Lysine methylation has primarily been described as a nuclear protein modification. Donlin et al. now identify the cytoplasmic protein chaperone Hsp90 as a target of lysine methylation by Smyd2. Hsp90 methylation in muscle cell cytoplasm contributes to the formation of a complex consisting of Smyd2, Hsp90, and the sarcomeric protein titin. This work describes the role of Smyd2 and Hsp90 in myofilament organization and muscle function.
Keywords: Smyd2, Hsp90, titin, lysine methylation, sarcomere, I-band
Abstract
Protein lysine methylation is one of the most widespread post-translational modifications in the nuclei of eukaryotic cells. Methylated lysines on histones and nonhistone proteins promote the formation of protein complexes that control gene expression and DNA replication and repair. In the cytoplasm, however, the role of lysine methylation in protein complex formation is not well established. Here we report that the cytoplasmic protein chaperone Hsp90 is methylated by the lysine methyltransferase Smyd2 in various cell types. In muscle, Hsp90 methylation contributes to the formation of a protein complex containing Smyd2, Hsp90, and the sarcomeric protein titin. Deficiency in Smyd2 results in the loss of Hsp90 methylation, impaired titin stability, and altered muscle function. Collectively, our data reveal a cytoplasmic protein network that employs lysine methylation for the maintenance and function of skeletal muscle.
Post-translational lysine methylation of histones plays a key role in the formation of chromatin-bound protein complexes that regulate a vast range of genome-related processes (Jenuwein and Allis 2001; Kouzarides 2007). Lysine methylation has unique characteristics that contribute to the distinct functional properties of methyl-lysine-dependent protein networks. In comparison with other modifications, such as phosphorylation and acetylation, lysine methylation is considered a stable mark (Byvoet et al. 1972; Duerre and Lee 1974; Rice and Allis 2001). In addition to its relative stability, the methyl mark can be found in one of three different forms. On histones, mono-, di-, or tri-methylation of individual lysines directs the association of functionally distinct chromatin proteins (Kouzarides 2007). In addition to histones, nonhistone proteins in the nucleus, including p53, G9a, and NF-κB, have recently been demonstrated to incur lysine methylation (Chuikov et al. 2004; Huang et al. 2006; Sampath et al. 2007; Ea and Baltimore 2009; Yang et al. 2009; Levy et al. 2010; Lu et al. 2010). Similar to histones, lysine methylation of nonhistone nuclear proteins contributes to formation of protein complexes (Sims and Reinberg 2008).
The role of lysine methylation in nuclear protein complex formation raises the question of whether methyl-lysine-dependent protein networks exist in the cytoplasm. The presence of the lysine methyltransferase (KMTase) Ezh2 in the cytoplasm and its role in cell surface receptor signaling have implicated a potential role for lysine methylation in the cytoplasm (Su et al. 2005). However, the lack of information regarding an Ezh2 cytoplasmic substrate has precluded further understanding of how lysine methylation impacts cytoplasmic pathways.
In the current study, we demonstrate that the KMTase Smyd2, which localizes predominantly to the cytoplasm, methylates the chaperone Hsp90. We identify a specific site for Hsp90 lysine methylation and show that methylation of this site critically depends on Smyd2. We provide evidence that Smyd2 and methylated Hsp90 form a complex with the sarcomeric protein titin and that Smyd2 is required for normal function of skeletal muscle in vivo.
Results and Discussion
The Smyd2 KMTase localizes predominantly to the cytoplasm
Our search for putative cytoplasmic substrates of lysine methylation began with in silico and experimental screens for cytoplasmic KMTases. We reasoned that identification of cytoplasmic KMTases could lead to the discovery of cytoplasmic networks regulated by lysine methylation. The in silico screening of 40 KMTases showed that 11 contain a nuclear export signal (NES) and lack a nuclear localization signal (NLS), suggesting the potential for nonnuclear localization (Supplemental Table 1; Supplemental Fig. 1A). These 11 KMTases were tagged with GFP and expressed in HEK 293 cells. We found that the enzymes Smyd2, Smyd4, and Set8/PR-Set7 predominantly localized to the cytoplasm (Supplemental Fig. 1A,B). Furthermore, endogenous Smyd2 was also found in the cytoplasm of HEK 293 and C2C12 myoblast cells, as well as adult human skeletal muscle tissue (Supplemental Fig. 1C–E), which expresses high levels of Smyd2 in comparison with other tissues (Brown et al. 2006; Kawamura et al. 2008; Sun et al. 2008).
Smyd2 methylates the cytoplasmic chaperone Hsp90
The high abundance of Smyd2 in the cytoplasm and its ability to methylate the structurally distinct nonhistone nuclear proteins p53 and Rb (Huang et al. 2006; Saddic et al. 2010) rendered Smyd2 a favorable candidate to catalyze lysine methylation of cytosolic proteins. We therefore developed an approach to screen for cytosolic substrates of Smyd2. Cytosolic extracts were fractionated based on molecular weight, and each fraction was then incubated with recombinant Smyd2 and radiolabeled cofactor S-adenosylmethionine (SAM). Using this method, we detected several Smyd2-methylated protein species that were then identified by MS/MS (Supplemental Fig. 2A,B). The most abundant Smyd2-methylated protein was found to be the chaperone Hsp90 (Fig. 1A; Supplemental Fig. 2B). Selectivity of Smyd2 for Hsp90 methylation was demonstrated by the inability of Smyd2 to methylate the Hsp70 chaperone or the endoplasmic reticular Hsp90 homolog gp96 (Fig. 1A; Supplemental Fig. 2C). Furthermore, the methyltransferases G9a and Set9, which have the capacity to methylate both histone and nonhistone proteins, failed to methylate Hsp90 in vitro (Supplemental Fig. 2D).
Figure 1.
Smyd2 methylates the Hsp90 chaperone. (A) Methyltransferase assay containing purified Hsp70, Hsp90, or histones and the radiolabeled cofactor SAM performed in the presence or absence of Smyd2. (B) Methyltransferase assays containing Smyd2 and C-terminal Hsp90 lysine-to-alanine (K–A) point mutants. Wild-type (Wt) and a dimerization domain-deleted (ΔDIM) Hsp90 were used as positive and negative controls, respectively. (His-F) His-Flag-tagged Hsp90 mutants; (e) endogenous copurifying Hsp90. (C) Whole-cell lysates from U20S and C2C12 cells stably integrated with Smyd2-specific shRNAs (sh1 and sh2) or two control vectors (empty vector [shE] and nonsilencing shRNA [shN]) analyzed by Western blot for methyl Hsp90 (anti-Hsp90K616me1 antibody), Hsp90, and Smyd2. HEK 293 cells with or without exogenous Smyd2 expression were used as positive and partial negative controls for methylated Hsp90, respectively. (D) Mouse tissue lysates analyzed by Western blot for methyl Hsp90, Hsp90, and Smyd2. (Br) Brain; (Thy) thymus; (Ht) heart; (Lu) lung; (Kid) kidney; (Spl) spleen; (SkM) skeletal muscle; (Liv) liver.
Using a panel of truncated Hsp90 proteins, as well as Hsp90 proteins in which individual lysine residues were replaced by alanine, we identified Lys 616 (K616) as the substrate for Smyd2 (Fig. 1B; Supplemental Fig. 3A,B). A mass spectrometry analysis of Hsp90 purified from HEK 293 cells overexpressing Smyd2 not only confirmed that K616 represents a Smyd2 substrate, but also demonstrated that, in vivo, a single methyl group is covalently attached to K616, rendering it monomethyl K616 (K616me1) (Supplemental Fig. 3C). K616 and the surrounding α-helix motif are highly conserved among Hsp90 homologs from mammals to yeast (Supplemental Fig. 3D). Using a custom antibody specific for monomethyl K616 (anti-Hsp90K616me1) (Supplemental Fig. 3E), endogenous Hsp90K616me1 was detected in various cell lines and ex vivo isolated cells (Fig. 1C,D). In particular, ex vivo isolated muscle tissues, which express the highest level of Smyd2 mRNA (Brown et al. 2006; Kawamura et al. 2008; Sun et al. 2008) and protein (Fig. 1D), also contained the highest levels of endogenous Hsp90K616me1 (Fig. 1D). Multiple lines of evidence support an essential role for Smyd2 in the generation of Hsp90K616me1. First, methylation of Hsp90 was detected with the methyl Hsp90 antibody upon ectopic expression of Smyd2, but not the four other Smyd2 family members (Supplemental Fig. 3F). Second, Hsp90K616me1 was undetectable in cells rendered Smyd2-deficient by expression of Smyd2-specific shRNAs (Fig. 1C).
Smyd2 and methylated Hsp90 colocalize on myofibrils
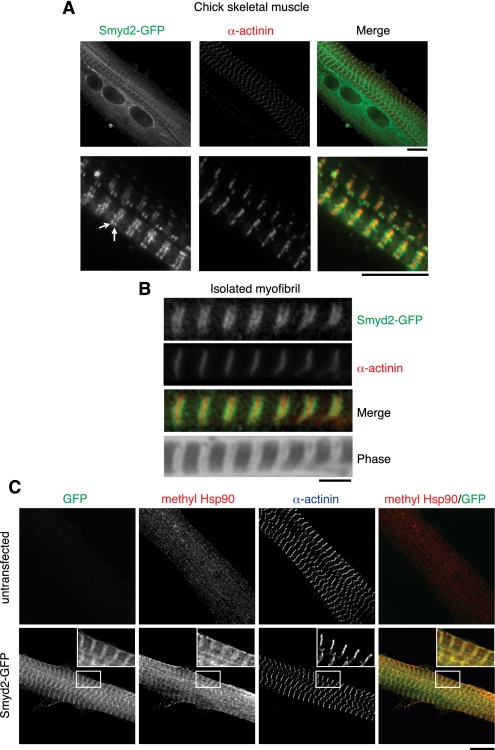
The abundance of Smyd2 and Hsp90K616me1 in striated muscle suggests that methylation of Hsp90 by Smyd2 may have a function specific to myocytes. A GFP-tagged Smyd2 protein expressed in primary chick skeletal myocytes demonstrated a highly ordered striated arrangement, suggestive of myofibril binding (Fig. 2A). Specifically, Smyd2 assembled as tightly spaced double bands flanking α-actinin striations that mark the sarcomeric Z-disks (Fig. 2A). Endogenous Smyd2 was also found in a similar pattern in human diaphragm muscle (Supplemental Fig. 4A). Incubation of purified human myofibrils with recombinant Smyd2-GFP protein, but not GFP alone, resulted in striations adjacent to Z-disks (Fig. 2B). These data implicated direct binding between Smyd2 and myofibrils at the sarcomeric region known as the I-band. Hsp90K616me1 colocalized with Smyd2-GFP at the sarcomeric I-band region, indicating that Smyd2 and methylated Hsp90 may function together in skeletal muscle (Fig. 2C).
Figure 2.
Smyd2 and methylated Hsp90 bind to muscle myofibrils. (A) Representative fluorescence images of embryonic chick skeletal myocytes expressing Smyd2-GFP. Arrows indicate a doublet of bands. Z-disks were visualized with an anti-α-actinin antibody. Bar, 10 μm. (B) Immunofluorescence images of isolated and stretched human cardiomyofibrils incubated with recombinant Smyd2-GFP and stained for α-actinin. Bar, 3 μm. (C) Immunofluorescence images of chick skeletal myocytes with or without Smyd2-GFP expression stained for methyl Hsp90 and α-actinin. Bar, 10 μm.
Smyd2 directly binds the sarcomeric protein titin
The main I-band components found immediately adjacent to Z-disks include actin filaments and a portion of the giant myofilament protein titin (Labeit and Kolmerer 1995). While actin and myosin filaments provide the active force for muscle contraction, titin confers elasticity and passive force under stretching (Furst et al. 1988). This function relies on the dynamic titin I-band region that operates as a “molecular spring” by extending and condensing as the muscle stretches and contracts, respectively (Li et al. 2002). Consequently, upon stretching, the titin I-band region extends away from the Z-disk, while other I-band components, such as actin, remain at a fixed distance (Linke et al. 1999; Cazorla et al. 2000). Upon myofibril stretching, we observed an increase in the distance between the Z-disk and Smyd2 striations (Fig. 3A). For any given magnitude of stretching, this distance precisely matched that between the Z-disk and the N2A domain of titin, implying that Smyd2 interacts with this titin domain (Fig. 3A). In support of an interaction, titin N2A striations colocalized with Smyd2-GFP (Supplemental Fig. 4B) and endogenous Smyd2 striations (Fig. 3B) in skeletal myocytes. A direct interaction between Smyd2 and the titin N2A domain was confirmed by yeast two-hybrid and GST pull-down assays, mapping the Smyd2-binding site to a unique sequence encoded by human titin exon 104 (Supplemental Fig. 4C,D). Despite the observed interaction, in vitro methyltransferase assays revealed no methylation of the titin N2A domain by Smyd2 (Supplemental Fig. 4E).
Figure 3.
Colocalization and stretch-induced movement of Smyd2 with the titin N2A domain. (A) Distance from the Z-disk center for both Smyd2-GFP and the titin N2A domain (detected with a titin N2A antibody) in isolated human myofibrils stretched to increasing sarcomere lengths. (Open triangle and gray line) Smyd2-GFP; (solid square and black line) Titin N2A; (F with arrow above) direction of stretch force. Bars, 3 μm. (B) Immunofluorescence images of chick skeletal myocytes stained for endogenous Smyd2, the titin N2A domain, and α-actinin. Bar, 10 μm. (C) Immunofluorescence images of human diaphragm muscle stained for endogenous methylated Hsp90 and the titin N2A domain (9D10 antibody). Bar, 5 μm.
Hsp90, Smyd2, and titin form a protein complex dependent on Hsp90 methylation
Methylated Hsp90 colocalized with the same titin domain where Smyd2 binds (Fig. 3C), suggesting the possibility that Smyd2, methylated Hsp90, and the titin N2A domain are found together within a complex. Supporting this hypothesis, we found near-stoichiometric quantities of Hsp90 associated with overexpressed Smyd2 (Fig. 4A), as well as an interaction between endogenously expressed Smyd2 and Hsp90 (Fig. 4B), in C2C12 myoblasts. Furthermore, an exogenously expressed titin N2A domain associated with the Smyd2–Hsp90 complex in C2C12 cells. This association was identified under conditions in which Hsp90 or Smyd2 was immunoprecipitated, demonstrating the existence of complexes containing all three proteins (Fig. 4C; Supplemental Fig. 4F, respectively). The association between Hsp90 and the titin N2A domain existed only in the presence of Smyd2 (Fig. 4C), demonstrating that Smyd2 is required for the Hsp90–titin interaction. In the absence of Smyd2-mediated methylation, the Smyd2–Hsp90 interaction was maintained, while titin binding to Hsp90 complexes was greatly reduced (Fig. 4C). These data suggest that Smyd2-mediated methylation of the chaperone Hsp90 promotes formation of a complex containing titin.
Figure 4.
Hsp90 associates with and stabilizes titin upon Smyd2-mediated methylation. (A) Flag immunoprecipitations (IP) from C2C12 myoblasts stably expressing Flag-tagged Smyd2 or containing an empty vector cassette (EV) analyzed by a Coomassie-stained gel. Proteins specifically associating with Smyd2 were identified by mass spectrometry (Mass Spec ID). (IgL) Immunoglobulin light chain; (IgH) immunoglobulin heavy chain. (B) Endogenous Smyd2 immunoprecipitations (IP) and whole-cell extracts (WCE) from control (shE) and Smyd2-shRNA expressing (sh1) C2C12 cells analyzed by Western blot for endogenous Hsp90β and Smyd2. The vertical line demarcates different exposure times for the same antibody. (C) Lysates and Flag immunoprecipitations from C2C12 myoblast cells transfected with Flag-tagged wild-type (Wt) and lysine point mutant (Kmt) Hsp90, and HA-tagged wild type (Wt) and enzymatic dead (ED) Smyd2, along with HA-tagged titin N2A and analyzed by Western blot for HA, methyl Hsp90, and Flag. All images were derived from the same blot, with vertical lines demarcating different exposure times for the same antibody. (D) Lysates from C2C12 myoblasts transfected with equal levels of titin N2A domain and increasing amounts of either wild-type or enzymatic dead (ED) Smyd2 cDNAs analyzed as in C. (E) Chicken skeletal myocytes transfected with control or Smyd2 siRNAs stained for α-actinin, titin N2A domain, and titin M-band. (Inset) RT–PCR of myocyte cDNA for Smyd2 and GAPDH. Bars, 10 μm.
Smyd2-mediated Hsp90 methylation impacts titin protein stability
Hsp90 has been shown to be critical in myofibril organization and function (Barral et al. 2002; Du et al. 2008; Hawkins et al. 2008; Codina et al. 2010). Although much of the evidence is related to its association with myosin, Hsp90 has also been implicated in titin function (Hawkins et al. 2008; Codina et al. 2010). Having observed that methylation of K616 is necessary for the interaction of Hsp90 with titin, we noted that this lysine lies within a putative chaperone–substrate interface (Harris et al. 2004; Fang et al. 2006). If titin represents a substrate of the chaperone Hsp90, one would predict that the interaction between methylated Hsp90 and titin would influence titin protein stability. C2C12 myoblasts expressing constant levels of titin N2A and increasing concentrations of Smyd2 cDNA showed increased methylation of endogenous Hsp90 and increasing concentrations of titin N2A protein (Fig. 4D). This finding pointed to a positive role for Smyd2 in the regulation of titin stability. Expression of an enzymatically inactive Smyd2 resulted in decreased concentrations of titin N2A protein (Fig. 4D), indicating compromised titin protein stability. These data suggest that the enzymatic capacity of Smyd2 to methylate Hsp90 impacts titin protein stability. In support of this concept, we found that reduced Smyd2 expression negatively affected the structural integrity of the titin N2A domain. Treatment of myocytes with the Smyd2-specific siRNA resulted in a diminished striated staining pattern for the titin N2A domain (59% of cells with Smyd2 siRNA and 31% of cells with control siRNAs; n = 100, P = 6 × 10−9), while α-actinin striations appeared normal (Fig. 4E). Reduced Smyd2 expression did not affect all regions of the giant titin filament (>1 μm) or induce titin protein degradation, as demonstrated by the wild-type titin patterning within the sarcomeric M-band region (Fig. 4E).
Smyd2 promotes stabilization of the sarcomeric I-band region
Our findings pointed to a potentially important role for Smyd2 in the regulation of muscle structure and function. Indeed, we found that reduced Smyd2 and methylated Hsp90 levels in zebrafish associated with skeletal and cardiac muscle defects (Fig. 5A). Zebrafish with reduced Smyd2 expression displayed severely impaired mobility and contracted tails (Fig. 5B,C). The impact on cardiac muscle performance was less severe, with reduced heart rates and fractional shortening decreased by ∼50% (data not shown). In an independent study on mouse cardiac development, Smyd2 was shown to be dispensable, suggesting a compensatory mechanism for the Smyd2 gene in higher-vertebrate heart development (Diehl et al. 2010). The zebrafish phenotypes were observed upon attenuation of both Smyd2 alleles together (smyd2a and smyd2b) or with reduced smyd2a expression alone (Fig. 5B,C). smyd2a expression was disrupted by two distinct nonoverlapping morpholino oligomers with the same phenotypic outcomes, but with variable penetrance (92.8% of n = 153 and 60% of n = 98). Histological analysis of zebrafish with reduced Smyd2 expression levels revealed disrupted skeletal muscle tissue (Supplemental Fig. 5A). Electron micrographs demonstrated a localized and consistent disorganization of sarcomeric structures in the Z-disk and I-band regions (Fig. 5D; Supplemental Fig. 5B). In contrast, the M-band regions maintained a normal ordered alignment in the absence of Smyd2 (Fig. 5D; Supplemental Fig. 5B). The disordered I-bands and Z-disks in Smyd2-deficient zebrafish indicate that Smyd2 stabilizes the precise sarcomeric region where the Smyd2–Hsp90 complex is found.
Figure 5.
Smyd2 regulates skeletal and cardiac muscle development and sarcomeric I-band structures in zebrafish. (A) Zebrafish lysates from control and smyd2a/b morpholino-treated animals analyzed by Western blot for Smyd2, methyl Hsp90, and Hsp90. (B,C) Zebrafish injected with control or smyd2 antisense morpholino oligonucleotides. smyd2a and smyd2b were targeted in B, and only smyd2a was targeted in C. Images represent fish at 72 h post-fertilization. (D) Transmission electron microscopy of zebrafish skeletal muscle. (Z) Z-disk; (M) M-band. Bars: main panels, 1 μm; insets, 0.1 μm. Graph represents a blind scoring for the structural integrity of I-band/Z-disk and M-band regions. The percentage of regions with characteristic structural appearance is plotted, with the error bars representing standard error. I-band/Z-disk: n = 42, P = 0.0002; M-band: n = 38, P = 0.3. (***) Statistical significance.
In summary, we identified the cytoplasmic chaperone Hsp90 as a previously unknown substrate for the methyltransferase Smyd2. In muscle, we found that Smyd2-mediated methylation of Hsp90 regulated the formation of a complex with the sarcomeric protein titin. In this context, the formation of complexes around lysine methylation at the titin filament could be viewed as functionally analogous to the formation of complexes around lysine methylation at the chromatin fiber. In vertebrates, several Smyd family KMTases are expressed most highly in muscle cells (Gottlieb et al. 2002; Fujii et al. 2003; Tan et al. 2006; Kawamura et al. 2008; Sun et al. 2008; Thompson and Travers 2008). Thus, it is attractive to speculate that the Smyd family of KMTases will prove to support sarcomeric contractile function through methylation of various muscle proteins.
Materials and methods
Methyltransferase assays
Reactions were performed in methyltransferase buffer (50 mM Tris at pH 8, 5 mM MgCl, 4 mM DTT) plus ∼10 mM 3H-S-adenosylmethionine (GE Healthcare or Perkin Elmer) and incubated for 45 min at 30°C. Protein gels were stained with Coomassie, incubated with liquid EN3HANCE (Perkin-Elmer), incubated in PEG solution (10% polyethylene glycol 8000, 7% methanol, 7% acetic acid, 1% glycerol), dried, and exposed to Biomax XAR film (Kodak) for 2–10 d at −80°C.
Antibodies
The antibodies used were Smyd2 (Sigma), Flag M2 (Sigma), Hsp90 (Stressgen), Hsp90β (Stressmarq), and HA.11 (Covance). The immunofluorescence antibodies used were α-actinin (Sigma), GFP (Abcam), Smyd2 (Eurogentec and Sigma), N2A-titin (Eurogentec), M-band titin (T114, custom-made), and Cy3- or Cy5-conjugated IgGs (Rockland).
Methyl Hsp90 antibody generation
A methyl Hsp90 peptide (NH2-NMERIMKme1AQALRDC) was synthesized by the Rockefeller University Proteomics Resource Center, injected into rabbits (Cocalico Biologicals), and affinity-purified (Open Biosystems).
Primary cultures of chick skeletal and cardiac myocytes
Breast muscle or heart from chicken embryos was minced and trypsinized, and nonadherent cells were plated on Matrigel-coated (BD) coverslips in growth medium (12.5% horse serum [HyClone], 12.5% chicken embryo extract, 5 mM glutamine in MEM [Cellgro]). After 2 d, growth medium was replaced by differentiation medium (12.5% horse serum, 2% chicken embryo extract) (Gregorio and Fowler 1995; Nawrotzki et al. 1995). Transfections were performed using Escort IV (Sigma) or Effectene (Qiagen).
Mechanical manipulation and fluorescence staining of isolated myofibrils
Myofibrils were isolated from human donor hearts, prepared for mechanical measurements as described (Neagoe et al. 2002), and incubated with recombinant Smyd2-GFP (1.5 μg/μL buffer). Distances from the Z-disk were measured under a Zeiss Axiovert-135 microscope in epifluorescence mode (100× objective) using a CCD camera (Sony) and ImageJ software.
Immunostaining of adult human diaphragm muscle
Muscle was fixed in 4% paraformaldehyde, permeabilized (0.5% Triton X-100), repeatedly washed, and incubated with primary and secondary antibodies. Samples were embedded in epoxy, thinly sectioned, and analyzed by confocal laser-scanning microscopy (Nikon, Eclipse Ti).
Smyd2 complex purification from myoblasts
C2C12 myoblast cells stably expressing His-Flag-tagged Smyd2 were lysed (0.1% NP-40, 20 mM HEPES, 100 mM NaCl, 1 mM MgCl2, 20 mM Na2MoO4, 2 mM Na3VO4, 30 mM NaF, protease inhibitor cocktail [Sigma]), incubated with Flag beads (Sigma), washed with increasing amounts of salt (final wash containing 500 mM NaCl), and eluted with Flag peptides (Sigma).
Immunofluorescence microscopy of cultured myocytes
Myocytes were incubated in relaxing buffer (150 mM KCl, 5 mM MgCl2, 10 mM MOPS, at pH 7.4, 1 mM EGTA, 4 mM ATP) and fixed in 2%–3% PFA. Images were captured with a Deltavision RT system (Applied Precision) and an inverted microscope (IX70, Olympus), a 100× objective, and a camera (CoolSNAP HQ; Photometrics) using SoftWoRx 3.5.1 software (Applied Precision).
siRNA Smyd2 silencing and analysis of titin N2A striations
Primary myocytes were transfected (Effectene, Qiagen) with 30 nM siRNAs (Ambion). One-hundred cells were scored for distinguishable N2A striations, and a binomial distribution test was used for statistical analyses.
Zebrafish (Danio rerio) injection procedures and analysis
Morpholino-modified oligonucleotide sequences are listed in the Supplemental Material. Animals were scored positive for skeletal or cardiac muscle defects if they exhibited pericardial edema, elongated hearts, inflow tract edema, reduced heart rate, curved tails, disorganized skeletal muscle fibers, and lack of myoseptum. Fish tail muscle was analyzed by transmission electron microscopy (Zeiss EM900) according to standard protocols. Three individuals blindly scored >30 sarcomeric I-band/Z-disk and M-band regions as either characteristic or disrupted in structure, with a Student's two-tailed t-test used for statistical analyses.
Acknowledgments
We thank I. Berger, J. Krysiak, P. Lang, S. Marquart, C. Li, and Y. Wang for technical work, and R. Chandwani, K. Jeffrey, T. Fang, and W. Obermann for manuscript review. We thank S. Smale for promoting the collaboration between the Tarakhovsky and Linke groups. This work was supported by grants from the Starr Cancer Consortium (13-A136 to A.T), the National Institutes of Health (RO1 AI068058 to A.T, NRSA AI066674 to L.T.D., RR00862 and RR022220 to B.T.C., HL083146 to C.C.G., and NRSA GM076989 to E.Y.J.), and the German Research Foundation (SFB 629 and Li 690/7-2 to W.A.L.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.177758.111.
References
- Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF 2002. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science 295: 669–671 [DOI] [PubMed] [Google Scholar]
- Brown MA, Sims RJ III, Gottlieb PD, Tucker PW 2006. Identification and characterization of Smyd2: A split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer 5: 26 doi: 10.1186/1476-4598-50-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byvoet P, Shepherd GR, Hardin JM, Noland BJ 1972. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys 148: 558–567 [DOI] [PubMed] [Google Scholar]
- Cazorla O, Freiburg A, Helmes M, Centner T, McNabb M, Wu Y, Trombitas K, Labeit S, Granzier H 2000. Differential expression of cardiac titin isoforms and modulation of cellular stiffness. Circ Res 86: 59–67 [DOI] [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, et al. 2004. Regulation of p53 activity through lysine methylation. Nature 432: 353–360 [DOI] [PubMed] [Google Scholar]
- Codina M, Li J, Gutierrez J, Kao JP, Du SJ 2010. Loss of Smyhc1 or Hsp90α1 function results in different effects on myofibril organization in skeletal muscles of zebrafish embryos. PLoS ONE 5: e8416 doi: 10.1371.journal.pone.0008416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Brown MA, van Amerongen MJ, Novoyatleva T, Wietelmann A, Harriss J, Ferrazzi F, Bottger T, Harvey RP, Tucker PW, et al. 2010. Cardiac deletion of Smyd2 is dispensable for mouse heart development. PLoS ONE 5: e9748 doi: 10.1371/journal.pone.0009748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SJ, Li H, Bian Y, Zhong Y 2008. Heat-shock protein 90α1 is required for organized myofibril assembly in skeletal muscles of zebrafish embryos. Proc Natl Acad Sci 105: 554–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerre JA, Lee CT 1974. In vivo methylation and turnover of rat brain histones. J Neurochem 23: 541–547 [DOI] [PubMed] [Google Scholar]
- Ea CK, Baltimore D 2009. Regulation of NF-κB activity through lysine monomethylation of p65. Proc Natl Acad Sci 106: 18972–18977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Ricketson D, Getubig L, Darimont B 2006. Unliganded and hormone-bound glucocorticoid receptors interact with distinct hydrophobic sites in the Hsp90 C-terminal domain. Proc Natl Acad Sci 103: 18487–18492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Tsunesumi S, Yamaguchi K, Watanabe S, Furukawa Y 2003. Smyd3 is required for the development of cardiac and skeletal muscle in zebrafish. PLoS ONE 6: e23491 doi: 10.1371/journal.pone.0023491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furst DO, Osborn M, Nave R, Weber K 1988. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: A map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J Cell Biol 106: 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, et al. 2002. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet 31: 25–32 [DOI] [PubMed] [Google Scholar]
- Gregorio CC, Fowler VM 1995. Mechanisms of thin filament assembly in embryonic chick cardiac myocytes: Tropomodulin requires tropomyosin for assembly. J Cell Biol 129: 683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SF, Shiau AK, Agard DA 2004. The crystal structure of the carboxy-terminal dimerization domain of htpG, the Escherichia coli Hsp90, reveals a potential substrate binding site. Structure 12: 1087–1097 [DOI] [PubMed] [Google Scholar]
- Hawkins TA, Haramis AP, Etard C, Prodromou C, Vaughan CK, Ashworth R, Ray S, Behra M, Holder N, Talbot WS, et al. 2008. The ATPase-dependent chaperoning activity of Hsp90a regulates thick filament formation and integration during skeletal muscle myofibrillogenesis. Development 135: 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL 2006. Repression of p53 activity by Smyd2-mediated methylation. Nature 444: 629–632 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD 2001. Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Kawamura S, Yoshigai E, Kuhara S, Tashiro K 2008. smyd1 and smyd2 are expressed in muscle tissue in Xenopus laevis. Cytotechnology 57: 161–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Labeit S, Kolmerer B 1995. Titins: Giant proteins in charge of muscle ultrastructure and elasticity. Science 270: 293–296 [DOI] [PubMed] [Google Scholar]
- Levy D, Kuo AJ, Chang Y, Schaefer U, Kitson C, Cheung P, Espejo A, Zee BM, Liu CL, Tangsombatvisit S, et al. 2010. Lysine methylation of the NF-κB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-κB signaling. Nat Immunol 12: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Linke WA, Oberhauser AF, Carrion-Vazquez M, Kerkvliet JG, Lu H, Marszalek PE, Fernandez JM 2002. Reverse engineering of the giant muscle protein titin. Nature 418: 998–1002 [DOI] [PubMed] [Google Scholar]
- Linke WA, Rudy DE, Centner T, Gautel M, Witt C, Labeit S, Gregorio CC 1999. I-band titin in cardiac muscle is a three-element molecular spring and is critical for maintaining thin filament structure. J Cell Biol 146: 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, Gudkov AV, Stark GR 2010. Regulation of NF-κB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci 107: 46–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrotzki R, Fischman DA, Mikawa T 1995. Antisense suppression of skeletal muscle myosin light chain-1 biosynthesis impairs myofibrillogenesis in cultured myotubes. J Muscle Res Cell Motil 16: 45–56 [DOI] [PubMed] [Google Scholar]
- Neagoe C, Kulke M, del Monte F, Gwathmey JK, de Tombe PP, Hajjar RJ, Linke WA 2002. Titin isoform switch in ischemic human heart disease. Circulation 106: 1333–1341 [DOI] [PubMed] [Google Scholar]
- Rice JC, Allis CD 2001. Histone methylation versus histone acetylation: New insights into epigenetic regulation. Curr Opin Cell Biol 13: 263–273 [DOI] [PubMed] [Google Scholar]
- Saddic LA, West LE, Aslanian A, Yates JR III, Rubin SM, Gozani O, Sage J 2010. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem 285: 37733–37740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath SC, Marazzi I, Yap KL, Sampath SC, Krutchinsky AN, Mecklenbrauker I, Viale A, Rudensky E, Zhou MM, Chait BT, et al. 2007. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell 27: 596–608 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Reinberg D 2008. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol 9: 815–820 [DOI] [PubMed] [Google Scholar]
- Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wulfing C, Tarakhovsky A 2005. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell 121: 425–436 [DOI] [PubMed] [Google Scholar]
- Sun XJ, Xu PF, Zhou T, Hu M, Fu CT, Zhang Y, Jin Y, Chen Y, Chen SJ, Huang QH, et al. 2008. Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS ONE 3: e1499 doi: 10.1371/journal.pone.0001499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Rotllant J, Li H, De Deyne P, Du SJ 2006. SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc Natl Acad Sci 103: 2713–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EC, Travers AA 2008. A Drosophila Smyd4 homologue is a muscle-specific transcriptional modulator involved in development. PLoS ONE 3: e3008 doi: 10.1371/journal.pone.0003008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XD, Huang B, Li M, Lamb A, Kelleher NL, Chen LF 2009. Negative regulation of NF-κB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J 28: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]