038/nature02653. [DOI] [PubMed] [Google Scholar]
- 19.Trombly MI, Su H, Wang X. A genetic screen for components of the mammalian RNA interference pathway in Bloom-deficient mouse embryonic stem cells. Nucleic Acids Res. 2009;37:e34. doi: 10.1093/nar/gkp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo G, Huang Y, Humphreys P, Wang X, Smith A. A PiggyBac-based recessive screening method to identify pluripotency regulators. PLoS One. 2011;6:e18189. doi: 10.1371/journal.pone.0018189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraser MJ, Ciszczon T, Elick T, Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, Liu P, Bradley A. Inducible gene trapping with drug-selectable markers and Cre/loxP to identify developmentally regulated genes. Mol. Cell Biol. 2004;24:9930–9941. doi: 10.1128/MCB.24.22.9930-9941.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joshi SK, Hashimoto K, Koni PA. Induced DNA recombination by Cre recombinase protein transduction. Genesis. 2002;33:48–54. doi: 10.1002/gene.10089. [DOI] [PubMed] [Google Scholar]
- 24.Peitz M, Pfannkuche K, Rajewsky K, Edenhofer F. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc. Natl Acad. Sci. USA. 2002;99:4489–4494. doi: 10.1073/pnas.032068699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li MA, Pettitt SJ, Yusa K, Bradley A. Genome-Wide Forward Genetic Screens in Mouse ES Cells. Methods Enzymol. 2010;477:217–242. doi: 10.1016/S0076-6879(10)77012-9. [DOI] [PubMed] [Google Scholar]
- 27.Hernandez D, Phillips IR, Shephard E. In: Methods in Molecular Biology. Phillips IR, Shephard E, editors. Vol. 320. Totwa, NJ: Humana Press; 2006. pp. 321–327. [Google Scholar]
- 28.Rabbitts P, Impey H, Heppell-Parton A, Langford C, Tease C, Lowe N, Bailey D, Ferguson-Smith M, Carter NP. Chromosome specific paints from a high resolution flow karyotype of the mouse. Nat. Genet. 1995;9:369–375. doi: 10.1038/ng0495-369. [DOI] [PubMed] [Google Scholar]
- 29.Chen YT, Bradley A. A new positive/negative selectable marker, puDeltatk, for use in embryonic stem cells. Genesis. 2000;28:31–35. doi: 10.1002/1526-968x(200009)28:1<31::aid-gene40>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 30.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, Wang X, Bradley A, Liu P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang Q, Kong J, Stalker J, Bradley A. Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis. 2009;47:404–408. doi: 10.1002/dvg.20508. [DOI] [PubMed] [Google Scholar]
- 33.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 34.Wang W, Bradley A, Huang Y. A piggyBac transposon-based genome-wide library of insertionally mutated Blm-deficient murine ES cells. Genome Res. 2009;19:667–673. doi: 10.1101/gr.085621.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 36.Jones ME, Thomas SM, Rogers A. Luria-Delbrück fluctuation experiments: design and analysis. Genetics. 1994;136:1209–1216. doi: 10.1093/genetics/136.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 39.Xu Y, Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- 40.Sugawara A, Goto K, Sotomaru Y, Sofuni T, Ito T. Current status of chromosomal abnormalities in mouse embryonic stem cell lines used in Japan. Comp. Med. 2006;56:31–34. [PubMed] [Google Scholar]
- 41.Liu X, Wu H, Loring J, Hormuzdi S, Disteche CM, Bornstein P, Jaenisch R. Trisomy eight in ES cells is a common potential problem in gene targeting and interferes with germ line transmission. Dev. Dyn. 1997;209:85–91. doi: 10.1002/(SICI)1097-0177(199705)209:1<85::AID-AJA8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 42.Liang Q, Conte N, Skarnes WC, Bradley A. Extensive genomic copy number variation in embryonic stem cells. Proc. Natl Acad. Sci. USA. 2008;105:17453–17456. doi: 10.1073/pnas.0805638105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adham IM, Khulan J, Held T, Schmidt B, Meyer BI, Meinhardt A, Engel W. Fas-associated factor (FAF1) is required for the early cleavage-stages of mouse embryo. Mol. Hum. Reprod. 2008;14:207–213. doi: 10.1093/molehr/gan009. [DOI] [PubMed] [Google Scholar]
- 44.Hickson I. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 45.Horie K, Kokubu C, Yoshida J, Akagi K, Isotani A, Oshitani A, Yusa K, Ikeda R, Huang Y, Bradley A, et al. A homozygous mutant embryonic stem cell bank applicable for phenotype-driven genetic screening. Nature Methods. 2011 doi: 10.1038/nmeth.1739. 23 October (doi:10.1038/nmeth.1739; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 46.Tate PH, Skarnes WC. Bi-allelic gene targeting in mouse embryonic stem cells. Methods. 2011;53:331–338. doi: 10.1016/j.ymeth.2010.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.