SMARCAL1 is an annealing helicase that helps maintain genomic integrity during DNA replication. Mutations in this protein cause SIOD disease. Using a combination of biochemical and structural approaches, Cortez and colleagues define the mechanism of SMARCAL1 action. They find that SMARCAL1 is a component of active replisomes and that it catalyzes Holliday junction migration and fork reversal. Mutations that cause SIOD disease lack these remodeling actives. SMARCAL1 has a vital role in surveying replication forks for damage and in remodeling forks to promote repair and restart.
Keywords: DNA repair, HARP, Holliday junction, fork reversal, SIOD, SAXS
Abstract
SMARCAL1 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A-like1) maintains genome integrity during DNA replication. Here we investigated its mechanism of action. We found that SMARCAL1 travels with elongating replication forks, and its absence leads to MUS81-dependent double-strand break formation. Binding to specific nucleic acid substrates activates SMARCAL1 activity in a reaction that requires its HARP2 (Hep-A-related protein 2) domain. Homology modeling indicates that the HARP domain is similar in structure to the DNA-binding domain of the PUR proteins. Limited proteolysis, small-angle X-ray scattering, and functional assays indicate that the core enzymatic unit consists of the HARP2 and ATPase domains that fold into a stable structure. Surprisingly, SMARCAL1 is capable of binding three-way and four-way Holliday junctions and model replication forks that lack a designed ssDNA region. Furthermore, SMARCAL1 remodels these DNA substrates by promoting branch migration and fork regression. SMARCAL1 mutations that cause Schimke immunoosseous dysplasia or that inactivate the HARP2 domain abrogate these activities. These results suggest that SMARCAL1 continuously surveys replication forks for damage. If damage is present, it remodels the fork to promote repair and restart. Failures in the process lead to activation of an alternative repair mechanism that depends on MUS81-catalyzed cleavage of the damaged fork.
SMARCAL1 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A-like1), also known as DNA-dependent ATPase A and HARP (Hep-A-related protein), is a member of the SNF2 family of ATPases (Flaus et al. 2006). Many of these proteins use the energy of ATP hydrolysis to translocate along DNA and thereby remodel DNA structures or DNA–protein interactions. They function in many cellular processes, including transcription, DNA replication, and DNA repair.
Biallelic mutations in SMARCAL1 cause the human disease Schimke immunoosseous dysplasia (SIOD) (Boerkoel et al. 2002). SIOD symptoms commonly include skeletal dysplasia, T-cell immunodeficiency, and kidney failure (Boerkoel et al. 2000). At the cellular level, SMARCAL1 deficiency causes increased DNA replication-associated damage (Bansbach et al. 2009, 2010; Postow et al. 2009; Yuan et al. 2009) and sensitizes cells to DNA-damaging agents that inhibit DNA replication (Bansbach et al. 2009; Ciccia et al. 2009; Yuan et al. 2009). SMARCAL1 localizes to damaged replication factories via an interaction with the ssDNA-binding protein replication protein A (RPA) (Bansbach et al. 2009; Ciccia et al. 2009; Yuan et al. 2009; Yusufzai et al. 2009), and this interaction is essential for its genome maintenance function (Bansbach et al. 2009; Yuan et al. 2009). SMARCAL1 is phosphorylated by checkpoint kinases in response to DNA damage (Bansbach et al. 2009; Postow et al. 2009). SMARCAL1 mutants derived from SIOD patients fail to rescue the genome maintenance defects caused by SMARCAL1 deficiency (Bansbach et al. 2009, 2010; Yuan et al. 2009). Thus, SMARCAL1 acts at damaged replication forks to maintain genome stability, and defects in this activity may underlie at least some of the phenotypes associated with SIOD (Bansbach et al. 2010).
The mechanism of how SMARCAL1 acts to repair damaged forks remains largely unknown. Biochemically, SMARCAL1 can bind to DNA that contains single- and double-stranded regions such as forks and DNA hairpins (Muthuswami et al. 2000; Yusufzai and Kadonaga 2008). DNA binding activates its ATPase activity, and this activity promotes DNA single-strand annealing even in the presence of RPA (Yusufzai and Kadonaga 2008). The N-terminal RPA-binding domain of SMARCAL1 is not necessary for this DNA strand-annealing activity (Bansbach et al. 2009; Yusufzai et al. 2009), but patient-derived mutants lack this function. The molecular basis for this activity may not be simply translocation along dsDNA, since the related protein RAD54 cannot perform this function despite being a robust translocase (Yusufzai and Kadonaga 2008).
SMARCAL1 is a multidomain protein. The ATPase domain, which lies in the C-terminal half of the protein, is split into two regions of primary amino acid sequence by a 115-amino-acid linker sequence. The N-terminal half of the protein contains a highly sequence conserved RPA-binding domain (Bansbach et al. 2009; Ciccia et al. 2009; Postow et al. 2009; Yuan et al. 2009; Yusufzai et al. 2009), a 200-amino-acid region of low sequence conservation without predicted domain structure, and two HARP domains. The HARP domains are 55 amino acids in length with high sequence similarity but unknown function and structure. They are separated by 40 amino acids, and the second HARP domain is linked to the ATPase domain by an additional 47 amino acids.
Fusing the HARP domains to the ATPase domain of the SNF2 proteins BRG1 or HELLS is sufficient to reconstitute DNA-dependent ATPase and annealing helicase activities, suggesting that the HARP domains are important determinants of the SMARCAL1 enzyme specificity (Ghosal et al. 2011). Paradoxically, the closest homolog of SMARCAL1 in humans, annealing helicase 2 (AH2, also known as ZRANB3), also has annealing helicase activity despite a different domain structure and no unambiguous HARP domains (Yusufzai and Kadonaga 2010).
In this study, we took genetic, biochemical, and biophysical approaches to understand how SMARCAL1 functions to maintain genome integrity. We found that SMARCAL1 travels with at least some elongating replication forks, and the MUS81 structure-specific endonuclease cleaves damaged forks in SMARCAL1-deficient cells. The HARP2 domain is essential for DNA binding, and both biochemical and small-angle X-ray scattering (SAXS) data indicate that the HARP2+SNF2 domains provide the minimal enzymatic unit. The HARP domain resembles the DNA-binding domain of the PUR-α protein and has limited ability to bind DNA on its own. Surprisingly, we found that SMARCAL1 can bind three-way and four-way DNA structures and model replication forks. Furthermore, SMARCAL1 branch-migrates the four-way junction and catalyzes extensive fork regression of model replication forks. These data provide mechanistic insight into how SMARCAL1 functions and suggest that it remodels stalled replication forks through fork regression and branch migration to promote replication fork restart and prevent replication-associated DNA double-strand breaks.
Results
SMARCAL1 is present at DNA replication forks during an unperturbed S phase and prevents MUS81-dependent double-strand breaks
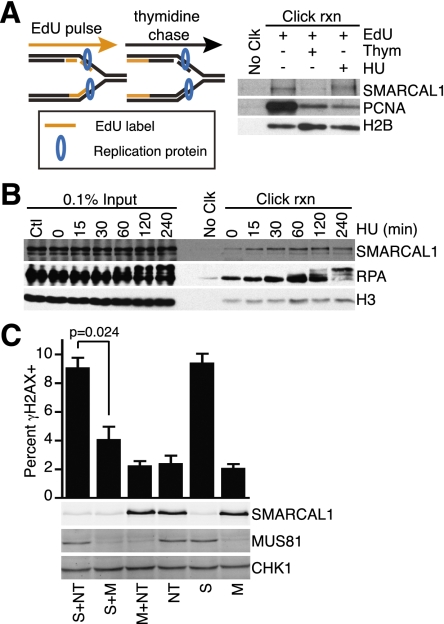
Previous analyses indicated that SMARCAL1 localizes to nuclear foci that colocalize with replisomes in response to agents that induce replication stress (Bansbach et al. 2009; Ciccia et al. 2009; Postow et al. 2009; Yuan et al. 2009; Yusufzai et al. 2009). This localization is dependent on an interaction with the replisome protein RPA. Silencing SMARCAL1 using RNAi causes elevated levels of γH2AX in replicating cells (Bansbach et al. 2009; Postow et al. 2009; Yuan et al. 2009). To determine whether SMARCAL1 actually is a component of active replisomes, we used the iPOND procedure (Sirbu et al. 2011) to purify active and stalled replication forks. SMARCAL1 is purified with nascent, 5-ethynyl-2′-deoxyuridin (EdU)-labeled DNA at elongating replication forks even when replication is not perturbed (Fig. 1A). It is not purified with the EdU-labeled DNA once the labeled DNA segment is no longer adjacent to the fork (after a chase in medium lacking EdU), indicating that it travels with at least some moving replisomes. As expected, SMARCAL1 is also found at forks stalled with hydroxyurea (HU) (Fig. 1A,B), and its mobility on SDS-PAGE gels is altered in these circumstances due to phosphorylation by checkpoint kinases (Bansbach et al. 2009).
Figure 1.
SMARCAL1 acts at replication forks to prevent MUS81-catalyzed double-strand breaks. (A) Cells were labeled with EdU for 10 min, the EdU was removed, and thymidine was added for 20 min or HU was added for 3 h prior to purifying the nascent DNA–protein complexes using the iPOND procedure. (B) EdU-labeled cells were treated with 2 mM HU for the indicated lengths of time prior to performing iPOND. The “No Clk” controls in A and B are samples treated with EdU only, but no biotin-azide was added during the click reaction. (C) U2OS cells were transfected with siRNAs targeting SMARCAL1 (S), MUS81 (M), or nontargeting (NT) as indicated. Three days after transfection, the cells were either stained with antibodies to γH2AX or harvested for immunoblotting with the indicated antibodies. The percentage of cells staining positive for γH2AX was determined by immunofluorescent imaging from three independent experiments. Cells with >10 foci were counted as positive. Error bars are the standard deviation (SD; n = 3).
The MUS81 endonuclease cleaves some blocked and damaged replication forks, generating a double-strand break and initiating recombination-based repair mechanisms (Osman and Whitby 2007). Therefore, we hypothesized that the high level of γH2AX found in SMARCAL1 silenced cells could be due to double-strand breaks catalyzed by MUS81. To address this question, we measured the proportion of cells containing γH2AX after SMARCAL1 and/or MUS81 depletion. As expected, silencing SMARCAL1 caused an induction of γH2AX, while silencing MUS81 had no effect (Fig. 1C). MUS81 silencing prevented γH2AX induction in SMARCAL1 silenced cells without significantly altering the efficiency of SMARCAL1 silencing (Fig. 1C). Thus, γH2AX induction after SMARCAL1 depletion is MUS81-dependent.
SMARCAL1 binds a wide variety of DNA substrates that combine ssDNA and dsDNA
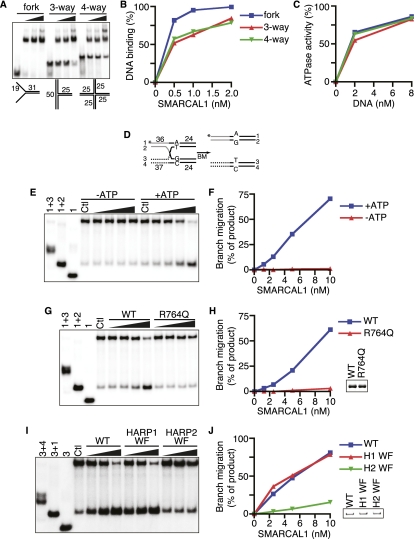
Our results suggest that SMARCAL1 either processes or prevents the formation of MUS81 substrates. Little is known about SMARCAL1 substrate specificity other than it prefers to bind DNA with both single- and double-stranded characteristics rather than ssDNA or dsDNA (Supplemental Fig. 1A; Muthuswami et al. 2000; Yusufzai and Kadonaga 2008), and its ATPase activity is activated upon DNA binding. To clarify the DNA determinants that mediate SMARCAL1 DNA binding and activation, we investigated a broad range of possible DNA substrates. We first evaluated how long the ssDNA arms of a fork need to be and found that significant SMARCAL1 binding is observable even with a fork length of only 5 nucleotides (nt) per arm (Supplemental Fig. 1B,C). Increasing the arm lengths beyond 5 nt increases the binding affinity. We also observe a second DNA–protein complex forming when the ssDNA region is lengthened to 20 nt or more. The second, higher-molecular-weight complex may contain more than one SMARCAL1 molecule.
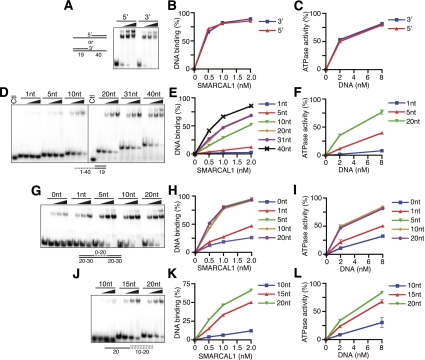
We next varied the length of one of the ssDNA arms while keeping the other constant and found that the length of the second arm did not influence binding affinity (Supplemental Fig. 1D,E). In fact, SMARCAL1 bound equivalently to a fork and an ssDNA overhang substrate. Both DNA substrates stimulated SMARCAL1 ATPase activity as well (Supplemental Fig. 1F). Furthermore, DNA substrates with either a 5′ or 3′ recessed end bind and stimulate SMARCAL1 ATPase activity equivalently (Fig. 2A–C).
Figure 2.
Characterization of the DNA-binding and DNA-stimulated ATPase activities of full-length SMARCAL1. (A,D,G,J) Increasing amounts of SMARCAL1 were incubated with the indicated oligonucleotide substrates prior to polyacrylamide gel electrophoresis. The control (Ctl) in D is an unhybridized single-stranded oligonucleotide. (B,E,H,K) Quantitation of a representative DNA-binding experiment. (C,F,I,L) Increasing amounts of DNA substrate were added to SMARCAL1, and ATPase activity was measured as the percentage of ATP hydrolyzed. Error bars represent the mean ± SD from three independent experiments. In cases in which no error bars are visible, the SD is smaller than the symbol size. The sequences of the oligonucleotides are listed in Supplemental Table 1, and a description of which oligonucleotides were used in each experiment is presented in Supplemental Table 2.
At a replication fork, the free 5′ end of the nascent nucleic acid on the lagging stand template would consist of a short RNA primer rather than DNA. To test whether SMARCAL1 can bind and be activated on the lagging strand, we examined a nucleic acid substrate that mimics this chimeric nucleic acid structure. A RNA–DNA primer substrate bound and stimulated SMARCAL1 equivalently to the DNA–DNA substrate (Supplemental Fig. 1G–I).
Next, we assessed how the length of ssDNA alters SMARCAL1-binding affinity. Five nucleotides are sufficient to allow some binding and elicit significant ATPase activity (Fig. 2D–F). SMARCAL1 binding and ATPase activity increase as the length of the ssDNA increases. SMARCAL1 also binds and is stimulated efficiently by a gapped DNA substrate. Maximum binding and activation require only a five single-stranded nucleotide gap, and even a nick can elicit some activity (Fig. 2G–I; Supplemental Fig. 1J,K). When a bubble replaces the gap, increasing the length of the mismatched nucleotides to 16 significantly increases affinity (Supplemental Fig. 1L,M). Thus, the length of ssDNA needed for optimal binding and activation of SMARCAL1 is shortest when it is presented in the context of a gap.
We also investigated how the length of dsDNA affects binding and ATPase stimulation of SMARCAL1. Optimal SMARCAL1 binding and ATPase activation requires 20 dsDNA nucleotides (Fig. 2J–L). Greater dsDNA lengths yield no further improvement in SMARCAL1 affinity (data not shown). Fifteen nucleotides of dsDNA on either side of a 5-nt gap are sufficient to elicit maximal SMARCAL1 binding (Supplemental Fig. 1N,O).
Taken together, these results show that SMARCAL1 binds and is activated by any nucleic acid structure that contains both single- and double-stranded regions, including an RNA–primer template. The optimal length of ssDNA that elicits binding depends on the structural context of the DNA, with 5 nt being sufficient for a gap and longer lengths promoting better binding to a forked or single-stranded overhang substrate. The optimal length of dsDNA is ∼15 nt. Finally, the dsDNA and ssDNA must be within the same molecule, since adding these separately to SMARCAL1 does not elicit any binding (Supplemental Fig. 1A).
HARP2 but not HARP1 is required for SMARCAL1 DNA-binding, ATPase, and annealing helicase activities
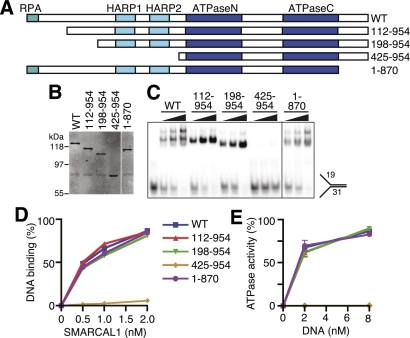
To understand how SMARCAL1 binds DNA, we examined the affinity of a series of truncated SMARCAL1 proteins for a forked DNA substrate (Fig. 3A). While deletion of the first 198 and the last 84 amino acids had no effect on SMARCAL1 DNA binding, deletion of the first 424 amino acids containing the HARP domains severely compromises the DNA-binding and ATPase activities of SMARCAL1 (Fig. 3B–E).
Figure 3.
The SMARCAL1 N terminus containing the HARP domains is necessary for DNA-binding and ATPase activity. (A) Diagram of the SMARCAL1 proteins used to identify domains required for function. Wild type (WT) in all figures is full-length SMARCAL1. (B) Overexpressed SMARCAL1 proteins were purified from HEK-293T cells and examined on an SDS-PAGE gel by immunoblotting. (C,D) Increasing amounts of purified SMARCAL1 proteins were incubated with the forked DNA substrate to measure DNA binding. (E) Increasing amounts of forked DNA were added to the SMARCAL1 fragments to measure DNA-stimulated ATPase activity. Error bars represent the mean ± SD from three independent experiments. In cases in which no error bars are visible, the SD is smaller than the symbol size. The DNA substrates corresponding to each symbol and line color are the same in D and E.
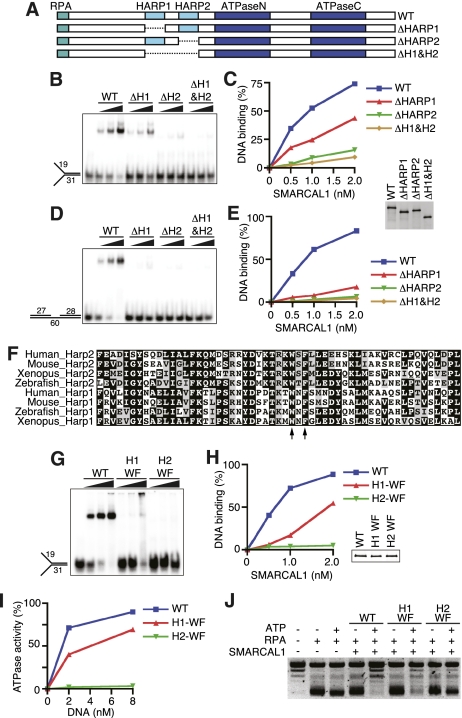
These results led us to hypothesize that the HARP domains may be essential for SMARCAL1 DNA binding. To test this hypothesis, we assessed the behavior of a series of HARP domain mutants (Fig. 4A). SMARCAL1 lacking the first HARP domain (ΔHARP1) binds to and is activated by a forked DNA substrate, although with slightly reduced affinity compared with wild-type SMARCAL1 (Fig. 4B,C; Supplemental Fig. 2A). In contrast, deleting the second HARP domain alone or in combination with the first HARP domain (SMARCAL1-ΔHARP2 and ΔHARP1+2) severely attenuated both DNA binding and ATPase activation. The effects of the deletions were even more severe when assayed with a 5-nt, single-stranded gap substrate (Fig. 4D,E).
Figure 4.
The HARP2 domain of SMARCAL1 is required for annealing helicase activity. (A) Diagram of the SMARCAL1 HARP domain deletion mutants purified after overexpression in HEK-293T cells. DNA binding was measured with increasing concentrations of a forked DNA substrate (B,C) or 5-nt gap DNA substrate (D,E). (F) Sequence alignment of the HARP1 and HARP2 domains of human, mouse, Xenopus laevis, and zebrafish SMARCAL1. The arrows point to the two residues mutated in the WF mutants used in G–J. (G,H) Forked DNA binding of the wild type and SMARCAL1 HARP-WF mutants purified from baculovirus-infected insect cells. Note that the HARP1-WF mutant reproducibly shifted much of the DNA substrate into the well of the gel at higher concentrations of protein. (I) Increasing amounts of forked DNA were added to the SMARCAL1 mutants to measure DNA-stimulated ATPase activity. Error bars represent the mean ± SD from three independent experiments. In cases in which no error bars are visible, the SD is smaller than the symbol size. (J) Annealing helicase activities of SMARCAL1 wild-type and mutant proteins. The concentration of the SMARCAL1 proteins in this assay is 15 nM. The insets in C and H are immunoblots confirming that equal concentrations of SMARCAL1 proteins were used.
To confirm the deletion results, we generated point mutants in HARP1 and HARP2. The HARP domains are evolutionarily conserved (Fig. 4F). We mutated two of the invariant residues within each domain to alanine (HARP1 W277A/F279A and HARP2 W372A/F379A). These mutants exhibit DNA-binding and ATPase activity similar to the corresponding complete deletion of the domain (Fig. 4G–I). Interestingly, we found that the decreased DNA-binding and ATPase activity of the HARP1-WF mutant yielded only a slight impairment of SMARCAL1 annealing helicase activity, while mutation of the HARP2 domain completely abolished the ability of SMARCAL1 to anneal an RPA-coated plasmid substrate (Fig. 4J). The complete deletion of HARP1 also had no effect on the SMARCAL1 annealing helicase activity (Supplemental Fig. 2B). These results suggest that HARP2 is critical for the DNA-binding, ATPase, and annealing helicase activities of SMARCAL1. HARP1 may have a supporting role in facilitating SMARCAL1 function.
Finally, we asked whether the HARP domains themselves have any DNA-binding activity. We found that a SMARCAL1 fragment encompassing both HARP domains (amino acids 198–425) is sufficient to bind forked DNA, albeit with much lower affinity than the full-length protein (Supplemental Fig. 3A–C). The HARP domain–DNA complex did not migrate as a discrete band in the electrophoretic mobility shift assay; however, we were able to supershift the DNA–protein complex with an antibody that recognizes the recombinant HARP1+2 protein fragment, confirming the complex was not due to a contaminant in the protein purification.
The HARP2-ATPase constitutes a structural core motor domain
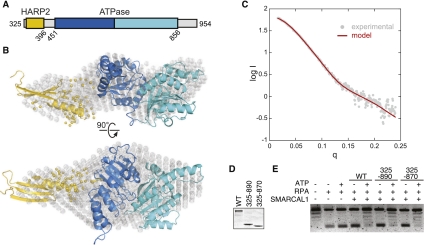
Our biochemical results demonstrate the importance of the HARP2 domain in SMARCAL1 function. To gain mechanistic insight into how SMARCAL1 might use this novel domain, SAXS experiments were performed to determine the spatial arrangement of the HARP2 and ATPase domains in solution (Fig. 5A). Limited proteolysis of the full-length protein purified from insect cells revealed a proteolytically resistant fragment consisting of the HARP2-ATPase regions (Supplemental Fig. 4A). Kratky analysis of SMARCAL1(325–954) revealed parabolic features, suggesting that the protein is globular with distinct domains (Supplemental Fig. 4B,C). The radius of gyration (rg) obtained from the Guinier region was 33.0 ± 0.3 Å (Supplemental Fig. 4D), indicating that the 75-kDa protein is elongated when compared with glucose isomerase, a spherical protein at 173 kDa with a similar Rg of 32 Å.
Figure 5.
HARP2-ATPase constitutes an active structural core domain. (A) Construct used for SAXS measurements. (B) The SAXS model constructed from HARP2 (residues 325–396, gold) and ATPase (residues 451–856, blue) homology models superimposed on the ab initio molecular envelope determined by GASBOR (gray spheres). The yellow spheres represent region 397–450 modeled in BUNCH. (C) The theoretical scattering curve (red) from the model shown in B is superimposed on the experimental SAXS data (gray circles) with a goodness of fit χ = 1.5. Coomassie-stained gel of wild-type or truncated SMARCAL1 proteins (D) used in an annealing helicase assay (E).
SAXS data provide complete structural information and can be used to distinguish between different conformations of a high-resolution model or build a complete atomistic model from known domains (Rambo and Tainer 2010). Therefore, we used the SAXS data of SMARCAL1 and homology models of both the HARP2 and ATPase domains to determine the solution state of the protein. To date, there are no known structural homologs of the HARP domain. However, we discovered by sequence–structure comparison (Shi et al. 2001) that there is good agreement between the predicted secondary structural elements of the HARP domains with tandem PUR repeats observed in the structure of the purine-rich element-binding protein PUR-α (Supplemental Fig. 5; Graebsch et al. 2009). PUR repeats are ∼140-residue motifs consisting of anti-parallel β-β-β-β-α topology that bind ssDNA and dsDNA and thus provide a reasonable structural model for the HARP domains. A model of the core ATPase domain was also created based on the crystal structure of Sulfolobus sulfotaricus (Sso) Rad54, which shares 23% sequence identity and 58% overall similarity with SMARCAL1 (Supplemental Fig. 6).
Preliminary normal mode analysis (Suhre and Sanejouand 2004) was performed on the core ATPase domain to produce a family of alternative conformations. Each conformation was then combined with the HARP2 model for partial ab initio modeling using a simulated annealing search algorithm. The models converged into an elongated structure that was independently validated by the close resemblance to the three-dimensional (3D) molecular envelope generated from the SAXS data using GASBOR (Fig. 5B), and the remarkable agreement between the experimental scattering curve and the theoretical curve calculated from the docking model (Fig. 5C). The resulting HARP2-ATPase model revealed that the HARP2 and ATPase motifs form one continuous domain in the absence of DNA, suggesting that their association constitutes a structural and functional core domain necessary to drive translocation. To test this idea, we assayed whether the HARP2-ATPase protein is sufficient to catalyze strand annealing. Indeed, SMARCAL1(325–870) and SMARCAL1(325–890) are both efficient ATP-dependent annealing helicases (Fig. 5D,E).
SMARCAL1 can bind and branch-migrate a four-way junction
The DNA-binding activities of SMARCAL1 characterized thus far suggest that SMARCAL1 may have dsDNA- and ssDNA-binding surfaces. Combined with the energy of ATP hydrolysis, SMARCAL1 may translocate along the DNA in a way that leads to single-strand annealing. To determine whether these properties could yield any other enzymatic consequences, we expanded our search for SMARCAL1 substrates to more complex DNA structures, including three-way and four-way Holliday junctions. Surprisingly, despite lacking any designed ssDNA regions, SMARCAL1 could bind these DNA substrates with only slightly reduced affinity compared with a fork substrate (Fig. 6A,B). Furthermore, both DNA substrates activated the SMARCAL1 ATPase (Fig. 6C).
Figure 6.
SMARCAL1 binds and branch-migrates Holliday junctions. The ability of SMARCAL1 to bind (A,B) and be activated (C) by forked, three-way, and four-way Holiday junctions was compared. The DNA substrates corresponding to each symbol and line color are the same in B and C. (D) Four-way branch migration substrate used in E–J. The 32P-labeled DNA strand (#1) for the experiments shown in E–H is indicated with an asterisk. Strand #3 was labeled for the experiment shown in I and J. (E) Increasing amounts of SMARCAL1 were incubated with the four-way branch migration substrate in the absence or presence of ATP as indicated. (G,I) Increasing amounts of wild-type (WT), R764Q, HARP1-WF, or HARP2-WF SMARCAL1 proteins were incubated with the DNA substrate in the presence of ATP. The first three lanes in E, G, and I are size standards generated by annealing the indicated oligonucleotides. The control (Ctl) samples are the annealed branch migration substrate in the absence of recombinant protein. (F,H,J) Quantitation of the reactions from E, G, and I, respectively. The amount of product in the control reactions (from spontaneous branch migration) was set at zero in each experiment, and all other samples are measured relative to the control sample. All reactions in E–J were performed for 20 min prior to termination and gel electrophoresis to characterize the products. The insets in H and J are Coomassie-stained gels confirming that equal concentrations of SMARCAL1 proteins were used.
Given that these structures bind SMARCAL1 and stimulate its ATPase activity, we asked whether SMARCAL1 could also induce branch migration like Rad54 (Bugreev et al. 2006). We prepared a synthetic Holliday junction consisting of two homologous and two heterologous arms, similar to those used in previous branch migration studies (Fig. 6D; Gari et al. 2008b). Indeed, SMARCAL1 catalyzed branch migration in an ATP-dependent manner (Fig. 6E,F). As expected, the SIOD patient-derived ATPase-defective mutant (R764Q) failed to promote branch migration despite having the ability to bind DNA (Fig. 6G,H; Yusufzai and Kadonaga 2008). To test the importance of the HARP domains in this process, we examined the branch migration properties of HARP1-WF and HARP2-WF SMARCAL1 mutants. While the HARP1 mutant was able to branch-migrate the Holliday junction as efficiently as the wild-type protein, the HARP2 mutant had severely attenuated activity (Fig. 6I,J).
SMARCAL1 can bind and branch-migrate a replication fork
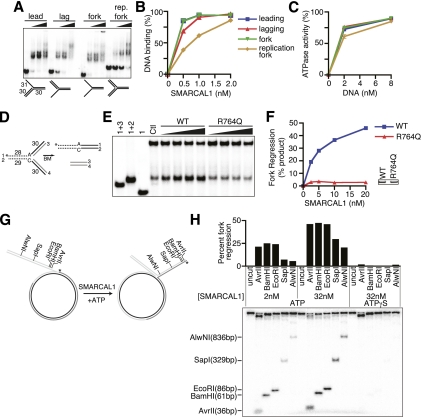
Previous studies indicate that SMARCAL1 acts at stalled replication forks but may not have an essential function in homology-directed double-strand break repair. Double-strand breaks are only thought to form in normal cells at persistently stalled forks (Petermann et al. 2010; Sirbu et al. 2011). Thus, we investigated whether SMARCAL1 could bind and process other branched structures that might exist at a transiently stalled fork. Specifically, we compared SMARCAL1 affinity to model forks with no nascent DNA strands, a leading strand, a lagging strand, or both. Strikingly, we found that SMARCAL1 binds to and is activated by each of these structures (Fig. 7A–C). To determine whether SMARCAL1 can catalyze remodeling of these replication fork structures, we prepared a substrate to monitor fork regression (Fig. 7D; Gari et al. 2008b). SMARCAL1 catalyzed displacement of the two “nascent” DNA strands and annealing of the parental strands (Fig. 7E,F). Again, the SIOD patient-derived R764Q mutation eliminated this activity.
Figure 7.
SMARCAL1 catalyzes fork regression of model replication forks. (A,B) Increasing amounts of SMARCAL1 were incubated with the indicated substrates to measure DNA binding. (C) ATPase activity of SMARCAL1 was measured in the presence of increasing amounts of leading, lagging, fork, and replication fork substrate. Symbols and line colors correspond to the same substrates as in B. Error bars represent the mean ± SD from three independent experiments. In cases in which no error bars are visible, the SD is smaller than the symbol size. (D) Diagram of the model replication fork substrates used to measure fork regression activity in E and F. A single mismatch is present at the fork junction to prevent spontaneous fork migration. The labeled strand (#1) is indicated by an asterisk. (E,F) Increasing amounts of SMARCAL1 (wild type [WT]) or R764Q SMARCAL1 were incubated with the annealed substrate for 20 min, the reaction was terminated, and products were separated by gel electrophoresis for analysis. The first three lanes in E are size standards generated by annealing the indicated oligonucleotides. The control (Ctl) sample is the annealed fork regression substrate in the absence of recombinant protein. The amount of product in the control reaction (from spontaneous regression of the model replication fork substrate) was set at zero in each experiment, and all other samples are measured relative to the control sample. The inset in F is a Coomassie-stained gel confirming that equal concentrations of SMARCAL1 proteins were used. (G) Diagram of the annealed gapped plasmid substrate used to measure SMARCAL1-catalyzed fork regression in H. The 32P-labeled DNA end is indicated with an asterisk. (H) Restriction digests with the indicated enzymes were completed following incubation of the plasmid substrate with the indicated concentrations of SMARCAL1 in the presence of ATP or ATPγS. The liberated, 32P-labeled DNA fragment was visualized on a polyacrylamide gel. The extent of fork regression was calculated as the amount of liberated fragment compared with the total radioactivity in the reaction. A representative experiment is shown.
SMARCAL1 does not possess any helicase activity (Yusufzai and Kadonaga 2008), so it is unlikely that it could unwind the nascent strands before annealing both parental and both nascent strands together. To confirm that the SMARCAL1 fork reversal activity is coordinated without the formation of ssDNA intermediates, we labeled the model nascent leading strand of the synthetic replication fork and performed a time-course assay. We found that only a double-stranded product consisting of the two nascent strands is formed without the appearance of any ssDNA intermediates (Supplemental Fig. 7A–C). We conclude that SMARCAL1 processes replication fork structures by coupling unwinding and annealing in a concerted manner to yield fork regression. As expected, the SMARCAL1 HARP1-WF mutant is able to regress the replication fork as efficiently as the wild-type protein, whereas mutations in the SMARCAL1 HARP2 domain eliminate fork regression activity (Supplemental Fig. 7D–F). Thus, HARP2 but not HARP1 is critical for SMARCAL1 fork regression activity.
Finally, we tested whether SMARCAL1 can catalyze fork regression and sustained migration on a plasmid-sized substrate that more closely models a stalled replication fork. We created a joint molecule by annealing gapped plasmids (Fig. 7G). This substrate mimics a stalled fork in which the lagging strand is 14 nt longer than the leading strand (Ralf et al. 2006; Blastyak et al. 2007). The extent of fork regression of this substrate was detected by restriction enzyme digestion to liberate a linear 5′-labeled lagging strand. SMARCAL1 efficiently catalyzed remodeling of this substrate, yielding substantial amounts of a regressed fork corresponding to movement of at least 836 base pairs (bp) (Fig. 7H). This reaction is dependent on the amount of SMARCAL1 added to the reaction and requires ATP hydrolysis, since ATPγS completely blocked remodeling of the substrate.
Discussion
Previous studies by our group and others defined SMARCAL1 as a replication stress response protein that acts to preserve genome integrity during DNA replication (Bansbach et al. 2009; Ciccia et al. 2009; Driscoll and Cimprich 2009; Postow et al. 2009; Yusufzai et al. 2009). Immunofluorescent imaging demonstrated that SMARCAL1 accumulates at damaged replication forks due to its interaction with RPA. We now report that SMARCAL1 associates with active, elongating replisomes, and its absence causes MUS81-dependent DNA damage. Significantly, we found that SMARCAL1 exhibits a much broader range of enzymatic activities than previously recognized, including an ability to promote branch migration of Holliday junctions and fork reversal of model replication forks. Concerted fork regression and branch migration coupled to DNA polymerization provides one mechanism to allow DNA damage bypass and replication restart (Petermann and Helleday 2010). SMARCAL1 depletion does not significantly slow the overall rate of DNA replication but is required for efficient DNA replication restart of stalled or collapsed replication forks (Ciccia et al. 2009). Thus, SMARCAL1 may continuously survey replisomes and promote efficient restart of stalled forks through its fork remodeling activity. In the absence of SMARCAL1, slowed or damaged forks are cleaved by MUS81, perhaps as an alternative mechanism of fork repair.
In addition, our results indicate that all SMARCAL1 activities require the HARP2 and SNF2-like ATPase domains. The HARP2 domain is required for DNA binding, and the HARP2-ATPase domains together form the functional enzymatic unit of SMARCAL1. Significant sequence similarity between the HARP domain and the DNA-binding domain of the PUR proteins combined with our SAXS data allowed us to derive a model of the solution state structure of the SMARCAL1 core enzyme. The HARP2 and ATPase motifs dock together and constitute a structural and functional core necessary to drive ATP-dependent translocation.
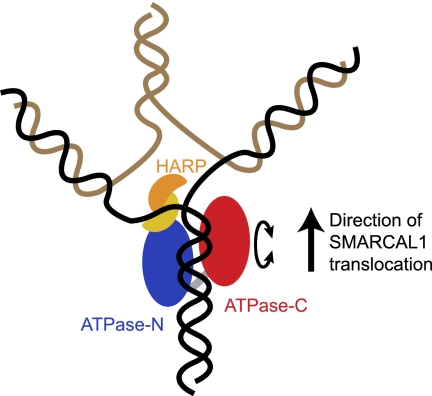
The HARP2 domain likely provides specificity to the action of the ATPase motor domain, thereby converting the energy of ATP hydrolysis into functional strand annealing, branch migration, and fork reversal. This type of activity could be facilitated by insertion of the HARP domain as a kind of wedge at the branch point within these structures (Fig. 8). One possibility supported by our data is that the compact HARP2-ATPase core enzyme contains both dsDNA- and ssDNA-binding surfaces encoded in the ATPase and HARP2 domains, respectively. DNA binding induces a conformational change, promoting ATP hydrolysis and protein translocation. A model for how this could function to promote fork regression is provided by the bacterial RecG protein, which shares some enzymatic activities with SMARCAL1 (Atkinson and McGlynn 2009). Further structural data, including high-resolution structures of SMARCAL1 with a bound DNA substrate, will be required to fully test this hypothesis.
Figure 8.
Model for how the translocase activity of the SMARCAL1 HARP2-ATPase core catalyzes fork regression. Existing structures of SNF2 translocases demonstrate that ATPase-N and ATPase-C lobes are capable of adopting different relative conformations and suggest that such conformational changes (depicted as a circular arrow) in response to the ATPase-binding and hydrolysis cycle may drive translocation along dsDNA (Durr et al. 2005; Thoma et al. 2005; Lewis et al. 2008). The SAXS model shows that the HARP2 domain in SMARCAL1 is physically associated with the ATPase-N lobe and may aid in the specialized annealing activity through ssDNA or junction binding. Translocation displaces the nascent DNA strands, induces fork regression, and promotes junction migration.
In contrast to the HARP2 domain, the HARP1 domain makes a modest contribution to the DNA-binding and ATPase activities of SMARCAL1 and is largely dispensable for its annealing, branch migration, and fork regression functions. While vertebrate SMARCAL1 proteins contain two HARP domains, invertebrate SMARCAL1 proteins contain only a single HARP domain adjacent to the ATPase domain, suggesting that only a single HARP domain is essential for its evolutionarily conserved functions.
Our conclusions about the important function of the HARP2 domain are generally consistent with a recent report that found that the HARP domains are important for the annealing helicase activity of SMARCAL1 (Ghosal et al. 2011). However, the Chen group (Ghosal et al. 2011) reported that deleting both HARP1 and HARP2 together did not impair either DNA-binding or ATPase activity despite eliminating the annealing helicase activity. In contrast, our data with both deletion and point mutants clearly point to a requirement for the HARP2 domain for all SMARCAL1 enzymatic functions. We tested multiple proteins purified from both insect and human cells using several different DNA substrates and always found that the HARP2 domain was critical for DNA binding, ATPase activity, strand annealing, and branch migration. We do not have an explanation for this discrepancy.
The ability of SMARCAL1 to efficiently bind to Holliday junctions and model replication forks that lack ssDNA regions was unexpected, since SMARCAL1 has very little affinity to dsDNA compared with the optimal substrates containing at least 15 nt of dsDNA and 5 nt of ssDNA. One possibility is that SMARCAL1 captures a small amount of these structures as the dsDNA regions near the junction or fork breath to expose ssDNA. Our data indicate that only a small amount of ssDNA or even just a nick is necessary for SMARCAL1 DNA binding when it is in the context of a gap. Likewise, only a small amount may be needed in the context of these more complicated structures. We also observed no significant specificity of human SMARCAL1 for 3′ or 5′ recessed junctions. Furthermore, a 5′ recessed junction containing a model RNA–DNA primer, as would be found during lagging strand replication, efficiently binds and activates SMARCAL1. This contrasts with a previous report that found a preference for a 3′-hydroxyl recessed end (Muthuswami et al. 2000). The origin of this difference may be because the previous report used a fragment of bovine SMARCAL1, whereas we used full-length human SMARCAL1 in our studies.
The ATP-dependent activity of SMARCAL1 to remodel Holliday junctions and replication forks and prevent DNA damage during S phase is reminiscent of the activities of other proteins, including FANCM, WRN, RAD5, BLM, and HLTF (Constantinou et al. 2000; Ralf et al. 2006; Blastyak et al. 2007; Franchitto et al. 2008; Gari et al. 2008a,b; Opresko et al. 2009; Achar et al. 2011). All of these proteins are thought to be recruited to damaged replication forks, but it is unclear whether any travel with active forks like SMARCAL1. In contrast to SMARCAL1, none of these proteins contain a HARP domain or exhibit annealing helicase activity. Instead, most are DNA helicases. Thus, the enzymatic mechanisms by which they remodel replication fork structures are likely to be different. Why there are so many different enzymes that can catalyze similar reactions on DNA is unclear. It is possible that some of these enzymes work coordinately at the same damaged fork. In this regard, it is interesting that the loss of WRN, like SMARCAL1, also causes MUS81-dependent fork cleavage (Franchitto et al. 2008), and we and others have found WRN in SMARCAL1 purifications, suggesting a possible physical interaction (Ciccia et al. 2009; data not shown). Coordination of their enzymatic activities might help remodel damaged forks in cells where many other replisome and repair proteins may be present. However, these proteins must also have distinct functions, since inactivating mutations cause different human diseases.
In summary, our data suggest that SMARCAL1 surveys DNA replication forks. When it detects a problem, it uses its DNA-stimulated ATPase motor to remodel the fork by catalyzing strand annealing, branch migration, and fork reversal to promote efficient fork repair. These activities are encoded within the HARP2-SNF2 ATPase domains, which form a functional enzyme flanked by regulatory sequences. Absence of SMARCAL1 forces the use of alternative fork repair mechanisms that involve MUS81-dependent DNA double-strand breaks.
Materials and methods
Cell culture
HEK-293T and U2OS cells were cultured in DMEM supplemented with 7.5% FBS. Sf9 cells were cultured in Insect XPRESS medium with 7.5% FBS at 27°C.
Antibodies
The antibodies used were as follows: Flag-M2 (Sigma), γH2AX and GAPDH (Millipore), RPA (Bethyl Laboratories), H3 (Abcam), and MUS81 (Novus). The SMARCAL1 antibody was described previously (Bansbach et al. 2009).
Detection of γH2AX
γH2AX foci were detected by indirect immunofluorescent imaging of fixed U2OS cells 72 h after transfection with siRNA as previously described (Lovejoy et al. 2009).
iPOND
The iPOND technique was performed as described previously (Sirbu et al. 2011). Briefly, cells were labeled for 10 min with EdU, then treated with 2 mM HU for increasing amounts of time. Alternatively, after the EdU labeling period, 10 μM thymidine was added to the growth medium for 20 min as a “chase” sample. This concentration of thymidine does not block replication but is sufficient to ensure that no additional EdU is incorporated. After cross-linking with formaldehyde and a click reaction to conjugate biotin to the EdU-labeled nascent DNA, protein–DNA complexes were isolated with streptavidin beads, cross-links were reversed, and the eluted proteins were analyzed by immunoblotting. The “no click” control omitted the biotin-azide during the click reaction.
Protein purification
Flag-SMARCAL1, His-SMARCAL1(325-954), HARP1-WF, and HARP2-WF were purified from baculovirus-infected cells essentially as described previously (Yusufzai and Kadonaga 2008) except that cells were lysed in TNT buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.2 mM PMSF, 1 mg/mL leupeptin, 1 mg/mL aprotinin, and 0.1% Triton X-100. Proteins for structural studies were purified by Ni-NTA affinity, ion exchange, and gel filtration chromatography. To purify SMARCAL1 proteins from human cells, HEK-293T cells were transfected with pLPCX-Flag-HA-SMARCAL1 plasmids using Lipofectamine 2000 (Invitrogen). Seventy-two hours after transfection, the cells were lysed in TNT buffer for 30 min on ice. After high-speed centrifugation, the cleared lysates were incubated with Flag-M2 beads (Sigma) for 3 h at 4°C. The beads were washed three times in wash buffer (TNT buffer containing 0.3 M LiCl) and twice in SMARCAL1 buffer (20 mM HEPES at pH 7.6, 20% glycerol, 0.1 M KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 0.2 mM PMSF, 0.01% IGEPAL CA-630). The bound proteins were eluted in SMARCAL1 buffer containing 0.25 mg/mL Flag peptide on ice, flash-frozen, and stored at −80°C.
DNA-binding, annealing helicase, and ATPase assays
The gel mobility shift assays for DNA-binding, annealing helicase, and SMARCAL1 ATPase assays were performed as described previously (Yusufzai and Kadonaga 2008) with the following modifications. For the gel mobility shift assay, increasing concentrations of purified SMARCAL1 (0, 0.5, 1, 2 nM final concentrations) were combined with radiolabeled oligonucleotide probe (1 nM final concentration) in binding buffer supplemented with 0.2% IGEPAL CA-630. The samples were loaded into a 5% polyacrylamide 0.5× TBE gel (82 × 28.5 cm, 1 mm thick), and subjected to electrophoresis in 0.5× TBE for 2 h and 30 min at 50 V at 4°C. The gels were dried and quantified using a Molecular Imager FX (Bio-Rad). DNA-binding reactions were performed at least twice, and a representative experiment is shown with quantitation. For the annealing helicase assay, the topoisomerase I was purchased (Invitrogen), and pBluescript was used as the plasmid substrate. For the ATPase assay, increasing concentrations of oligonucleotides (0, 2, or 8 nM final concentration) were incubated with purified SMARCAL1 (8 nM final concentration) in a final volume of 10 μL, and the reactions were incubated for 30 min at 30°C. The results are presented as the percent of ATP hydrolyzed to ADP during the reaction. ATPase assays were performed a minimum of three times each, and graphs depict means and standard deviation error bars. All oligonucleotide sequences are described in Supplemental Table 1, and all DNA substrates are described in Supplemental Table 2. All figures show a representative experiment from at least two replicates.
Homology modeling
The HARP repeats were identified as evolutionary structural homologs to PUR-α repeats using the FUGUE sequence–structure homology recognition server (Shi et al. 2001). The HARP2 (amino acids 325–396) homology model was constructed using the crystal structure of PUR-α (Protein Data Bank [PDB] ID 3K44) (Graebsch et al. 2009) residues 41–185 as a template. The ATPase model (SMARCAL1 residues 451–856) was generated from residues 455–891 of the SsoRad54 crystal structure (PDB ID 1Z63) (Durr et al. 2005). In both cases, the SMARCAL1 sequences were threaded onto the crystal structure using Swiss PDB Viewer, and the model was optimized using Swiss Model (http://swissmodel.expasy.org).
SAXS data collection and model building
SAXS data were collected at the SIBYLS beamline at the Advanced Light Source and prepared as described (Hura et al. 2009). Specifically, SAXS data were collected on SMARCAL1(325–954) in buffer containing 20 mM HEPES (pH 7.6), 200 mM NaCl, 2 mM MgCl2, 0.5 mM TCEP, 5% glycerol, and 1% sucrose. The protein sample was prepared for SAXS as described (Kazantsev et al. 2011) using a Shodex KW402.5 size exclusion column. The peak fraction was analyzed for SAXS as a 2/3 dilution series starting from 3 mg/mL. Three exposure times (0.5, 1, and 6 sec) were taken at 25°C and 12 keV. Guinier and Kratky analysis was performed as described (Putnam et al. 2007; Rambo and Tainer 2011). Linearity of the Guinier region for each exposure demonstrated a lack of radiation damage and aggregation (Supplemental Fig. 4D). SAXS profiles were overlaid, inspected for concentration-dependent scattering, and merged (Hura et al. 2009). For modeling, the composite scattering curve was generated from data from 1-sec exposures of 2 and 3 mg/mL samples. The maximum dimension (116 Å) was determined using GNOM (Svergun 1992). Atomistic-based modeling of the SAXS data was achieved with the program BUNCH (Petoukhov and Svergun 2005) using HARP and ATPase homology models. The models were treated as independent domains in a simulated annealing algorithm to determine their relative spatial arrangements. Missing residues between the HARP and ATPase domains (397–450) were modeled as dummy residues as described (Petoukhov and Svergun 2005). Ab initio modeling was performed with GASBOR using 630 dummy residues. Ten independent modeling runs were performed and averaged (Volkov and Svergun 2003) to produce a final macromolecular envelope. The final model targeted residues 325–856, consistent with a Porod volume of 91,148 Å3 calculated from the SAXS data. The missing C-terminal 99 residues were not included in the modeling based on proteolytic sensitivity of the C terminus (Supplemental Fig. 4A).
Branch migration and fork regression assays
Oligonucleotide #48 was end-labeled with [γ-32]ATP and T4 polynucleotide kinase (New England Biolabs) and purified through a G25 column (GE Healthcare). To prepare tailed or forked intermediates, 250 nM complementary ssDNA oligonucleotides (#48/#54 and #55/#56 for the branch migration, and #48/#50 and #53/#54 for the fork regression) were annealed in 20 μL of SSC buffer (15 mM NaCitrate at pH 7.0, 150 mM NaCl) in a PCR machine. To prepare the branch migration and the fork regression substrate, 32 nM 32P-labeled and 48 nM nonlabeled DNA intermediates were incubated in reaction buffer (40 mM Tris at pH 7.5, 20 mM KCl, 5 mM MgCl2, 100 μg/mL BSA, 2 mM ATP, 2 mM DTT) for 30 min at 37°C. The DNA substrates were diluted threefold in reaction buffer and mixed with increasing amounts of SMARCAL1 in a 20-μL reaction volume. The reaction was completed for 20 min at 37°C and terminated by the addition of 3× stop buffer (0.9% SDS, 50 mM EDTA, 40% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol). Samples were loaded into 8% polyacrylamide 1× TBE gels (82 × 28.5 cm, 1 mm thick) and subjected to electrophoresis in 1× TBE for 90 min at 80 V at room temperature. The gels were dried and quantified using a Molecular Imager FX (Bio-Rad).
The plasmid-sized replication fork model substrate was generated and purified as described (Blastyak et al. 2007). Recombinant SMARCAL1 purified from insect cells was incubated with 0.5 nM substrate for 20 min at 37°C in reaction buffer (20 mM Tris at pH 7.5, 50 mM KCl, 5 mM MgCl2, 100 μg/mL BSA, 2 mM ATP or ATPγS, 1 mM DTT). The reaction was quenched by the addition of 10 mM ATPγS and 10 mM MgCl2. One microliter (2–20 U, depending on the enzyme) of the indicated restriction enzymes was added to the reaction and further incubated for 30 min at 37°C. The reaction products were then separated on a 6% polyacrylamide gel. The gel was dried and quantified using a Molecular Imager FX (Bio-Rad).
Acknowledgments
This work was supported by NIH grant R01CA136933 to D.C. R.B. is supported in part by a Department of Defense Breast Cancer Research Program post-doctoral fellowship (W81XWH-10-1-0581). A.C.M. and C.B. are supported in part by the Vanderbilt Training Program in Environmental Toxicology (T32 ES07028). The SAXS analysis was made possible by the core facilities supported by the SBDR NIH grant P01CA092584 and a Center in Molecular Toxicology (P30 ES000267) pilot project grant to B.F.E.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.178459.111.
References
- Achar YJ, Balogh D, Haracska L 2011. Coordinated protein and DNA remodeling by human HLTF on stalled replication fork. Proc Natl Acad Sci 108: 14073–14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J, McGlynn P 2009. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res 37: 3475–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D 2009. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev 23: 2405–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansbach CE, Boerkoel CF, Cortez D 2010. SMARCAL1 and replication stress: An explanation for SIOD? Nucleus 1: 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell 28: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel CF, O'Neill S, Andre JL, Benke PJ, Bogdanovic R, Bulla M, Burguet A, Cockfield S, Cordeiro I, Ehrich JH, et al. 2000. Manifestations and treatment of Schimke immuno-osseous dysplasia: 14 new cases and a review of the literature. Eur J Pediatr 159: 1–7 [DOI] [PubMed] [Google Scholar]
- Boerkoel CF, Takashima H, John J, Yan J, Stankiewicz P, Rosenbarker L, Andre JL, Bogdanovic R, Burguet A, Cockfield S, et al. 2002. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet 30: 215–220 [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Mazina OM, Mazin AV 2006. Rad54 protein promotes branch migration of Holliday junctions. Nature 442: 590–593 [DOI] [PubMed] [Google Scholar]
- Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ 2009. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev 23: 2415–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC 2000. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep 1: 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R, Cimprich KA 2009. HARPing on about the DNA damage response during replication. Genes Dev 23: 2359–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr H, Korner C, Muller M, Hickmann V, Hopfner KP 2005. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 121: 363–373 [DOI] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T 2006. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 34: 2887–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, Pichierri P 2008. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J Cell Biol 183: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A 2008a. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci 105: 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A 2008b. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell 29: 141–148 [DOI] [PubMed] [Google Scholar]
- Ghosal G, Yuan J, Chen J 2011. The HARP domain dictates the annealing helicase activity of HARP/SMARCAL1. EMBO Rep 12: 574–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graebsch A, Roche S, Niessing D 2009. X-ray structure of Pur-α reveals a Whirly-like fold and an unusual nucleic-acid binding surface. Proc Natl Acad Sci 106: 18521–18526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL II, Tsutakawa SE, Jenney FE Jr, Classen S, Frankel KA, Hopkins RC, et al. 2009. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat Methods 6: 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Rambo RP, Karimpour S, Santalucia J Jr, Tainer JA, Pace NR 2011. Solution structure of RNase P RNA. RNA 17: 1159–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis R, Durr H, Hopfner KP, Michaelis J 2008. Conformational changes of a Swi2/Snf2 ATPase during its mechano-chemical cycle. Nucleic Acids Res 36: 1881–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CA, Xu X, Bansbach CE, Glick GG, Zhao R, Ye F, Sirbu BM, Titus LC, Shyr Y, Cortez D 2009. Functional genomic screens identify CINP as a genome maintenance protein. Proc Natl Acad Sci 106: 19304–19309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswami R, Truman PA, Mesner LD, Hockensmith JW 2000. A eukaryotic SWI2/SNF2 domain, an exquisite detector of double-stranded to single-stranded DNA transition elements. J Biol Chem 275: 7648–7655 [DOI] [PubMed] [Google Scholar]
- Opresko PL, Sowd G, Wang H 2009. The Werner syndrome helicase/exonuclease processes mobile D-loops through branch migration and degradation. PLoS ONE 4: e4825 doi: 10.1371/journal.pone.0004825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Whitby MC 2007. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair (Amst) 6: 1004–1017 [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T 2010. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11: 683–687 [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T 2010. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37: 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI 2005. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J 89: 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Woo EM, Chait BT, Funabiki H 2009. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem 284: 35951–35961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA 2007. X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys 40: 191–285 [DOI] [PubMed] [Google Scholar]
- Ralf C, Hickson ID, Wu L 2006. The Bloom's syndrome helicase can promote the regression of a model replication fork. J Biol Chem 281: 22839–22846 [DOI] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA 2010. Bridging the solution divide: Comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr Opin Struct Biol 20: 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA 2011. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95: 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Blundell TL, Mizuguchi K 2001. FUGUE: Sequence–structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol 310: 243–257 [DOI] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev 25: 1320–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Sanejouand YH 2004. ElNemo: A normal mode Web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res 32: W610–W614 doi: 10.1093/nar/gkh368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI 1992. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25: 495–503 [Google Scholar]
- Thoma NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP 2005. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat Struct Mol Biol 12: 350–356 [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI 2003. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr 36: 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ghosal G, Chen J 2009. The annealing helicase HARP protects stalled replication forks. Genes Dev 23: 2394–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT 2008. HARP is an ATP-driven annealing helicase. Science 322: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT 2010. Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif. Proc Natl Acad Sci 107: 20970–20973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kong X, Yokomori K, Kadonaga JT 2009. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev 23: 2400–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]