Eukaryotic DNA replication initiates are multiple genomic loci. In this study, Masai and colleagues identify the conserved telomere-binding protein Rif1 as a crucial factor in determining origin firing and timing. Deletion of Rif1, which is found to bind nontelomeric chromosome arms independently of Taz1, leads to extensive deregulation of origin firing. Rif1 is thus identified as an essential regulator of the replication timing program during the cell cycle.
Keywords: replication origin, replication timing, late-firing origin, Cdc7 kinase, telomere-binding protein, ChIP–chip assay
Abstract
One of the long-standing questions in eukaryotic DNA replication is the mechanisms that determine where and when a particular segment of the genome is replicated. Cdc7/Hsk1 is a conserved kinase required for initiation of DNA replication and may affect the site selection and timing of origin firing. We identified rif1Δ, a null mutant of rif1+, a conserved telomere-binding factor, as an efficient bypass mutant of fission yeast hsk1. Extensive deregulation of dormant origins over a wide range of the chromosomes occurs in rif1Δ in the presence or absence of hydroxyurea (HU). At the same time, many early-firing, efficient origins are suppressed or delayed in firing timing in rif1Δ. Rif1 binds not only to telomeres, but also to many specific locations on the arm segments that only partially overlap with the prereplicative complex assembly sites, although Rif1 tends to bind in the vicinity of the late/dormant origins activated in rif1Δ. The binding to the arm segments occurs through M to G1 phase in a manner independent of Taz1 and appears to be essential for the replication timing program during the normal cell cycle. Our data demonstrate that Rif1 is a critical determinant of the origin activation program on the fission yeast chromosomes.
Replication of genomic DNA occurs only once in a cell cycle, and its accuracy ensures genome integrity. As in prokaryotes, eukaryotic DNA replication is also believed to initiate at specific genomic loci called replication origins (ori). Although ori in budding yeast are known to be associated with a specific 11-base-pair (bp) sequence termed ACS (ARS [autonomous replicating sequence] consensus sequence), no such sequence has been identified in ori of other eukaryotes (for review, see Sclafani and Holzen 2007; Masai et al. 2010). However, it is now well established that DNA replication is initiated at selected pre-RC (prereplicative complex)-binding sites (BSs) on the chromosomes, and timing of the firing of these pre-RCs is coordinately regulated during S phase (for review, see Takeda and Dutta 2005). Here, we call the profiles of the sites and timing of origin firing the “replication program.” Recent reports indicate that the replication program is under developmental regulation, just like gene expression (Hiratani and Gilbert 2009). Dramatic changes of the replication program or replication domain structures are observed during induction of differentiation of mouse embryonic stem cells or between different cell types (for review, see Masai et al. 2010). Studies in yeasts showed that chromatin structures (histone modification) near the origins can affect the firing efficiency (Aparicio et al. 2004). The checkpoint also regulates the sites and timing of firing. Dormant or late origins are activated in the absence of checkpoint regulators (Santocanale and Diffley 1998; Shirahige et al. 1998; Matsumoto et al. 2011). The checkpoint represses late origins also in mammalian cells (Aladjem 2007). However, factors that determine the genome-wide replication program or replication domain structures are still elusive. The mechanisms of dramatic reorganization of replication domains during differentiation are not clear.
pre-RC is generated during early G1, and this step may be under chromatin regulation; the timing and efficiency of pre-RC assembly were reported to contribute to early firing of ori in fission yeast (Wu and Nurse 2009). pre-RC is activated by phosphorylation mediated by CDK (cyclin-dependent kinase) and Cdc7–Dbf4 kinase followed by loading of replisome factors, including Cdc45 and DNA polymerases. Phosphorylation of Mcm by CDK and Cdc7–Dbf4 is crucial for initiation of DNA replication and may be the critical step for selection of active origins (Masai et al. 2000, 2006; Walter 2000; Sclafani et al. 2002; Sheu and Stillman 2006; Yabuuchi et al. 2006). However, how Cdc7 phosphorylates the selected pre-RC at different timings during S phase is totally unknown.
We studied the Hsk1 kinase, the fission yeast homolog of Cdc7, and found that deletion of a checkpoint adaptor protein, mrc1, can bypass the requirement of Hsk1 function for viability (Matsumoto et al. 2011). We further showed that firing of some late/dormant origins was deregulated in mrc1Δ, which may contribute to the bypass of hsk1Δ. This finding led to the idea that a search for other hsk1Δ suppressors could enable us to identify novel regulators of the replication program.
Rif1 was originally discovered in budding yeast as a telomer-binding protein, and its orthologs have been identified in fission yeast and in humans. Budding yeast Rif1 negatively regulates telomere length via interaction with the Rap1 C-terminal segment (Hardy et al. 1992; Levy and Blackburn 2004; Hirano et al. 2009). In fission yeast, Rif1 binds to the dsDNA in telomeres through interaction with Taz1, an ortholog of human TRF1/TRF2. Rif1 is required for telomere maintenance in a Taz1-dependent manner, although its role in telomere maintenance is minor, and Rif1 shows diffuse nuclear localization even in taz1Δ (Cooper et al. 1997; Kanoh and Ishikawa 2001; Miller and Cooper 2003).
Findings in this study show that Rif1 may selectively bind to not only telomeres, but also arm regions, and may regulate the sites and timing of origin firing throughout chromosomes in fission yeast. It is important to note that, in addition to activation of dormant/late ori, some active, early-firing origins are suppressed in rif1Δ cells, indicating that Rif1 is not simply a repressor of origin activation, but rather a critical determinant of the genome-wide origin activation program in fission yeast. Potential roles of Rif1 in regulation of origin selection and timing are discussed.
Results
Rif1 bypasses Hsk1 functions for viability
We recently reported that growth of hsk1-null cells can be restored by deletion of Mrc1. Further studies indicated that the selection and efficiency of origin activation were affected in mrc1Δ cells (Hayano et al. 2011; Matsumoto et al. 2011). These findings prompted us to conduct screening of novel mutants that may rescue the growth of hsk1Δ cells, with a hope that the identified genes may regulate the origin activation in fission yeast.
Since Hsk1 is essential for DNA replication and cell survival under normal growth conditions at 30°C, the hsk1Δ mutant carrying the ura4+ plasmid containing hsk1+ is not able to grow on the SD (synthetic defined medium) plate containing 5-FOA, which eliminates ura4+ plasmid DNA (Supplemental Fig. S1). We randomly integrated a budding yeast LEU2 marker gene into chromosomes to conduct insertion mutagenesis in hsk1Δ and screened for the LEU2 transformants on the SD plate containing 5-FOA at 30°C. As a result, we isolated 16 LEU2 clones that could survive in the absence of Hsk1. mrc1::LEU2 was among the 16 strains, confirming that the procedure is working. Out of the 16 clones, the four most vigorous suppressors were identified as rif1 mutants.
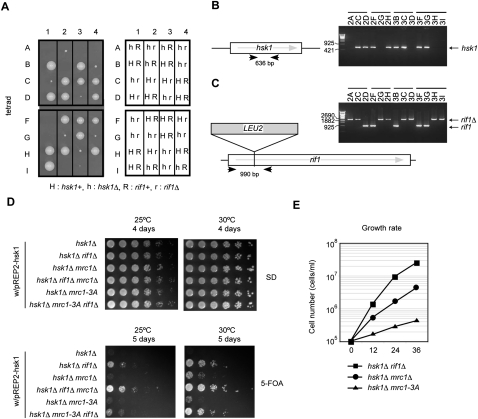
In order to confirm the bypass of hsk1+ function by rif1Δ, a diploid strain, h+/h− hsk1+/hsk1::kan rif1+/rif1::LEU2, was constructed and subjected to tetrad analysis. Among 48 tetrads dissected, 24 tetrads gave more than two viable segregants, supporting our notion that hsk1Δ rif1Δ is viable (Fig. 1A). To further verify this, genotypes of spores were determined by colony PCR (Fig. 1B,C). While hsk1Δ rif1+ spores gave no visible colonies, small colonies emerged from hsk1Δ rif1Δ spores (Fig. 1A). These data clearly demonstrate that rif1Δ can restore the growth of hsk1Δ, albeit with a reduced growth rate, and exclude the possibility that unknown mutations or gene rearrangements are responsible for the viability of hsk1Δ rif1Δ cells. Bypass was observed in spo4Δ encoding a second, meiosis-specific Cdc7-like kinase (Supplemental Fig. S2), showing that aberrant expression of spo4 is not responsible for the suppression.
Figure 1.
rif1Δ recovers growth defect of hsk1Δ. (A) Tetrads derived from h+/h− hsk1::kan/hsk1+ rif1::LEU2/rif1+ diploid cells were microdissected by using a Singer MSM micromanipulator onto YES agar plates and were photographed after 7 d of growth at 30°C. Colonies resulting from eight such tetrads are shown. Genotypes of the spores from the eight tetrads are shown in the right panel. (H) hsk1+; (h) hsk1::kan; (R) rif1+; (r) rif1::LEU2. The genotypes of these colonies were determined by colony PCR as shown in B and C. Locations of the primers and parts of the results of colony PCR detecting the Hsk1-coding sequence (B) or LEU2 insertion disruption of the rif1 gene (C) are shown. The primer sets used were Sal-hsk1, 5′-TTTAGTCGACAAGTAGTCCTGCTAGAATATACAATG-3′; and seq-03R, 5′-CTATCTCCATGAGTGCATCCACGTCGTCCT-3′ (B); and rif1-1fw, 5′-ATGACAAAAGAAATTGCTGTGAAGGAGGCT-3′; and rif1-99rv, 5′-AAATGCAGTTCCCTTAATATCACTTCTAAA-3′ (C). (D) hsk1Δ and its derivatives with the ura4+ plasmid carrying hsk1+ were cultured in YES medium. Fivefold serial dilutions of exponentially growing cells as indicated were spotted onto SD plates with or without 5-FOA. Cells were incubated for 4 or 5 d at 25°C or 30°C. Rescue of growth of hsk1Δ by mrc1Δ is known to occur more efficiently at 30°C than at 25°C (Matsumoto et al. 2011). (E) Exponentially growing cells were diluted to 1 × 105 cells per milliliter and cultured in YES medium at 30°C. The numbers of cells were counted at the indicated time points.
rif1Δ restored the growth of the hsk1Δ mutant more efficiently than mrc1Δ or mrc1-3A (S604AT645AT653A) (Fig. 1D), specifically defective in replication checkpoint activation (Zhao et al. 2003; Xu et al. 2006). Indeed, the rif1Δ hsk1Δ mutant grew better than mrc1Δ hsk1Δ or mrc1-3A hsk1Δ in liquid culture (Fig. 1E). Furthermore, the rif1Δ mrc1Δ hsk1Δ triple mutant grew better than the rif1Δ hsk1Δ or mrc1Δ hsk1Δ double mutant, suggesting that rif1Δ and mrc1Δ suppress the hsk1 mutation through independent mechanisms (Fig. 1D).
Deletion of rif1 bypasses Hsk1 function in a manner independent of the checkpoint, taz1, or rap1
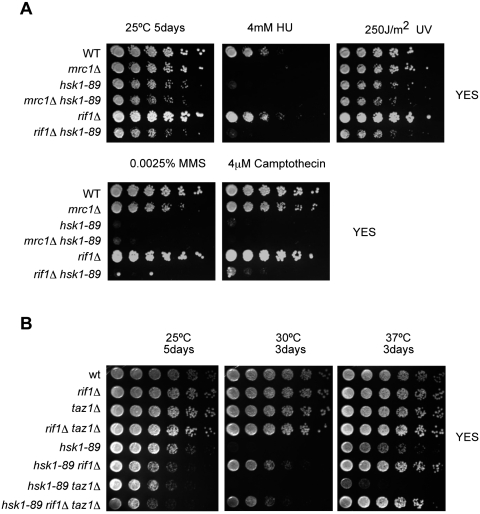
We examined whether Rif1 is required for activation of DNA replication or the DNA damage checkpoint. In contrast to mrc1Δ or cds1Δ, the rif1Δ mutant was not sensitive to hydroxyurea (HU), MMS (alkylating agent), camptothecin (an inhibitor of topoisomerase I), or UV irradiation (Fig. 2A), indicating that Rif1 is not necessary for cellular responses to replication stresses. The levels of Cds1 kinase activity after HU treatment were similar between the wild-type and rif1Δ cells (Fig. 2; Supplemental Fig. S3). Thus, Rif1 is likely to affect the selection of ori through a checkpoint-independent manner. It should also be noted that rif1Δ does not bypass the sensitivity of hsk1-89 to various DNA-damaging agents (Fig. 2A). Growth of the hsk1-89 mutant at 30°C was not recovered by deletion of taz1 and rap1, which is required for telomere maintenance. On the contrary, taz1Δ showed a synthetic growth defect with hsk1-89 at 37°C (Fig. 2B; Supplemental Fig. S4), a semipermissive temperature for hsk1-89 (Matsumoto et al. 2005, 2011). These results suggest that rif1Δ-mediated bypass of Hsk1 function is independent of its telomere-related functions.
Figure 2.
rif1Δ is resistant to inhibitors of DNA replication or DNA damage treatment, and taz1Δ is not able to bypass Hsk1 function. (A) The cells with the indicated genotypes were spotted onto YES plates containing no drug, 4 mM HU, 0.0025% MMS, or 4 mM camptothecin. UV was irradiated at 250 J/m2 onto the cells spotted on YES plates. After incubation for 5 d at 25°C, pictures were taken. (B) rif1Δ, taz1Δ, and rif1Δ taz1Δ were generated in wild-type and hsk1-89 backgrounds. Exponentially growing strains were spotted onto YES plates and grown for 3–5 d at 25°C, 30°C, and 37°C. hsk1-89 cells are able to grow at 37°C but not at 30°C (Matsumoto et al. 2005).
Rif1 regulates initiation of DNA replication both positively and negatively throughout the genome in fission yeast
To examine the effect of rif1Δ on the selection of replication initiation sites, we performed chromatin immunoprecipitation (ChIP) and microarray (ChIP–chip) assays in the cells released from M phase in the presence of HU. BrdU-incorporating DNA was immunoprecipitated with anti-BrdU antibody, and purified DNA was hybridized to the Schizosaccharomyces pombe tiling array (Hayano et al. 2011). Some late-firing origins, including AT2080 and pARS745, fired in rif1Δ (Supplemental Table S2) as well as in mrc1Δ and cds1Δ (Kim and Huberman 2001; Hayashi et al. 2007; Matsumoto et al. 2011), as indicated by ChIP–chip and two-dimensional (2D) assays.
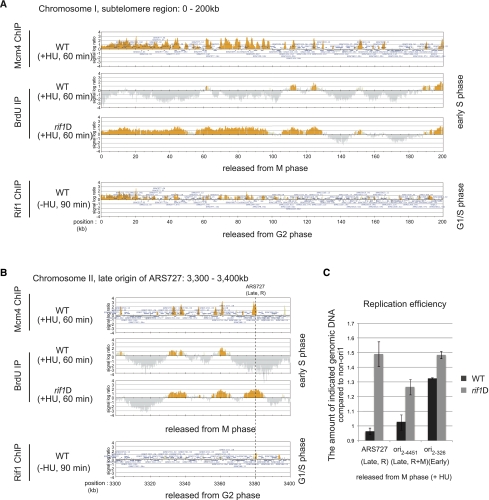
The subtelomeric regions form heterochromatin and are replicated late in S phase (Hayashi et al. 2009), and BrdU is not incorporated into this region in the presence of HU in wild-type cells (Fig. 3A). However, significant and extensive BrdU incorporation was detected across the subtelomeric region (chromosome I, 0–98 kb [left arm] and 5500–5582 kb [right arm]; chromosome II, 0–118 kb [left arm] and 4467–4558 kb [right arm]) in rif1Δ (Fig. 3A; Supplemental Fig. S5), indicating that Rif1 suppresses initiation of DNA replication in the subtelomeric segments. We evaluated replication efficiency also by measuring the copy number of chromosome DNA. Genomic DNA was isolated from the cells released from M phase in the presence of HU, and then the copy number of genomic DNA was measured at each origin (Fig. 3C). Two late-firing origins—ori2–4451 and ARS727, present on chromosome II and repressed by both the DNA replication checkpoint and Rif1 and by Rif1 alone, respectively—replicated more efficiently in rif1Δ than in wild type (Fig. 3B,C). ori2–326, an early-firing origin less efficient than ars2004 (data not shown), replicated slightly more efficiently in rif1Δ than in wild type (Fig. 3C; see also Fig. 7B, below). These results indicate that Rif1 suppresses initiation of DNA replication from some late-firing origins and low-efficiency early-firing origins.
Figure 3.
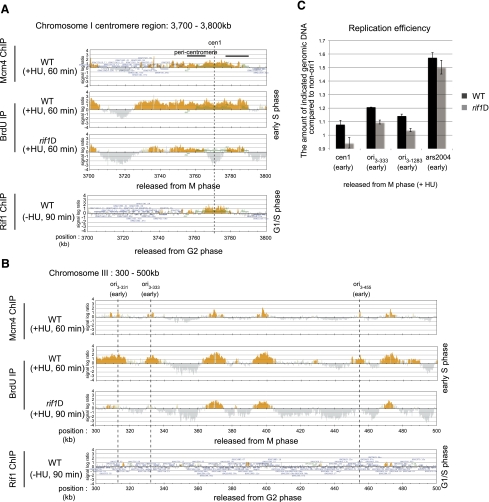
Dormant or inefficient origins at the subtelomeric region and in the arm region are activated in rif1Δ. nda3-KM311 mutants in the wild-type and rif1Δ backgrounds were incubated for 5 h at 20°C (arrest in M phase) and then released into the cell cycle for 60 min at 30°C in the presence of 25 mM HU and 200 mM BrdU. Mcm4-3Flag-bound DNA and BrdU-incorporating DNA were immunoprecipitated with anti-Flag antibody and anti-BrdU antibody, respectively. For Rif1 ChIP–chip assays, cells in the cdc25-22 background were incubated for 3 h at 36.5°C (arrest in G2 phase) and then released into the cell cycle for 90 min at 25°C (G1/S-phase boundary). Rif1-10Flag was immunoprecipitated with anti-Flag antibody. Collected DNA was purified and analyzed by using Gene Chip S. pombe tiling array (Affymetrix). (A) The 0- to 200-kb region of chromosome I containing the subtelomeric region. (B) The 3300- to 3400-kb region of chromosome II containing a dormant origin, ARS 727. (C) Quantification of genomic DNA by quantitative PCR. Genomic DNA was extracted from the cells arrested in M phase and from those released from M phase in the presence of 25 mM HU for 60 min (S phase). Amounts of genomic DNA at indicated locations were quantified by real-time PCR. The ratio of the amount of genomic DNA in S phase to that in M phase was calculated for all of the locations. The non-ori1 region that is not replicated under this condition was used for normalization. (ARS727) A dormant origin that is fired in rif1Δ, but not in checkpoint mutants (Late, R); (ori2–4451) a dormant origin that is fired in both rif1Δ and mrc1Δ (Late, R+M); (ori2–326) an early-firing origin on chromosome II that initiates replication less efficiently than ars2004. Error bars indicate the SD from three independent experiments.
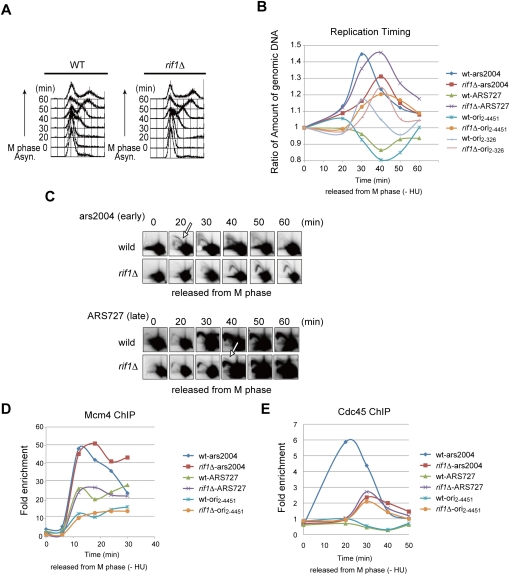
Figure 7.
Rif1 regulates replication timing and loading of Cdc45 during S-phase progression in the absence of HU. Cells arrested in M phase at 20°C were released into the cell cycle at 30°C in the absence of HU as described in the legend for Figure 3. (A) Cell cycle progression was monitored by FACS analysis of DNA content. (B) Replication timing at ars2004 (strong early-firing origin), ARS727 (late-firing/dormant origin), ori2–4451 (late-firing/dormant origin), and ori2–326 (weak early-firing origin) was determined by quantification of genomic DNA at each location and normalization to the DNA level at the non-ori1 loci. (C) At each time point after release, replication intermediates were prepared and analyzed in 2D gel electrophoresis, as described before (Hayano et al. 2011). The arrows indicate the bubble arc. Loading of Mcm4 (D) and Cdc45 (E) was analyzed using the Mcm4-3Flag- and Cdc45-3Flag-tagged strains, respectively, at the indicated loci.
Centromeres and their neighboring segments replicate efficiently in the presence of HU (Fig. 4A), as reported previously (Kim and Huberman 2001). Interestingly, BrdU incorporation into the centromere segment decreased in rif1Δ (chromosome I, 3756–3785 kb; chromosome II, 1600–1645 kb; chromosome III, 1065–1143 kb). Especially, DNA replication at the central core segments was completely suppressed in rif1Δ cells (Fig. 4A; Supplemental Fig. S6). On chromosome III, most of the ori are early-firing, and loss of rif1 resulted in down-regulation at most of these origins (Fig. 4B; Supplemental Table S2). DNA replication at ars2004, one of the most efficient early-firing origins, was also reduced in rif1Δ (Supplemental Fig. S7). Measurement of copy numbers at cen1, ori3–333, ori3–1283, and ars2004 in rif1Δ also showed inhibition of DNA replication at these early-firing origins (Fig. 4C). These results indicate that Rif1 is required also for efficient and early S-phase replication at some early-firing origins.
Figure 4.
DNA replication at some early-firing origins in the arm region and at centromeres is repressed in rif1Δ. BrdU incorporation and BSs of Mcm4 and Rif1 were analyzed as described in the legend for Figure 3. (A) The 3700- to 3800-kb region of chromosome I containing the centromere. Rif1 binds specifically to the centromere core region, and DNA replication in this specific segment is repressed in rif1Δ. Black bars indicate the locations of the pericentromeric regions. (B) The 300- to 500-kb region on chromosome III. Most of the ori on chromosome III are early-firing and are repressed in rif1Δ. Rif1 does not bind to these repressed origins. (C) Replication efficiency was determined at the indicated locations by quantitative PCR as described in the legend for Figure 3. cen1 and ori3–333 (an early-firing origin) are indicated in A and B, respectively. ori3–1283 is another early-firing origin on chromosome III, and ars2004 is a well-characterized efficient early-firing ori. Replication efficiency of these origins decreased in rif1Δ. Error bars indicate the SD from three independent experiments.
The numbers of origins affected by rif1Δ
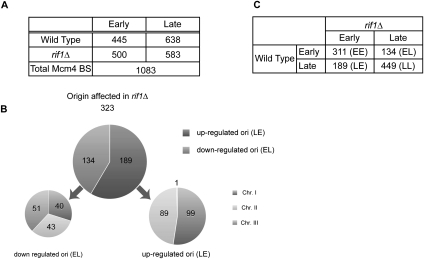
The peaks of Mcm4 binding and BrdU incorporation were counted throughout the chromosomes, except for subtelomeric and centromeric regions that contained too many Mcm4-binding peaks to be distinguished into single peaks. We chose to define the numbers of ori (Mcm4-BSs) in hsk1-89 cells, since in these cells, the peak pattern is clearer (because Mcm4 does not move from the ori due to the failure of Cdc45 loading) (see Supplemental Fig. S12). We detected 1083 ori with a false discovery rate (FDR) threshold of 0.1% using rMAT (Droit et al. 2010) (in contrast to 693 in the wild-type cells) (Supplemental Table S1). Detailed comparison of pre-RC localization and BrdU-incorporating sites throughout the chromosome arm regions indicated that 445 (41%) out of 1083 ori were actually activated to incorporate BrdU in the presence of HU in the wild-type cells (“early” ori). The numbers of the initiation sites increased to 500 in rif1Δ cells (Fig. 5A). In total, 323 origins were identified to be affected in rif1Δ cells in BrdU ChIP–chip assays (Fig. 5B). Genomic copy number assays detected small changes occurring in rif1Δ cells (e.g., ori2–326 and ars2004) (Figs. 3C, 4C), suggesting that the numbers of Rif1-regulated origins may be underestimated in BrdU ChIP–chip assays. One-hundred-eighty-nine dormant origins (99, 89, and 1 on chromosomes I, II, and III, respectively) fired in rif1Δ. On the other hand, 134 origins (40, 43, and 51 on chromosomes I, II, and III, respectively) were down-regulated in rif1Δ cells (Fig. 5B). We classified ori into EE (“early to early”; early in wild-type and early in rif1Δ), LL (“late to late”), EL (“early to late”), and LE (“late to early”). The numbers of EE, LL, EL, and LE were 311, 449, 134, and 189 (Fig. 5C). These results indicate that Rif1 regulates the firing of ori both negatively and positively across the chromosomes. This is in sharp contrast to checkpoint mutants, which did not affect the firing at efficient, early-firing origins (Hayano et al. 2011; Matsumoto et al. 2011).
Figure 5.
pre-RC and BrdU-incorporating sites detected in the wild-type and rif1Δ cells by ChIP–chip assays. (A) Cells were harvested at 60 min after release from M-phase arrest in the presence of HU as described in the legend for Figure 3. The number of Mcm4-BSs in hsk1-89 cells (at 30°C) and those of BrdU-incorporation sites in wild-type and rif1Δ cells are presented. (B) The numbers of origins up-regulated or down-regulated in rif1Δ cells are indicated. The numbers of up-regulated or down-regulated origins on each chromosome are also shown. (C) The identified ori were classified into four categories according to their overlap with BrdU domains: EE (“early to early”; early in wild-type and early in rif1Δ), LL (“late to late”), EL (“early to late”) and LE (“late to early”) The numbers of EE, LL, EL, and LE are presented.
Rif1 binds to selected arm segments in addition to telomeres and centromeres
To clarify the mode of Rif1-mediated replication regulation, we determined localization of Rif1 on fission yeast chromosomes by ChIP–chip assays. Cells were released from G2 phase and collected at 90 min (G1/S phase). Rif1 binds to the subtelomeric regions, and some of these Rif1-BSs colocalized with Mcm4-BSs (Fig. 3A). ARS727, which is silent in the wild type but is efficiently fired in the presence of HU in rif1Δ, is bound with Rif1 (Fig. 3B). Rif1 binds also to the centromere core segment (Fig. 4A). Interestingly, replication of this segment is completely suppressed in rif1Δ, whereas the pericentromere heterochromatin regions (dh/dg repeats) to which Swi6 binds and where transcription is silenced due to heterochromatin formation (Partridge et al. 2000; for review, see Buhler and Moazed 2007) are not bound with Rif1 (Fig. 4A). However, rif1Δ cells did not show a growth defect in the presence of the anti-microtubule drug thiabendazole (TBZ), which is an inhibitor of mitosis (Supplemental Fig. S8), suggesting no distinct role of Rif1 in chromosome segregation.
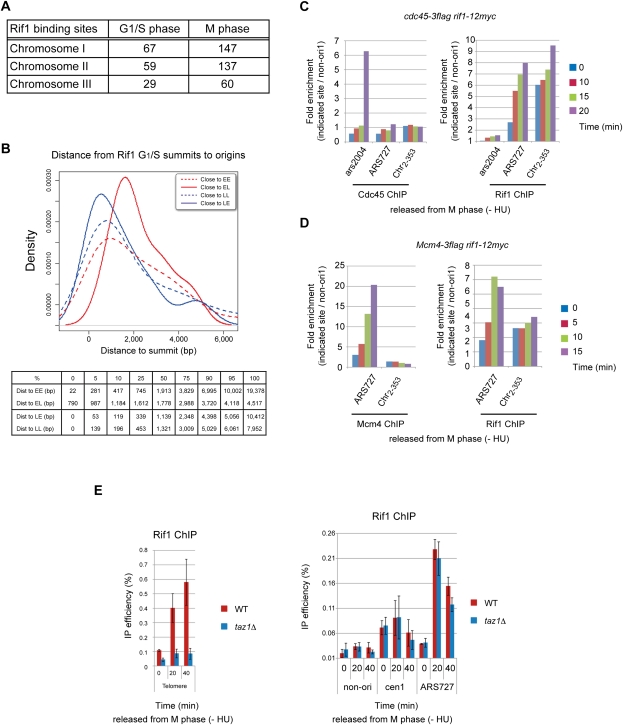
We identified 155 Rif1-BSs in the G1/early-S-phase cells (Fig. 6A). We compared the distribution of these Rif1-BSs and ori locations. Among the 155 BSs, 63 BSs (41%) overlapped with ori, which were further divided into 38 late ori (60%) and 25 early ori (40%). Next, we examined the closest ori for all of the BSs, and 77 and 78 BSs were close to early and late ori, respectively. The distance between a BS and the nearest ori was shorter with late ori than with early ori (Supplemental Fig. S9). This difference (distance to closest ori between late ori and early ori) appeared to be significant (P-value = 0.008 with a Wilcoxon test).
Figure 6.
Rif1-BSs detected by ChIP–chip assays: cell cycle regulation and dependence on Taz1. (A) The numbers of Rif1-BSs on each chromosome are presented (total numbers: 155 BSs in G1/S and 344 in M phase). (B) Distribution of distances between Rif1-90min-BSs (Rif1-BSs in G1/S) and the closest ori plotted for each category of ori. (Broken red line) EE; (red line) EL; (broken blue line) LE; (blue line) LL. The table below shows the distribution (quantiles) of distances between Rif1-90min-BSs and the closest ori, summarized for each category. The size in each column shows the maximum distance for the fraction indicated to be contained; 50% corresponds to median value. (C,D) Dually tagged cells (cdc45-3Flag rif1-12myc [C] and mcm4-3Flag rif1-12myc [D]) were arrested in M phase and synchronously released into G1–S phase in the absence of HU. After harvest, cells were split into two parts, and Cdc45 and Rif1 (C) or Mcm4 and Rif1 (D) were immunoprecipitated with anti-Flag antibody or anti-Myc antibody, respectively. Cdc45-, Mcm4-, and Rif1-binding DNA were amplified with primer sets of non-ori1 (not bound by Mcm4 or Rif1), ars2004 (early-firing origin), ARS727 (late-firing origin bound by Rif1), and Chr2–353 (a non-ori bound by Rif1 but not by Mcm4). (E) Rif1-12myc cells in the wild-type or taz1Δ background arrested in M phase at 20°C were released into the cell cycle at 30°C in the absence of HU. Binding of Rif1 at TEL (the telomere sequence close to telomere repeats) (left) and at arm regions (including centromeres and dormant origins) (right) was quantified at each time point after release. ARS727 is described in the legend for Figure 3. (cen1) Centromere core regions on chromosome I.
Among 155 Rif1-90min-BSs, 69, 8, 46, or 32 BSs were located close to EE, EL, LE, or LL, respectively. Fifty percent of the 189 LE ori are associated with Rif1-90min-BSs within 1.14 kb from the ori (Fig. 6B). The value was 1.5 kb for Rif1-Mphase-BSs, which is also significantly shorter than that for other ori (P-value < 1 × 10−4 with a Kruskall-Wallis test) (Supplemental Fig. S10). On the other hand, the distance between the EL ori and Rif1-BSs appears to be the farthest. Summit distance profiles of Mcm4-BSs around Rif1-90min-BS and Rif1-Mphase-BS summits also indicated most centered clustering at LE and most dispersed clustering at EL (Supplemental Fig. S11).
In summary, Rif1 is generally located close to the late ori compared with early ori. It is especially closer to LE ori that are up-regulated in rif1Δ. Thirty-one (67%) out of 46 LE associated with 90min-BSs are also Rif1-Mphase-BSs, indicating that Rif1 bound to chromatin may regulate the firing timing of late ori. The results also suggest a possibility that Rif1 could directly regulate the firing of nearby late ori.
Rif1 associates with arm segments at early G1 phase in a Taz1-independent manner
We then examined the timing of the arm binding of Rif1 protein in more detail. Cells were released from M phase in the absence of HU, and binding of Rif1 to ARS727 (ori) and Chr2–353 (non-ori), both of which are Rif1-BSs, was examined at various times after release. The timing of binding was compared with that of Mcm or Cdc45 by using dually tagged strains. Cdc45 was loaded onto ars2004 at 20 min after release, but significant amounts of Rif1 bound to Chr2–353 and ARS727 even at the time of M-phase arrest. The binding at ARS727 kept increasing until 20 min after release (G1/S) (Fig. 6C,D), whereas that at Chr2–353 was more or less constant during G1 and increased at S phase (Fig. 6C,D). The timing and kinetics of Rif1 binding to ARS727 are similar to those of Mcm4 binding.
Rif1 is known to bind to telomeres through Taz1 in fission yeast (Kanoh and Ishikawa 2001). Therefore, we examined whether binding of Rif1 to various locations on chromosomes depends on Taz1. At different time points after synchronous release of Rif1-12myc cells from M phase, Rif1 was immunoprecipitated with anti-Myc antibody, and coimmunoprecipitated DNAs were quantified by real-time PCR. Binding of Rif1 to telomeres significantly increases as cells progress from M to S phase, and this binding is not detected in taz1Δ, as expected (Fig. 6E). Rif1 binds to centromeres during M phase and stays bound there until S phase completes. Binding of Rif1 to the arm origin (ARS727) increases at the G1/S boundary and decreases as S phase proceeds (Fig. 6E). Binding of Rif1 to centromere and arm segments was not affected by taz1Δ, indicating that Rif1 binds to nontelomere chromosomes in a Taz1-independent manner (Fig. 6E). These results suggest that Rif1 binds to telomere and nontelomere segments through distinct mechanisms.
Regulation of replication timing by Rif1 in an unperturbed cell cycle
The experiments described above show that Rif1 affects origin firing profile in the presence of HU. Next, we addressed whether Rif1 affects the origin firing timing in an unperturbed cell cycle. Cells were arrested in M phase and released into S phase in the absence of HU. Overall progression of the cell cycle, as detected by FACS analyses of DNA content, is similar between wild-type and rif1Δ cells (Fig. 7A). Replication timing was determined by measuring the amount of genomic DNA at various origins and normalizing them with those at non-ori1. ars2004 (efficient early-firing origin) replicated at 30 min, and replication timing at ARS727 (late/dormant origin) was later than the non-ori1 region, which is located 30 kb away from ars2004, in wild-type cells (Fig. 7B). However, in rif1Δ cells, replication timing of ars2004 was delayed and its efficiency was also reduced. On the other hand, ARS727 replicated slightly earlier than ars2004, and its replication efficiency exceeded that of ARS2004 (Fig. 7B).
In order to determine whether replication timing is deregulated in rif1Δ, we conducted 2D assays in synchronously growing wild-type and rif1Δ cells. A strong bubble arc was detected at ars2004 at 20 min after release in wild type, whereas its appearance was delayed by 10–20 min in rif1Δ. In contrast, a bubble arc was not detected at ARS727 in wild type, but was detected at 30 min after release in rif1Δ (Fig. 7C). These results clearly show that Rif1 regulates replication timing during the normal course of S-phase progression.
ori2–326 is an early-firing origin, but its replication efficiency increases in rif1Δ (Fig. 3C). DNA replication at ori2–326 occurred at the timing that is the same as that of ars2004 in the wild type, although the efficiency was lower. In rif1Δ, the timing of replication at ori2–326 was delayed, while the efficiency was higher than in wild type (Fig. 7B). This clearly indicates that the firing timings of both early- and late-firing origins are greatly affected by rif1Δ and are compressed into a smaller segment of S phase. Replication efficiency is also affected by rif1Δ, but could be affected by other factors as well, including chromatin context.
Next, we examined which stage of DNA replication is affected in rif1Δ cells. We first examined the timing of pre-RC assembly, since Wu and Nurse (2009) reported that timing and efficiency of pre-RC assembly may determine the timing of firing of origins in the following S phase. We examined the timing of Mcm4 binding to various origins (Fig. 7D). The results indicate that the timing and levels of Mcm4 binding are almost identical between wild-type and rif1Δ cells, indicating that Rif1 does not affect the process of pre-RC assembly. ChIP–chip assays of Mcm4 binding also indicated that locations of Mcm4-BSs are basically identical between the wild type and rif1Δ (Supplemental Fig. S12). On the other hand, Cdc45 bindings to the same origins were affected by rif1Δ (Fig. 7E). They basically correlated with timing and levels of BrdU incorporation. ChIP–chip assays of Cdc45 binding in wild type and rif1Δ also showed that the origins that are activated in rif1Δ do have Cdc45 protein bound (Supplemental Fig. S13). Thus, Rif1 protein may influence the steps after pre-RC assembly, most likely the steps regulating the loading of Cdc45.
Discussion
Deletion of rif1 restores the growth of hsk1Δ
Although Hsk1 is essential for initiation of DNA replication and viability at 30°C in fission yeast, we recently discovered that deletion of mrc1 can bypass the requirement of hsk1+ for cell growth (Matsumoto et al. 2011). Further studies showed that precocious initiation of DNA replication takes place in mrc1Δ cells (Hayano et al. 2011). These results prompted us to search for other mutations that may bypass the requirement of hsk1+ for viability, since we predicted that it may lead to the identification of novel factors that regulate the replication program. Such screening resulted in isolation of several novel genes, the loss of which may alleviate the requirement of hsk1+ for growth. One of these genes was rif1, known to interact with telomeres and regulate telomere maintenance. The growth defect and aberrant morphology of hsk1-89 mutations were largely corrected by rif1Δ (data not shown). The S phase proceeds and completes in hsk1-89 rif1Δ cells at a nonpermissive temperature.
Rif1 regulates origin firing both positively and negatively
It was reported previously that cds1Δ partially recovers growth of the hsk1-1319 mutant, another allele of hsk1 (Snaith et al. 2000). We also reported that cds1Δ and mrc1-3A, a checkpoint-defective mutant of mrc1, can partially rescue the growth of hsk1-89 at a nonpermissive temperature and weakly restore the growth of hsk1Δ cells (Matsumoto et al. 2005, 2011). It has been well established that late-firing or dormant origins are activated in checkpoint mutants in budding yeast (Santocanale and Diffley 1998; Shirahige et al. 1998). In fission yeast, it has been reported that mutations in rad3 or cds1Δ activate some of the dormant origins (Heichinger et al. 2006; Hayashi et al. 2007). Thus, abrogation of replication checkpoint functions can reduce the requirement of hsk1 for DNA replication, presumably by activating the firing at inefficient origins. However, rif1Δ is completely resistant to various genotoxic agents, including HU and MMS, and can activate Cds1 kinase in response to HU (Fig. 2A; Supplemental Fig. S3), and thus it is unlikely that rif1Δ bypasses the Hsk1 function in a checkpoint-dependent manner.
Nevertheless, rif1Δ restored efficient growth of hsk1Δ cells. ChIP–chip analyses showed deregulation of firing at many dormant origins, including those at the clusters of pre-RC in the subtelomeric regions (Figs. 3, 5). One-fourth of the deregulated origins in rif1Δ did not overlap with those activated in either of the checkpoint mutants (Supplemental Fig. S14), consistent with the speculation that Rif1 can regulate origin firing in a manner distinct from that by mrc1Δ or a checkpoint mutant. In fact, mrc1Δ and rif1Δ exhibited an additive effect on the restoration of the growth of hsk1Δ, indicating that the two mutations are in different epistasis groups in terms of the hsk1 bypass (Fig. 1D).
The replication of the centromeric region, which efficiently occurs in the presence of HU, is reduced in rif1Δ cells. Particularly, replication of the centromere core region completely depends on Rif1 (Fig. 4A; Supplemental Fig. S6). We also noticed that some very active early-firing origins are repressed in rif1Δ cells. Especially, most of the origins on chromosome III that are efficient and early firing are suppressed in rif1Δ (Fig. 4B; Supplemental Table S2). These observations led us to conclude that Rif1 can both positively and negatively regulate the firing events on the whole genome.
Rif1 binds to telomere, centromere, and arm regions
Rif1 binds extensively to subtelomeric segments, and this binding peaks at the G1/S boundary (data not shown). Replication in the subtelomeric region is extensively deregulated in the absence of Rif1 protein (Fig. 3B), indicating that Rif1 negatively regulates origin firing in this segment. Similar repression of replication was reported to be mediated by the Ku complex, which binds to the telomeres (Cosgrove et al. 2002; Lian et al. 2011). Thus, the protein machinery assembled at telomeres may function to suppress replication initiation in early S phase. Rif1 also binds to the centromere core region, and this binding is observed at a relatively constant level throughout the cell cycle (Figs. 4A, 6E). It is noteworthy that replication of the very segment bound by Rif1 is silenced in rif1Δ, suggesting that bound Rif1 may play an essential role in early replication of the centromere core segment. It was previously reported, however, that no bubble arc is observed in the centromere core segments, leading to the conclusion that replication initiation does not take place in this segment despite the presence of pre-RC (Kim et al. 2003). The firing of the pericentromeric region is also reduced in rif1Δ, suggesting that the efficient replication of the entire centromere region may depend on Rif1.
In addition to subtelomeric and centromeric segments, 344 Rif1-binding loci were identified on the entire chromosomes during M phase. In the arm segment, Rif1 binding at some loci increases at early G1 at about the same timing as Mcm loading, peaking at the G1/S boundary, and decreases as S phase progresses. The relationship between Rif1 binding and origin regulation in the arm segments is complex. Rif1 binds to ARS727, a dormant origin deregulated specifically in rif1Δ, but not to all of the Rif1-suppressed origins (Figs. 3B, 6B). Rif1 binds also to non-pre-RC sites as well as to early-firing origins (Fig. 6; Supplemental Fig. S15). In contrast to binding to telomeres, binding of Rif1 to centromere and arm segments is independent of Taz1 (Fig. 6E). Consistent with this, taz1Δ did not bypass hsk1Δ (Fig. 2B). Thus, Rif1 binds to telomere and arm regions with distinct mechanisms, and loss of Rif1 binding to the arm segment may be crucial to the rescue of hsk1Δ as well as for deregulation of the genome-wide origin firing program. Indeed, the rif1 mutant lacking the C-terminal 140 amino acids does not bind to the telomere but does bind to arm regions, and this mutant is not able to rescue hsk1Δ or change the origin firing pattern (Y Kanoh, M Hayano, S Matsumoto, and H Masai, unpubl.).
Potential conserved sequences for chromatin binding of Rif1
A late consensus sequence (LCS) derived from ARS727 was reported to have a potential to regulate replication timing; the presence of this sequence rendered the nearby origins late-replicating on a plasmid (Yompakdee and Huberman 2004). Since Rif1 binds to ARS727, it is an interesting possibility that Rif1 may directly recognize the LCS.
We searched for a potential consensus motif enriched in Rif1-BSs, using the MEME software. We could not find any motif present in all of the sites in common, but we did find a motif specifically enriched in around half of LE Rif1-90min-BSs and in a few LL sites. It is CNWWGTGGGGG (with W being A or T), which is similar to the LCS motif (RKKGGGGGAW, with R being G or A, and K being G or T) (Supplemental Table S3). Using an E-value threshold at 1 × 10−4 and a FDR threshold (Q-value) of 1%, we found with FIMO software that 20 of the 46 LE Rif1-90min-BSs contain at least one occurrence of the consensus, and 11 out of the 32 LL-BSs do as well. A similar number of BSs with this motif were found within Rif1-Mphase-BSs (22 out of 66 LE and eight out of 144 LL). These results may suggest a possibility that the recognition of a LCS-like motif might be involved in regulation of origin firing by Rif1. Even if Rif1 may recognize a LCS-like motif, that would not be the only mechanism by which Rif1 is recruited to chromatin, since only a portion of BSs contain such motifs. The purified Rif1 protein does bind to DNA, but we did not detect any difference in its affinity between ARS727 and other sequences to which Rif1 does not bind in ChIP assays (S Kudo, M Hayano, and H Masai, unpubl.). Thus, Rif1 may bind to arm regions through other proteins. Identification of such factors will clarify the mechanism by which Rif1 binds to chromatin and regulates replication timing.
Forty-one percent (63 out of 155; 90min-BSs) or 23% (80 out of 344; Mphase-BSs) of the Rif1-BSs overlapped with the pre-RC sites (Fig. 6B; Supplemental Fig. S15). These potential origins, constituting 7.6% (80 out of 1083) of the total pre-RC sites, included both early-firing origins (25 and 23) and dormant/late-firing origins (38 and 57). These overlaps were statistically significant. Analyses of distances between each BS and the nearest ori indicated that the late origins, especially the LE category of ori, tend to be associated with Rif1-BSs within a short distance. These results support a possibility that Rif1 may regulate the late origin firing through association with the vicinity of the target ori. The remaining nonorigin Rif1-BSs appear to overlap with RNA polymerase-BSs (Y Kanoh and H Masai, unpubl.), suggesting a possibility that Rif1 may be loaded onto the arm segments in two distinct mechanisms: one through replication-related mechanisms, and the other through transcriptional machinery. Indeed, the transcriptional levels of many genes changed in rif1Δ cells (M Hayano, Y Kanoh, S Matsumoto, and H Masai, upubl.; data not shown).
Potential mechanisms of Rif1-mediated regulation of the replication program in fission yeast
Rif1 affects the genome-wide replication profile. The most striking effect of rif1Δ is the timing of origin firing. In each case tested, late-firing origins are fired at an earlier timing, and firing of early-firing origins is delayed. In contrast, the efficiency of DNA replication (of early-firing origins) can decrease or increase, depending on the ori in question. Thus, we conclude that Rif1 is a critical regulator of replication timing. In the absence of Rif1, replication timing regulation is virtually lost, and all of the origins are fired more less in the mid-S-phase stage or randomly during S phase.
It was reported that early recruitment of pre-RC factors may contribute to early and efficient firing during the following S phase (Wu and Nurse 2009). Therefore, Rif1 may regulate pre-RC assembly. We examined this possibility by measuring the timing and extent of Mcm4 binding to different origins by quantitative PCR as well as by ChIP–chip assays (Fig. 7D; Supplemental Fig. S13). We did not observe any significant difference in the timing and efficiency of binding of Mcm4 to origins. In contrast, binding of Cdc45 advanced/increased or delayed/reduced in rif1Δ cells in accordance with the mutation's effect on the timing of BrdU incorporation (Fig. 7E). Thus, Rif1 does not affect the pre-RC assembly, but affects the loading of Cdc45.
A simple explanation for the bypass of hsk1Δ by rif1Δ is that Rif1 is an inhibitor of DNA replication, and Hsk1 counteracts the inhibition. Rif1 itself does not appear to be a target of Hsk1. Hsk1/Cdc7 is required for efficient loading of Cdc45 at ori (Masai et al. 2006; Sheu and Stillman 2006; Yabuuchi et al. 2006; Heller et al. 2011), and Rif1 inhibits loading of Cdc45 at dormant origins. Thus, it is plausible that the absence of Rif1 stimulates loading of Cdc45 at normally silent ori, and under this condition, Cdc45 is loaded onto chromatin, albeit with a reduced efficiency, in an amount sufficient for initiation of DNA replication without Hsk1 kinase. Although Hsk1 kinase facilitates loading of Cdc45 at ori, it is apparently not essential for Cdc45 loading under some conditions (Matsumoto et al. 2011).
Histone modification has been reported to influence the origin firing timing in budding yeast. The lack of rpd3, encoding histone H4K12 deacetylase, led to early firing of late origins (Knott et al. 2009), and tethering of Gcn5, the histone H3, and the H2B acetyltransferase in the vicinity of a late-firing origin ARS1412 caused H3K18 acetylation and early firing of this origin (Vogelauer et al. 2002). Thus, Rif1 could regulate origin firing timing by affecting the histone modification status near its BSs.
In both mammals and yeasts, it has been known that the establishment of a late-firing program requires events that occur between mitosis and the subsequent G1 phase. In budding yeast, sequences flanking late-firing origins facilitate their targeting to the nuclear periphery in G1, where a modified chromatin structure can be established and the late-replicating program is determined (Raghuraman et al. 1997; Heun et al. 2001). In mammalian cells, it was proposed that a major nuclear reorganization that happens during early G1 may serve as a timing decision point (TDP) that will determine the timing of DNA replication (Dimitrova and Gilbert 1999). Therefore, it is tempting to speculate that Rif1 may be involved in this step, where chromatin organization is reset at early G1 phase. Binding of Rif1 to the arm segments at late M/early G1 phase (Fig. 6E) is consistent with this prediction.
Although Rif1 tends to be located near ori, in most cases, they do not completely overlap. We failed to detect physical interactions between Rif1 and replisome components such as Mcm and Cdc45. Thus, Rif1 may regulate both replication and transcription through affecting the global nuclear architecture of chromatin. The absence of Rif1 would result in the loss of intricate nuclear architecture, which may be required for coordinated activation of early and late origin firing. This would lead to a loss of distinction between early- and late-firing origins and compression of replication timing into smaller segments of S phase in rif1Δ cells. Detailed analyses of chromatin organization in nuclei in rif1Δ and wild-type cells would be required to test these hypotheses.
Rif1 is a conserved factor, and its mammalian homolog has been shown to play minimum roles in telomere maintenance, but to play roles in response to DNA damage (Silverman et al. 2004). It would be very important to analyze the potential functions of Rif1 in the regulation of the DNA replication program in higher eukaryotes and to determine whether regulation of the genome-wide replication program by Rif1 is evolutionarily conserved.
Materials and methods
Media and general methods for yeast genetics
The details of the strains used in this study and their genotypes are described in Supplemental Table S1. YES medium containing 0.5% yeast extract, 2% glucose, and 0.1 mg/mL each adenine, uracil, and leucine (Sigma) was used for cell culture, and YES plates were made by adding 2% agar to YES medium. The “SD without leucine” plate contained 0.67% (w/v) yeast nitrogen without amino acid, 2% glucose, 100 mg/mL adenine, 100 mg/mL uracil, and 2% agar. Tetrad analyses were conducted using a Singer MSM manual micromanipulator (Singer Instrument Co. Ltd.). For FACS analyses, cells were fixed with 70% ethanol. After treatment with RNaseA for 2 h at 37°C, DNA was stained with 4 mg/mL propidium iodide.
Screening for mutants that are viable in a hsk1Δ background.
The Saccharomyces cerevisiae LEU2 gene was amplified with a primer set of LEU2 1F, CGGGATCCTTGTCCTGTACTTCCTTGTT; and Leu2 R, GCTCTAGACCTCCTCCTTGTCAATATTA. To randomly disrupt fission yeast genes in a hsk1Δ background, a LEU2 DNA fragment was introduced into hsk1Δ cells carrying the ura+ plasmid harboring the hsk1 gene (MS143). Cells were grown on SD without leucine plates for several days at 30°C, and then growing cells were replica-plated onto SD plates containing 0.1% (w/v) 5-FOA at 30°C to remove the plasmid containing the hsk1 gene. Survivors were cultured in YES medium and harvested. Genomic DNA was isolated with the DNeasy Blood and Tissue kit (Qiagen). Deep sequencing (Illumina) was conducted to identify the genes that were disrupted by the LEU2 marker insertion.
ChIP assays and real-time PCR
ChIP assays were performed as previously described (Hayano et al. 2011). The following primers were used for amplification: ARS727, 5′-AACATATACGGTGAGATGGGAT-3′ and 5′-ATTCGTATTTTCCAATGCTTCCT-3′; ori2–4451, 5′-ACGCTTAACTTCGCACCCTTT-3′ and 5′-GGCACTCAACTACATGTAACTT-3′; ori2–326, 5′-TCTCTCAGTAACGCATCA-3′ and 5′-CGAAATTAAGTCTCACCC-3′; cen1, 5′-GGTCGGTTTTCTTTTTGTTG-3′ and 5′-GATCCTTTTGTTACCGGC-3′; ori3–333, 5′-AAACAAGCGGTACAACAG-3′ and 5′-CAGCTTACCGAAACTAACAAC-3′; ori3–1283, 5′-ACCAAAAGAAGGAAGCAAGA-3′ and 5′-ACCTGTGTTGATTTTGGC-3′; ars2004, 5′-CTTTTGGGTAGTTTTCGGATCC-3′ and 5′-ATGAGTACTTGTCACGAATTC-3′; non-ori1 (30 kb away from ars2004), 5′-TCGAAGATCCTACCGCTTTC-3′ and 5′-GATTCACATAACCCGCTAGC-3′; Chr2–353, 5′- AACCCCCTTAAAACTATCTC-3′ and 5′-GACGGCAAAGGAAAAGAA-3′; TEL, 5′-CGGCTGACGGGTGGGGCCCAATA-3′ and 5′-GTGTGGAATTGAGTATGGTGAA-3′. The TEL primer set was as described (Kanoh and Ishikawa 2001). Real-time PCR was performed by using SYBR Premix Ex Taq (Takara) and Light Cycler 480 (Roche Diagnostics).
ChIP–chip and data analysis
Chromatin-immunoprecipitated DNA was amplified by the invitro translated (IVT) amplification method. Briefly, chromatin-immunoprecipitated DNA was treated with calf intestine alkaline phosphatase (New England Biolabs) and TdT (Roche Diagnostics) to add a polyT tail to the DNA ends. DNA synthesis was conducted on the polyT-tailed chromatin-immunoprecipitated DNA by using a polyA primer containing the T7 RNA polymerase promoter DNA sequence. The resulting dsDNA was used as a template for T7 RNA polymerase-mediated transcription. Then, the complementary DNA was synthesized from the resulting RNA with SuperScript II RT (Invitrogen) to obtain the amplified chromatin-immunoprecipitated DNA. Chromatin-immunoprecipitated DNA was again converted to RNA by T7 RNA polymerase and then to DNA by RT as described above (second cycle). The amplified chromatin-immunoprecipitated DNA was labeled and hybridized with a Gene Chip S. pombe tiling array (Affymetrix) as described previously (Voineagu et al. 2009).
The BSs of Mcm4 and Rif1 were detected using the rMAT R package (http://www.r-project.org; Droit et al. 2010). With the MAT normalization and the robust option, 1083 Mcm4-BSs were detected in the hsk1-89 strain with a FDR threshold of 0.1%, and 155 and 344 Rif1-BSs (at G1/S phase [90 min] and M phase, respectively) were detected with a FDR threshold of 5%.
BrdU accumulation domains were defined using the method previously described (Schwartz et al. 2006; http://www.r-project.org; an unpublished R package kindly provided by Richard Bourgon). Briefly, after rMAT normalization, probe signals were smoothed over 1.4-kb windows. P-values were assigned to each window by a nonparametric procedure that estimates the null distribution as the left part of the distribution of smoothed log ratio (IP over input). Then, windows significantly different from this “null distribution” (corrected P-value < 0.01) were extracted and continuous windows were gathered to determine BrdU accumulation domains. This led to the detection of 316 and 374 BrdU domains in wild-type and rif1Δ cells, respectively. The raw and processed microarray data are accessible through Gene Expression Omnibus (GEO) data sets under accession numbers GSE34369, GSE31650, and GSE28182.
Definition of origin timing and distance to origin
Mcm4-BSs were considered as proxies for ori. An origin was set as firing early when it overlapped with a BrdU domain and was set as late or dormant otherwise. As a result, 445 (out of 1083 origins) were defined as early in wild type, and 500 were defined as early in the rif1Δ strain. Origins were further classified into EE, LL, EL, and LE types.
The fraction of Rif1-BSs overlapping with origins was compared with the proportion of overlap observed with 10,000 repetitions of a data set of 155 or 344 random sequences with origins. The P-value was computed as the number of random fractions of overlap greater than or equal to the observed proportion.
For each Rif1-BS and each origin, a summit was defined as the position of the probe with the highest log ratio. The distance from each Rif1 summit to the origin summits was re-evaluated so that each Rif1-BS was paired with its closest origin. The random distance distribution was estimated with the distance of 100 data sets of 155 or 344 random points to their closest origin summit.
Motif detection
The occurrences of the LCS consensus motif RKKGGGGGAW (where R represents G or A, K represents G or T, and W represents A or T) (Yompakdee and Huberman 2004) was searched on the genome using regular expressions.
Then, sequences of Rif1-BSs were searched for enriched motifs with MEME (from the MEME suite; threshold P-value 1 × 10−4) (Bailey et al. 2009), separating BSs according to the class of their closest origin (EE, EL, LE, and LL). The following motif was found to be overrepresented in BSs close to LE and LL origins: CNWWGTGGGGG (with W being A or T, and N being any base). This potentially new motif was further validated by searching it with FIMO (from the MEME suite) (Bailey et al. 2009) using an E-value threshold at 1 × 10−4 and a FDR threshold (Q-value) of 1%.
Acknowledgments
We thank Naoko Kakusho and Michie Shimmoto for excellent technical assistance in some of the experiments. We thank all of the members of the laboratory for helpful discussion. This work was supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by the Takeda Science Foundation, and by the Astellas Foundation for Research on Metabolic Disorders (to H.M.). K.S. is supported by Grant-in-Aid for Scientific Research (S). C.R.G. is supported by Promotion of Independence for Young Investigators.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.178491.111.
References
- Aladjem MI 2007. Replication in context: Dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet 8: 588–600 [DOI] [PubMed] [Google Scholar]
- Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM 2004. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol 24: 4769–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Bodén M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS 2009. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 doi: 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Moazed D 2007. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol 14: 1041–1048 [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747 [DOI] [PubMed] [Google Scholar]
- Cosgrove AJ, Nieduszynski CA, Donaldson AD 2002. Ku complex controls the replication time of DNA in telomere regions. Genes Dev 16: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM 1999. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell 4: 983–993 [DOI] [PubMed] [Google Scholar]
- Droit A, Cheung C, Gottardo R 2010. rMAT—an R/Bioconductor package for analyzing ChIP–chip experiments. Bioinformatics 26: 678–679 [DOI] [PubMed] [Google Scholar]
- Hardy CF, Sussel L, Shore D 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev 6: 801–814 [DOI] [PubMed] [Google Scholar]
- Hayano M, Kanoh Y, Matsumoto S, Masai H 2011. Mrc1 marks early-firing origins and coordinates timing and efficiency of initiation in fission yeast. Mol Cell Biol 31: 2380–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Katou Y, Itoh T, Tazumi A, Yamada Y, Takahashi T, Nakagawa T, Shirahige K, Masukata H 2007. Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J 26: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MT, Takahashi TS, Nakagawa T, Nakayama J, Masukata H 2009. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat Cell Biol 11: 357–362 [DOI] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bahler J, Nurse P 2006. Genome-wide characterization of fission yeast DNA replication origins. EMBO J 25: 5171–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP 2011. Eukaryoticorigin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 146: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Laroche T, Raghuraman MK, Gasser SM 2001. The positioning and dynamics of origins of replication in the budding yeast nucleus. J Cell Biol 152: 385–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Fukunaga K, Sugimoto K 2009. Rif1 and rif2 inhibit localization of tel1 to DNA ends. Mol Cell 33: 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Gilbert DM 2009. Replication timing as an epigenetic mark. Epigenetics 4: 93–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11: 1624–1630 [DOI] [PubMed] [Google Scholar]
- Kim SM, Huberman JA 2001. Regulation of replication timing in fission yeast. EMBO J 20: 6115–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Dubey DD, Huberman JA 2003. Early-replicating heterochromatin. Genes Dev 17: 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SR, Viggiani CJ, Tavare S, Aparicio OM 2009. Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev 23: 1077–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Blackburn EH 2004. Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol 24: 10857–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian HY, Robertson ED, Hiraga S, Alvino GM, Collingwood D, McCune HJ, Sridhar A, Brewer BJ, Raghuraman MK, Donaldson AD 2011. The effect of Ku on telomere replication time is mediated by telomere length but is independent of histone tail acetylation. Mol Biol Cell 22: 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Matsui E, You Z, Ishimi Y, Tamai K, Arai K 2000. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a critical threonine residue of Cdc7 by Cdks. J Biol Chem 275: 29042–29052 [DOI] [PubMed] [Google Scholar]
- Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, Kim JM, Ishii A, Tanaka T, Kobayashi T, et al. 2006. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem 281: 39249–39261 [DOI] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M 2010. Eukaryotic chromosome DNA replication: Where, when, and how? Annu Rev Biochem 79: 89–130 [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Ogino K, Noguchi E, Russell P, Masai H 2005. Hsk1–Dfp1/Him1, the Cdc7–Dbf4 kinase in Schizosaccharomyces pombe, associates with Swi1, a component of the replication fork protection complex. J Biol Chem 280: 42536–42542 [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Hayano M, Kanoh Y, Masai H 2011. Multiple pathways can bypass the essential role of fission yeast Hsk1 kinase in DNA replication initiation. J Cell Biol 195: 387–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Cooper JP 2003. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol Cell 11: 303–313 [DOI] [PubMed] [Google Scholar]
- Partridge JF, Borgstrom B, Allshire RC 2000. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev 14: 783–791 [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Brewer BJ, Fangman WL 1997. Cell cycle-dependent establishment of a late replication program. Science 276: 806–809 [DOI] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V 2006. Genome-wide analysis of Polycomb target in Drosophila melanogaster. Nat Genet 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM 2007. Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Tecklenburg M, Pierce A 2002. The mcm5-bob1 bypass of Cdc7p/Dbf4p in DNA replication depends on both Cdk1-independent and Cdk1-dependent steps in Saccharomyces cerevisiae. Genetics 161: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B 2006. Cdc7–Dbf4 phosphorylates Mcm proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell 24: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618–621 [DOI] [PubMed] [Google Scholar]
- Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T 2004. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev 18: 2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith HA, Brown GW, Forsburg SL 2000. Schizosaccharomyces pombe Hsk1p is a potential cds1p target required for genome integrity. Mol Cell Biol 20: 7922–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda DY, Dutta A 2005. DNA replication and progression through S phase. Oncogene 24: 2827–2843 [DOI] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M 2002. Histone acetylation regulates the time of replication origin firing. Mol Cell 10: 1223–1233 [DOI] [PubMed] [Google Scholar]
- Voineagu I, Surka CF, Shishkin AA, Krasilnikova MM, Mirkin SM 2009. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat Struct Mol Biol 16: 226–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter JC 2000. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J Biol Chem 275: 39773–39778 [DOI] [PubMed] [Google Scholar]
- Wu PY, Nurse P 2009. Establishing the program of origin firing during S phase in fission yeast. Cell 136: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YJ, Davenport M, Kelly TJ 2006. Two-stage mechanism for activation of the DNA replication checkpoint kinase Cds1 in fission yeast. Genes Dev 20: 990–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuuchi H, Yamada Y, Uchida T, Sunathvanichkul T, Nakagawa T, Masukata H 2006. Ordered assembly of Sld3, GINS and Cdc45 is distinctly regulated by DDK and CDK for activation of replication origins. EMBO J 25: 4663–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yompakdee C, Huberman JA 2004. Enforcement of late replication origin firing by clusters of short G-rich DNA sequences. J Biol Chem 279: 42337–42344 [DOI] [PubMed] [Google Scholar]
- Zhao H, Tanaka K, Nogochi E, Nogochi C, Russell P 2003. Replication checkpoint protein Mrc1 is regulated by Rad3 and Tel1 in fission yeast. Mol Cell Biol 23: 8395–8403 [DOI] [PMC free article] [PubMed] [Google Scholar]