The migration and invasive potential of cancer cells are critical determinants of metastasis. The RNA-binding protein IGF2BP1 is implicated in regulating motility of such tumor-derived cells. In this study, Hüttelmaier and colleagues shed light on the molecular pathways underlying this motility regulation. They find that IGF2BP1 promotes the velocity and directionality of tumor-derived cell migration via the post-transcriptional regulation of two new target mRNAs: MAPK4 and PTEN. They detail the mechanisms by which modulating these mRNAs influences actin dynamics and regulates phospholipid-dependent signaling networks, leading to fast, polarized migration of tumor-derived cells.
Keywords: IGF2BP1, ZBP1, MAPK4, PTEN, HSP27, cell migration
Abstract
In primary neurons, the oncofetal RNA-binding protein IGF2BP1 (IGF2 mRNA-binding protein 1) controls spatially restricted β-actin (ACTB) mRNA translation and modulates growth cone guidance. In cultured tumor-derived cells, IGF2BP1 was shown to regulate the formation of lamellipodia and invadopodia. However, how and via which target mRNAs IGF2BP1 controls the motility of tumor-derived cells has remained elusive. In this study, we reveal that IGF2BP1 promotes the velocity and directionality of tumor-derived cell migration by determining the cytoplasmic fate of two novel target mRNAs: MAPK4 and PTEN. Inhibition of MAPK4 mRNA translation by IGF2BP1 antagonizes MK5 activation and prevents phosphorylation of HSP27, which sequesters actin monomers available for F-actin polymerization. Consequently, HSP27–ACTB association is reduced, mobilizing cellular G-actin for polymerization in order to promote the velocity of cell migration. At the same time, stabilization of the PTEN mRNA by IGF2BP1 enhances PTEN expression and antagonizes PIP3-directed signaling. This enforces the directionality of cell migration in a RAC1-dependent manner by preventing additional lamellipodia from forming and sustaining cell polarization intrinsically. IGF2BP1 thus promotes the velocity and persistence of tumor cell migration by controlling the expression of signaling proteins. This fine-tunes and connects intracellular signaling networks in order to enhance actin dynamics and cell polarization.
Cell migration and the invasive potential of cancer cells are essential determinants of metastasis (Gertler and Condeelis 2011). Various studies indicate that the post-transcriptional fine-tuning of gene expression by microRNAs provides multilayered regulatory networks directing various aspects of tumor cell migration and metastasis (Ma and Weinberg 2008; Valastyan and Weinberg 2009). Surprisingly, little is known about how RNA-binding proteins (RBPs) control the motility of tumor cells.
IGF2 mRNA-binding proteins (IGF2BPs) were previously suggested as prometastatic factors (Yaniv and Yisraeli 2002; Vikesaa et al. 2006; Oberman et al. 2007; Vainer et al. 2008). They constitute a group of three oncofetal proteins that are expressed during embryogenesis and become de novo synthesized in various human malignancies in which their expression correlates with poor prognosis (Yaniv and Yisraeli 2002; Yisraeli 2005; Kobel et al. 2007; Findeis-Hosey and Xu 2010). IGF2BPs reside almost exclusively in the cytoplasm, where they associate with mRNPs and direct the fate of specific target mRNAs. In tumor-derived cells, IGF2BP1 was shown to interfere with degradation of the MYC mRNA upon associating with the coding region instability determinant (CRD) in the ORF (Lemm and Ross 2002; Kobel et al. 2007; Weidensdorfer et al. 2009). This suggests that IGF2BP1 enhances tumor cell proliferation by sustaining elevated MYC expression (Kobel et al. 2007). A role of IGF2BPs in controlling cell migration was revealed initially in primary cells. Studies in Xenopus demonstrate that the IGF2BP3 homolog Vg1-RBP promotes directed migration of neuronal crest cells (Yaniv et al. 2003). In primary neurons and fibroblasts, IGF2BP1, also termed zipcode-binding protein 1 (ZBP1), directs localization of the β-actin (ACTB) mRNA to exploratory growth cones or lamellipodia, respectively (Kislauskis et al. 1994; Ross et al. 1997; Zhang et al. 2001). This asymmetric sorting of the ACTB mRNA essentially relies on IGF2BP1-facilitated inhibition of ACTB mRNA translation via a cis element, the so-called “zipcode,” in the ACTB-3′ untranslated region (UTR) (Huttelmaier et al. 2005). The control of ACTB mRNA translation by IGF2BP1 allows the spatiotemporal fine-tuning of ACTB protein synthesis, a process presumed to direct growth cone guidance in primary neurons (Leung et al. 2006; Yao et al. 2006; Sasaki et al. 2010). In tumor-derived cells, IGF2BP1 and IGF2BP3 promote the formation of invadopodia by preventing degradation of the CD44 mRNA, supporting their role as prometastatic factors (Vikesaa et al. 2006). Moreover, IGF2BPs were shown to facilitate the formation of polarized lamellipodia and regulate migration of tumor-derived cells in vitro (Lapidus et al. 2007; Oberman et al. 2007; Vainer et al. 2008). These findings indicate that IGF2BP1 is an essential regulator of cell migration. However, it has remained elusive via which target mRNAs and mechanisms IGF2BP1 regulates the motility of tumor-derived cells.
We reveal that IGF2BP1 promotes the directed movement of tumor-derived cells by fine-tuning intracellular signaling networks. The protein modulates actin dynamics in order to enhance the velocity of cell migration and promotes intrinsic cell polarization by regulating MK5- and phospholipid-dependent signaling networks. IGF2BP1 facilitates these regulatory roles by the post-transcriptional control of MAPK4 and PTEN expression.
Results
IGF2BP1 promotes tumor cell migration and F-actin integrity
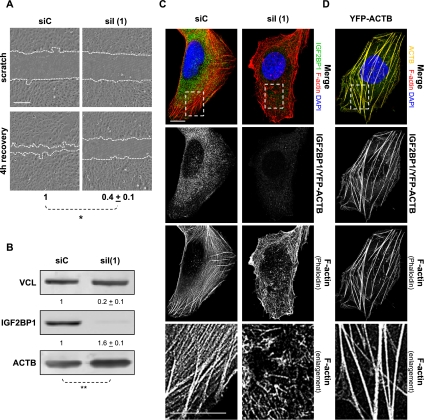
The role of IGF2BP1 in controlling the migration of tumor-derived cells was evaluated by “scratch” analyses. Wound closure was severely impaired in osteosarcoma-derived U2OS and ovarian carcinoma-derived ES-2 cells upon IGF2BP1 knockdown (Fig. 1A; Supplemental Fig. S1A). In U2OS cells, IGF2BP3 expression was beyond detection limits, and IGF2BP2 mRNA levels remained largely unaffected by IGF2BP1-directed siRNA (Supplemental Fig. S2A). This suggested an IGF2BP1-directed control of cell migration. To test this further, cell migration was monitored by scratch analyses in U2OS cells stably expressing GFP-chZBP1, the chicken ortholog of IGF2BP1. Cell motility remained unaffected by the exclusive knockdown of endogenous IGF2BP1 [Supplemental Fig. S2B–D, siI(2)], but was severely impaired upon the knockdown of both IGF2BP1 and transgenic GFP-chZBP1 [Supplemental Fig. S2B–D, siI(1)].
Figure 1.
IGF2BP1 knockdown disturbs the actin cytoskeleton and inhibits cell motility. (A) U2OS cells were transfected with IGF2BP1-directed [siI(1)] or control (siC) siRNAs for 48 h before scratching confluent (∼90%) cell layers. Wound closure was monitored by light microscopy immediately and 4 h after wounding, as indicated by dashed lines. The velocity of wound closure was determined relative to controls (siC). Bar, 250 μm; Student's t-test, (*) P < 0.05. (B) U2OS cells were transfected as in A. Protein levels were analyzed by Western blotting with the indicated antibodies 72 h post-transfection. Protein abundance was determined relative to controls using VCL (vinculin) for normalization, as indicated above the panels. Student's t-test, (**) P < 0.005. Note that ACTB mRNA levels remained unaffected by IGF2BP1 knockdown (data not shown), indicating that IGF2BP1 inhibits ACTB protein synthesis. (C,D) F-actin organization and IGF2BP1 knockdown (immunostaining) were monitored by confocal microscopy upon phalloidin labeling of U2OS cells transfected with the indicated siRNAs for 72 h (C) or YFP-ACTB for 48 h (D). Enlargement of the boxed regions in the top panels are shown in the bottom panels. Bars, 10 μm.
Whether the observed regulation of cell migration was correlated with the role of IGF2BP1 in controlling ACTB mRNA translation (Huttelmaier et al. 2005) was analyzed by IGF2BP1 knockdown. This resulted in a significant increase of ACTB protein levels in both U2OS and ES-2 cells due to the release of translational inhibition by IGF2BP1 (Fig. 1B; Supplemental Fig. S1B). Surprisingly, however, the knockdown of IGF2BP1 also induced a severe loss of stress fibers and increase of needle-like F-actin structures in both cell lines (Fig. 1C; Supplemental Fig. S1C,D). In cells expressing GFP-chZBP1, this change in F-actin organization was only observed upon the knockdown of endogenous IGF2BP1 and transgenic GFP-chZBP1 [Supplemental Fig. S2E, siI(1)]. In contrast, the actin cytoskeleton appeared unaffected by the exclusive knockdown of endogenous IGF2BP1 [Supplemental Fig. S2E, siI(2)]. This indicated that ZBP1 complemented the depletion of endogenous IGF2BP1 and thus confirmed the protein as an essential regulator of both the actin cytoskeleton and cell migration. To test whether F-actin reorganization observed upon IGF2BP1 knockdown was due to up-regulated ACTB protein synthesis, cells were transfected with YFP-ACTB. Stress fibers were thickened and stabilized (Fig. 1D), but the redistribution of actin filaments was not observed. Hence, reorganization of the actin cytoskeleton induced by IGF2BP1 knockdown was largely independent of the up-regulated ACTB protein synthesis resulting from the release of translational control on the mRNA. This suggested that IGF2BP1 “stabilizes” F-actin architecture by other means and possibly promotes cell migration by controlling the fate of additional target mRNAs.
Identification of novel IGF2BP1 target transcripts
Candidate target mRNAs of IGF2BP1 were identified by a loss-of-function screen in stressed U2OS cells. In these, IGF2BP1 exclusively stabilizes its target mRNAs, whereas it inhibits translation or prevents turnover of these transcripts in nonstressed cells (Stohr et al. 2006). Hence, one screening criterion, the reduction of mRNA levels upon IGF2BP1 knockdown during cellular stress, was expected to identify target mRNAs regulated at the level of translation or turnover by IGF2BP1 in unperturbed cells.
Comparative microarray analyses identified 74 annotated transcripts decreased by at least eightfold in stressed U2OS cells upon IGF2BP1 knockdown (Supplemental Table S1). Classification of these by the Database for Annotation, Visualization and Integrated Discovery (DAVID) revealed protein kinase cascade (PKC) as the most significantly affected cluster (Supplemental Fig. S3A–C). This cluster was comprised of PTEN, MAPK4, STAT3, RPS6KA5, RAPGEF2, SHANK3, STK17A, TIAF1, and F2R. A role in modulating the actin cytoskeleton and cell migration was sufficiently indicated only for the first three of these proteins (Supplemental Table S1). However, a selective decrease of STAT3 mRNA levels upon IGF2BP1 knockdown in stressed U2OS cells could not be confirmed by quantitative RT–PCR (qRT–PCR) (Supplemental Table S1).
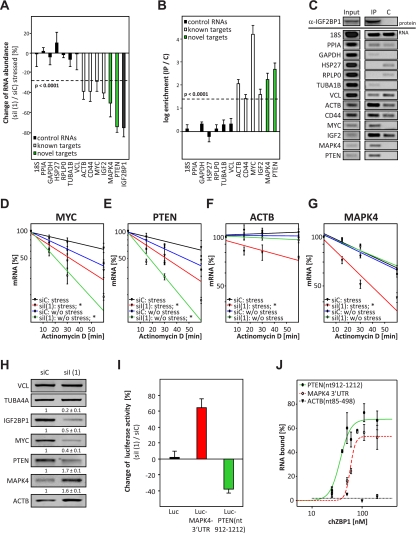
MAPK4 and PTEN were previously proposed to modulate the actin cytoskeleton and, potentially, cell migration by regulating the phosphorylation of HSP27 (Gerits et al. 2007; Tak et al. 2007; Kostenko et al. 2009) or cellular PIP3/PIP2 ratios (Kolsch et al. 2008; Cain and Ridley 2009; Berzat and Hall 2010), respectively. To validate PTEN and MAPK4 as target transcripts, their abundance was determined in stressed cells using qRT–PCR (Fig. 2A). The amount of seven control transcripts (Fig. 2A, black), including HSP27, remained largely unaffected by IGF2BP1 knockdown. In contrast, the levels of four known (ACTB, CD44, MYC, and IGF2) (Fig. 2A, white) and the two novel (PTEN and MAPK4) (Fig. 2A, green) target transcripts were decreased. Association of these mRNAs with IGF2BP1 was monitored by RT–PCR upon immunopurification of IGF2BP1 from nonstressed cells. Only the known and novel target mRNAs were selectively copurified (Fig. 2B,C). Notably, decreased mRNA levels upon IGF2BP1 knockdown in stressed U2OS cells and association with IGF2BP1 in nonstressed cells was also confirmed for RAPGEF2 and RPS6KA5 (Supplemental Table S1). Taken together, these findings confirmed the suitability of the screening approach and indicated MAPK4, PTEN, RAPGEF2, and RPS6KA5 as novel target mRNAs of IGF2BP1. To address the role of IGF2BP1 in modulating both actin dynamics and cell migration, we focused on the most promising candidates: MAPK4 and PTEN.
Figure 2.
IGF2BP1 inhibits MAPK4 mRNA translation and prevents PTEN mRNA degradation. (A) U2OS cells transfected with the indicated siRNAs for 72 h were stressed by arsenate (2.5 mM) for 2 h before analyzing RNA abundance by qRT–PCR. Changes in RNA amounts upon IGF2BP1 knockdown were determined relative to controls transfected with a nontargeting siRNA (siC) by the ΔΔCt method using seven control RNAs (black bars) for cross-normalization. Significance of reduced RNA abundance is indicated by a dashed line (P < 0.0001). (B,C) The association of indicated RNAs with IGF2BP1 protein (IP) in nonstressed U2OS cells was analyzed by coimmunopurification followed by quantitative (B) or semiquantitative (C; 40 cycles) RT–PCR. In B, the coenrichment of RNAs with IGF2BP1 (IP) was determined relative to controls (C) incubated with protein-G beads by the ΔΔCt method using PPIA for normalization. The dashed line indicates significant copurification (P < 0.0001). (C, top panel) Immunopurification of IGF2BP1 was confirmed by Western blotting. (D–G) U2OS cells transfected as in A were treated with actinomycin D (5 μM; without stress) or actinomycin D and arsenate (2.5 mM; stress) for the indicated time. RNA decay was monitored by qRT–PCR using the ΔΔCt method and cross-normalization to PPIA and RPLP0 mRNAs, since these were barely degraded during the analyzed time frame. RNA decay is shown in semilogarithmic scale and was fitted by linear regression. Significance was determined by paired Student's t-test; (*) P < 0.05. (H) Protein levels in U2OS cells transfected as in A were analyzed by Western blotting with the indicated antibodies. Protein abundance upon IGF2BP1 knockdown was determined relative to controls (siC) by cross-normalization to VCL and TUBA4A, as indicated above the panels. (I) U2OS cells were transfected with the indicated siRNAs for 48 h before the transfection of Firefly and Renilla luciferase reporters for 24 h. The activity of Firefly reporters upon IGF2BP1 knockdown was determined relative to controls (siC) by normalization to Renilla activities. The firefly reporters used were as follows: (1) firefly with vector-encoded BGH-3′ UTR (black), (2) firefly with MAPK4-3′ UTR (red), and (3) firefly fused in-frame to the rare codon fragment (912–1212 nt) of the PTEN ORF (green). (J) Binding of the indicated in vitro transcribed Atto680-labeld RNAs to increasing amounts of recombinant chZBP1, the chicken homolog of IGF2BP1, was analyzed by filter binding using infrared (IR) scanning (data not shown). The percentage of bound RNA was determined by input-normalized fluorescence intensities plotted against the protein concentration using the Hill equation for fitting as previously described (Kohn et al. 2010). Error bars indicate standard deviation of at least three independent analyses.
IGF2BP1 controls PTEN mRNA turnover and MAPK4 mRNA translation
How IGF2BP1 controls PTEN and/or MAPK4 mRNA fate was first evaluated by monitoring the turnover of both mRNAs. ACTB and MYC served as controls, since IGF2BP1 prevents MYC mRNA turnover, whereas it inhibits ACTB mRNA translation without affecting the decay of this transcript in unperturbed cells (Huttelmaier et al. 2005; Kobel et al. 2007; Weidensdorfer et al. 2009). MYC and PTEN mRNAs decayed faster in stressed and nonstressed cells upon IGF2BP1 knockdown (Fig. 2D,E). In contrast, the degradation of MAPK4 and ACTB mRNAs was more pronounced exclusively in stressed cells (Fig. 2F,G). This suggested that IGF2BP1 promoted PTEN expression by preventing decay of the PTEN mRNA and that it inhibited MAPK4 protein synthesis in nonstressed cells. In agreement, the abundance of PTEN and MYC protein was reduced, whereas MAPK4 and ACTB protein levels were increased, by IGF2BP1 knockdown (Fig. 2H; Supplemental Fig. S4B). The amount of MAPK4 and PTEN protein essentially recovered to control levels when expressing siRNA-insensitive Flag-ZBP1 (Supplemental Fig. S4A). Notably, RAPGEF2 protein levels were reduced, whereas RPS6KA5 protein amounts were increased, upon IGF2BP1 knockdown in U2OS cells, supporting a role of IGF2BP1 in controlling the expression of both factors (Supplemental Fig. S4B).
Regulation of PTEN and MAPK4 mRNA fate by IGF2BP1 was analyzed further by the use of luciferase reporters. The activity of a reporter transcript comprising the MAPK4-3′ UTR was increased, whereas its turnover remained unaffected, by IGF2BP1 knockdown (Fig. 2I, red; Supplemental Fig. S4D). The opposite was observed for a reporter mRNA comprising a rare codon stretch of the PTEN ORF (nucleotides 912–1212, corresponding to amino acids 304–stop) in-frame to the luciferase coding sequence (CDS) (Supplemental Fig. S4C). This decayed faster, and thus reporter activity was reduced upon IGF2BP1 knockdown (Fig. 2I, green; Supplemental Fig. S4E). Whether IGF2BP1 binds to the MAPK4-3′ UTR or rare codon region of the PTEN ORF was analyzed in vitro by filter-binding studies. Recombinant chZBP1 protein associated with both in vitro transcribed RNAs, whereas essentially no binding was observed for an RNA comprising the 5′ region of the ACTB ORF (Fig. 2J). Thus, IGF2BP1 inhibited MAPK4 expression by interfering with mRNA translation upon associating with the MAPK4-3′ UTR. In contrast, IGF2BP1 promoted PTEN expression by interfering with PTEN mRNA degradation upon associating with a rare codon-comprising fragment of the PTEN ORF.
IGF2BP1 antagonizes the phosphorylation of HSP27 by interfering with MK5 activation
The identification of MAPK4 and PTEN as novel target mRNAs of IGF2BP1 suggested that IGF2BP1 controls intracellular signaling. MAPK4 activates MK5 by phosphorylating the latter at T182, resulting in cytoplasmic accumulation of MK5 (Kant et al. 2006; Coulombe and Meloche 2007; Deleris et al. 2008; Perander et al. 2008; Aberg et al. 2009). MK5, in turn, phosphorylates HSP27 (HSPB1) and was suggested to thereby induce altered microfilament organization (Kostenko and Moens 2009; Doshi et al. 2010). PTEN interferes with PIP3-directed activation of various signaling molecules, including small GTPases like RAC1 or protein kinases of the AKT family. The latter were proposed to modulate microfilament organization by phosphorylating downstream targets that strikingly include HSP27 (Read and Gorman 2009a,b).
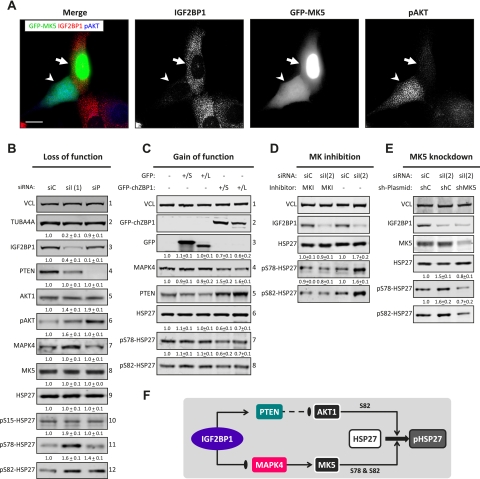
Whether IGF2BP1 controls the activation of AKTs and/or MK5 was analyzed by IGF2BP1 knockdown. This induced phosphorylation of AKTs and promoted cytoplasmic accumulation of GFP-MK5 (Fig. 3A, arrowhead), suggesting activation of both kinases. Up-regulated phosphorylation of AKTs, indicating kinase activation, was confirmed by Western blotting and was correlated with increased PIP3 levels upon both IGF2BP1 and PTEN knockdown (Fig. 3B, panel 6; Supplemental Fig. S5A–C). Cytoplasmic accumulation of GFP-MK5 was induced in a concentration-dependent manner by the up-regulation of MAPK4 protein levels in response to IGF2BP1 knockdown as well as the overexpression of Flag-MAPK4 (Fig. 3B, panel 7; Supplemental Fig. S5D,E). Activation of MK5 and AKTs was finally evaluated by monitoring phosphorylation of their substrate, HSP27. The knockdown of PTEN induced phosphorylation of HSP27 at S82 due to the activation of AKTs, which exclusively phosphorylate this residue (Fig. 3B, panels 10–12; Supplemental Fig. S6; Kostenko and Moens 2009). In contrast, phosphorylation of HSP27 was up-regulated at S78 and S82 upon IGF2BP1 knockdown due to increased MAPK4-directed activation of MK5, which phosphorylates at both residues (Fig. 3B, panels 10–12; Supplemental Fig. S6). This suggested that IGF2BP1 antagonized phosphorylation of HSP27 by inhibiting MAPK4 but promoting PTEN protein synthesis. To test this further, GFP-chZBP1 was stably expressed (S) or transduced (L) by lentiviruses (Fig. 3C). In both cases, MAPK4 protein levels (Fig. 3C, panel 4) decreased, PTEN protein abundance (Fig. 3C, panel 5) increased, and phosphorylation of HSP27 (Fig. 3C, panels 6–8) was down-regulated at S78 and S82. Whether IGF2BP1-facilitated inhibition of MK5 activation was sufficient to antagonize phosphorylation of HSP27 was tested by the pharmacological inhibition of MKs (MKI) or MK5 knockdown (Fig. 3D,E). Both abolished the up-regulation of HSP27 phosphorylation observed upon IGF2BP1 knockdown. Hence, IGF2BP1 antagonized the activation of AKTs and MK5 by promoting PTEN but inhibiting MAPK4 expression. However, in U2OS cells, IGF2BP1-facilitated inhibition of MK5 activation was sufficient to antagonize phosphorylation of HSP27 at S78 and S82 (Fig. 3F).
Figure 3.
IGF2BP1 antagonizes MK5-directed phosphorylation of HSP27 by inhibiting MAPK4 mRNA translation. (A) U2OS cells were transfected with IGF2BP1-directed siRNAs for 48 h followed by the transfection of GFP-MK5 for 24 h. Cells were processed for immunostaining of IGF2BP1 and phosphor-AKTs (pAKT). Images were acquired by confocal microscopy. Arrowheads indicate a cell showing IGF2BP1 depletion with GFP-MK5 equally distributed between cytoplasm and nucleus as well as up-regulated phosphorylation of AKTs. Arrows indicate a control cell with unaffected IGF2BP1 expression in which GFP-MK5 is enriched in the nucleus and phosphorylation of AKTs is barely observed. Bar, 10 μm. (B) U2OS cells were transfected with control (siC), IGF2BP1-directed [siI(1)], or PTEN-directed (siP) siRNAs for 72 h. The abundance of the indicated proteins upon IGF2BP1 or PTEN knockdown was determined relative to controls (siC) by Western blotting. Relative protein levels determined by cross-normalization to VCL and TUBA4A are indicated above the panels. Phosphorylation of AKTs and HSP27 was determined by normalization to total AKT1 or HSP27 levels, respectively. Representative Western blots of three independent analyses are shown. (C) Protein abundances or HSP27 phosphorylation in U2OS cells stably transfected (S) or transduced by lentiviruses (L) encoding GFP or GFP-chZBP1 were determined by Western blotting as described in B. (D) U2OS cells transfected with the indicated siRNAs for 24 h were cultured in the presence of MKI (10 μM) for 48 h. Protein abundance and HSP27 phosphorylation were analyzed by Western blotting 72 h after the transfection of siRNAs as described in B. (E) U2OS cells transfected with the indicated siRNAs and/or MK5-directed shRNA (shMK5) for 72 h were analyzed by Western blotting using the indicated antibodies. HSP27 phosphorylation, indicated above the panels, was determined relative to the respective controls (siC and shC) as described in B. (F) Scheme suggesting that IGF2BP1 antagonizes the phosphorylation of HSP27 by promoting PTEN expression but inhibiting MAPK4 protein synthesis. Enhanced expression of PTEN reduces PIP3-dependent AKT activation and thus interferes with phosphorylation of HSP27 at S82. Decreased expression of MAPK4 limits MK5 activation and thus antagonizes phosphorylation of HSP27 at S78 and S82.
IGF2BP1 controls actin dynamics by antagonizing the phosphorylation of HSP27
The abundance of HSP27 modulates the actin cytoskeleton and influences cell migration (Mounier and Arrigo 2002; Salinthone et al. 2008; Kostenko and Moens 2009; Doshi et al. 2010). In vitro, HSP27 was proposed to sequester actin monomers and weakly cap F-actin in a phosphorylation-dependent manner (An et al. 2004; During et al. 2007; Doshi et al. 2010; Jia et al. 2010). This suggested that IGF2BP1 could control actin dynamics by antagonizing MK5-directed phosphorylation of HSP27.
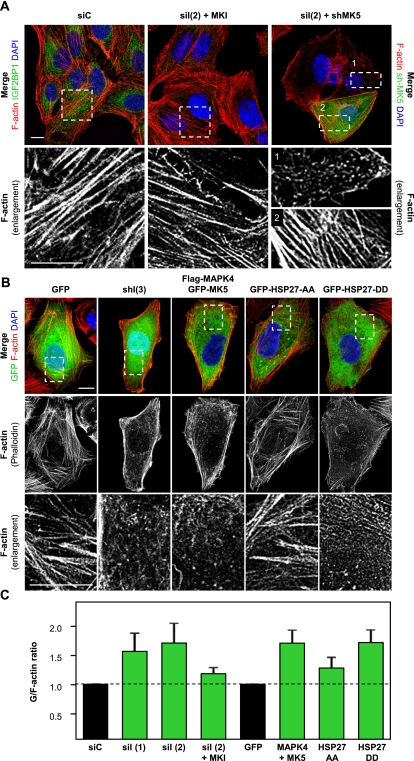
IGF2BP1 knockdown-induced reorganization of the actin cytoskeleton was abolished by both MKI and MK5 knockdown (Fig. 4A). To analyze how deregulation of F-actin architecture was correlated with cellular G-actin/F-actin ratios, the latter were determined by the labeling of monomeric G-actin using DNaseI-Alexa594 and F-actin staining by phalloidin-TRITC. Redistribution of the actin cytoskeleton correlated with up-regulated G-actin/F-actin ratios upon (Fig. 4B,C; Supplemental Fig. S7A–C): (1) the shRNA- or siRNA-mediated knockdown of IGF2BP1; (2) the forced expression of MAPK4, either alone or in combination with MK5; and (3) the expression of a phosphor-mimicking HSP27 mutant (HSP27-DD: S78,82D). In all these cases, the phosphorylation of HSP27 was increased or mimicked, respectively (see Fig. 3B; Supplemental Fig. S6). In contrast, the actin cytoskeleton and G-actin/F-actin (G/F) ratios appeared largely unaffected by a nonphosphorylatable HSP27 mutant (HSP27-AA: S78,82A) or the expression of MK5 (Fig. 4B,C; Supplemental Fig. S7A–C). The latter was insufficient to significantly promote phosphorylation of HSP27 when expressed alone (Supplemental Fig. S6). This suggested that the IGF2BP1-facilitated control of HSP27 phosphorylation by MK5 at both residues (S78 and S82) is essential for regulating the cellular G/F equilibrium. In agreement, G/F ratios remained largely unaffected upon modulating phosphorylation of HSP27 exclusively at S82 by the knockdown of PTEN or the overexpression of AKT1 (Supplemental Fig. S7B,C). Moreover, the G-actin/F-actin equilibrium and thus F-actin organization remained essentially unchanged upon IGF2BP1 knockdown when MK signaling was inhibited pharmacologically [Fig. 4A; Supplemental Fig. S7C, siI(2)+MKI]. Taken together, these findings indicated that IGF2BP1 modulates actin dynamics by antagonizing the phosphorylation of HSP27 at S78 and S82.
Figure 4.
IGF2BP1 modulates F-actin organization by antagonizing MK5-directed phosphorylation of HSP27. (A) U2OS cells were transfected with the indicated siRNAs or shRNAs for 72 h before F-actin labeling by phalloidin-TRITC, immunostaining of IGF2BP1, and labeling of nuclei by DAPI. Where indicated, cells were treated with MKI (+MKI; 10 μM) for 48 h. Representative images acquired by confocal microscopy are shown. Dashed boxes in the top panels indicate regions shown as enlargements in the bottom panels. Note that enlargement 1 shows residual F-actin in an IGF2BP1 knockdown cell, whereas enlargement 2 shows restored F-actin upon IGF2BP1 and MK5 knockdown. Cells transfected with shRNA-encoding plasmids are identified by the coexpression of GFP. Bars, 10 μm. (B) U2OS cells were transfected with the indicated GFP-tagged proteins for 48 h or IGF2BP1-directed shRNA [shI(3)] for 72 h. Cells were processed for staining of F-actin and nuclei as described in A. Enlargements (bottom panels) of boxed areas (top panel) show representative images of F-actin organization observed in the majority of cells. Note that the used shRNA plasmid encodes GFP. Bars, 10 μm. (C) U2OS cells were transfected with indicated siRNAs (72 h) or plasmids (48 h) and incubated with MKI for 48 h where indicated. G-actin/F-actin ratios were analyzed by fluorescence spectrometry using phalloidin-TRITC to label F-actin and DNaseI-Alexa594 to label G-actin. G-actin/F-actin ratios were determined relative to siC or GFP transfected controls (black bars) using DAPI staining for normalization, as indicated by the dashed line. Error bars indicate standard deviation of at least three independent analyses. Note that disturbed stress fiber organization correlates with increased G-actin/F-actin ratios. A comprehensive analysis of G/F ratios is shown in Supplemental Figure S7C.
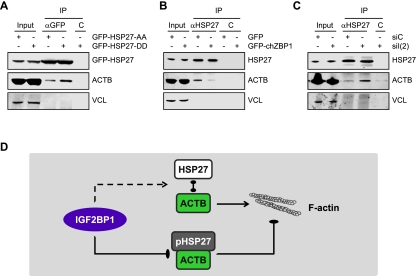
Whether the phosphorylation of HSP27 could affect actin sequestering and thus actin dynamics by modulating HSP27–ACTB association was analyzed by immunopurification. Compared with GFP-HSP27-AA, the association of ACTB with GFP-HSP27-DD was enhanced in U2OS cells (Fig. 5A). To analyze whether IGF2BP1 thus modulates ACTB association by antagonizing HSP27 phosphorylation, endogenous HSP27 was immunopurified from U2OS cells stably expressing GFP-chZBP1 or transfected with IGF2BP1-directed siRNAs. Compared with the respective controls, HSP27–ACTB association was decreased by the overexpression of GFP-chZBP1, whereas it was increased upon IGF2BP1 knockdown (Fig. 5B,C). This was in good agreement with the inverse correlation of HSP27 phosphorylation and IGF2BP1 expression as well as changes of G-actin/F-actin ratios observed upon down-regulation or up-regulation of IGF2BP1 expression (Fig. 3B,C; Supplemental Fig. S7C). Hence, by antagonizing MK5-directed phosphorylation of HSP27, IGF2BP1 decreased G-actin sequestering by HSP27 and thus modulated the cellular G-actin/F-actin equilibrium (Fig. 5D).
Figure 5.
IGF2BP1 modulates HSP27–ACTB association by antagonizing MK5-directed phosphorylation of HSP27. (A–C) Phosphorylation-dependent association of ACTB and HSP27 was analyzed by coimmunopurification of ACTB with transiently transfected GFP-tagged HSP27 mutants (A, αGFP) or of endogenous HSP27 from cells stably transfected with GFP-chZBP1 (B, αHSP27) or transiently transfected with IGF2BP1-directed siRNAs (C, αHSP27). (C) Protein-G beads served as controls. (D) Scheme suggesting that IGF2BP1 antagonizes the association of HSP27 with monomeric ACTB by inhibiting MK5-directed phosphorylation of HSP27. This presumably interferes with the sequestering of actin monomers by HSP27 and thus promotes F-actin polymerization.
IGF2BP1 enhances the velocity of tumor cell migration by antagonizing MK5 activation
The regulation of actin dynamics by IGF2BP1 implied that the protein modulates cell migration by antagonizing MK5-mediated phosphorylation of HSP27. Wound healing analyses confirmed that IGF2BP1 knockdown as well as HSP27-DD overexpression severely reduced cell motility, whereas HSP27-AA did not (Supplemental Fig. S8A). In agreement, the stable expression of GFP-chZBP1 promoted cell migration due to down-regulated phosphorylation of HSP27 (see Fig. 3C; Supplemental Fig. S8B). Migration defects induced by IGF2BP1 knockdown were essentially abolished by MK5 knockdown (sh-MK5) in U2OS cells or by MKI in ES-2 cells (Supplemental Fig. S8A,C).
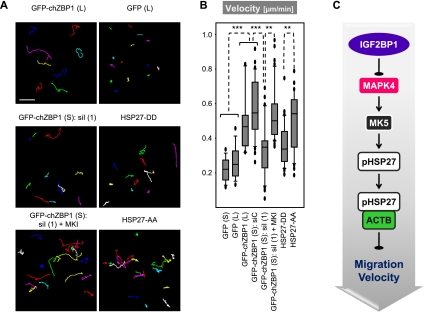
To analyze the role of IGF2BP1 in controlling migration at the single-cell level, cell motility was monitored by time-lapse microscopy. Migration velocity, determined as the mean displacement of individual cells, was significantly increased by stable (S) or lentiviral (L) overexpression of GFP-chZBP1 (Fig. 6A,B; Supplemental Fig. S9A,B; Supplemental Movie M1). The velocity of cell migration was reduced to the level of GFP-expressing controls by IGF2BP1/GFP-chZBP1 knockdown, whereas it remained increased by concurrently blocking MK signaling (Fig. 6A,B; Supplemental Fig. S9A,B; Supplemental Movie M2). To analyze the role of HSP27 phosphorylation in controlling cell motility in an IGF2BP1-independent manner, U2OS cells were transiently transfected with GFP-tagged HSP27-AA or HSP27-DD. Migration velocity was significantly increased by nonphosphorylatable HSP27-AA compared with the phosphor-mimicking HSP27-DD (Fig. 6A,B; Supplemental Fig. S9A,B; Supplemental Movie M3). These findings indicated that IGF2BP1 promoted the velocity of cell migration by inhibiting MK5-directed phosphorylation of HSP27 (Fig. 6C).
Figure 6.
IGF2BP1 promotes cell migration velocity by antagonizing MK5-directed phosphorylation of HSP27. (A,B) Single-cell motility of U2OS cells stably expressing the indicated proteins (S), transduced by lentiviruses (L), or transiently transfected (GFP-HSP27-AA and GFP-HSP27-DD) was monitored on collagen-coated glass-bottomed dishes. Where indicated, cells were transfected with the indicated siRNAs (72 h) and/or treated with MKI (48 h) before single-cell tracking using time lapse microscopy (3-min interval; 5 h). (A) Cell tracks for 15–50 cells were determined by the “manual tracking” plugin for ImageJ in at least three independent analyses. Representative cell tracks are shown in A, and representative single-cell movies are provided as Supplemental Movies M1–M3. (B) The mean displacement of cells (velocity) is shown as a box plot. Bar, 75 μm. Statistical significance, (**) P < 0.005; (***) P < 0.0005. A comprehensive summary of determined cell migration parameters is shown in Figure S9, A and B. (C) Scheme suggesting that IGF2BP1 promotes the velocity of cell migration by antagonizing MK5-directed phosphorylation of HSP27 and thus limiting sequestering of ACTB monomers.
IGF2BP1 stimulates directed cell migration by PTEN-modulated cell polarization
Visual inspection of individual cell tracks suggested that IGF2BP1/GFP-chZBP1 enhanced not only the velocity of cell migration, but also its directionality (Fig. 6A). This was assessed in a quantitative manner by determining the ratio of net distance (D) to total track distance (T). The D/T ratio and thus the directionality of migration were significantly increased by the stable expression of GFP-chZBP1 in the absence of external guidance cues (Supplemental Fig. S9A,B). However, the directionality of movement remained largely unaffected by the overexpression of HSP27 mutants when compared with cells stably expressing GFP (Supplemental Fig. S9A,B). This suggested that IGF2BP1 promoted the persistence of directed cell migration in a HSP27-independent manner.
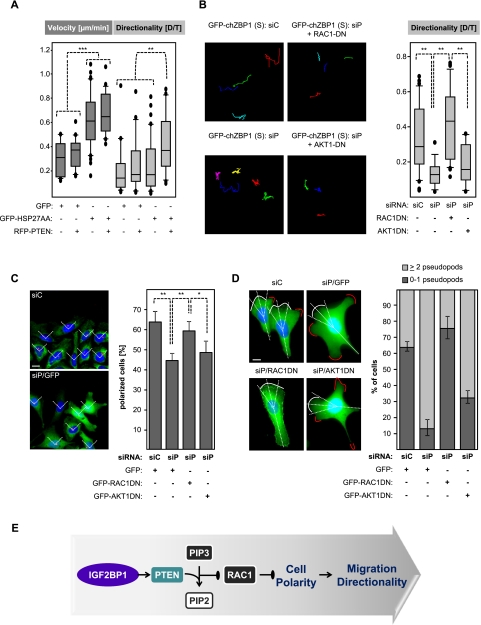
The stable or lentiviral overexpression of chZBP1 enhanced PTEN protein abundance (see Fig. 3C). However, the knockdown of IGF2BP1 and GFP-chZBP1 in these cells led to an only modest reduction of PTEN protein amounts, which remained significantly above levels observed in cells stably expressing GFP (Supplemental Fig. S9C). In correlation with this, D/T ratios and thus the directionality of cell migration were barely reduced upon IGF2BP1/GFP-chZBP1 knockdown in stably transfected U2OS cells (Supplemental Fig. S9A,B). These observations suggested that IGF2BP1 could control the directionality of cell migration by enhancing the expression of PTEN and thus antagonizing PIP3-directed signaling. This was analyzed by the knockdown of PTEN in naive and U2OS cells stably expressing GFP-chZBP1. F-actin architecture, G-actin/F-actin ratios, and the velocity of cell migration remained largely unaffected by the knockdown of PTEN (Supplemental Figs. S7B,C, S9A,B). Strikingly, however, the directionality of cell movement was reduced to levels observed in cells stably transfected with GFP upon the knockdown of PTEN in cells stably expressing GFP-chZBP1 (Supplemental Fig. S9A,B; Supplemental Movie M4). To test the role of PTEN in an IGF2BP1-independent manner, RFP-PTEN was expressed alone or in combination with HSP27-AA (Fig. 7A). The latter enhanced only the velocity of cell migration, whereas its coexpression with RFP-PTEN promoted both the velocity and directionality of cell migration (Fig. 7A; Supplemental Movie M5). Notably, velocity and directionality of migration remained largely unaffected by the expression of PTEN alone. Taken together, these analyses indicated that IGF2BP1 enforces the persistence of directed cell migration in the absence of external guidance cues by enhancing PTEN expression.
Figure 7.
IGF2BP1 promotes cell polarity and migration directionality in a PTEN-dependent manner. (A,B) Naive U2OS cells (A) or U2OS cells stably expressing GFP-chZBP1 (S) (B) were transfected with plasmids (48 h) and/or siRNAs (72 h) as indicated. Single-cell migration was monitored essentially as described in Figure 6, A and B. The velocity of cell migration was determined as “mean displacement,” whereas directionality was determined by the ratio of net distance (D) to total track distance (T). Migration data are shown as box plots. Statistical significance was determined by Student's t-test; (**) P < 0.005; (***) P < 0.0005. A comprehensive summary of determined cell migration parameters is shown in Supplemental Figure S9, A and B. (C) U2OS cells stably expressing GFP-chZBP1 were cotransfected with the indicated siRNAs and plasmids (60 h) encoding GFP or dominant-negative variants of RAC1 (RAC1DN) or AKT1 (AKT1DN). Cell polarity was determined by immunostaining of the MTOC using an anti-pericentrin antibody 2 h after scratching of ∼80% confluent cell layers seeded on a collagen matrix. Cells were scored as “polarized” (right panel) when the MTOC was localized within a 90° sector (left panel; indicated by dashed lines) facing directly toward the wound. Errors indicate standard deviation determined for >50 cells analyzed in three independent experiments. Bar, 10 μm. Statistical significance was determined by Student's t-test; (*) P < 0.05; (**) P < 0.005. Note that the number of polarized cells is significantly reduced by PTEN knockdown but remains largely unaffected when coexpressing RAC1DN. (D) Cells transfected as in C were cultured on collagen-coated coverslips at low density for 3 h before fixation and DAPI staining. Pseudopods (indicated by red line) outside a 45° sector centered by the longest cell axis crossing the nucleus (indicated as dashed lines) were classified as lateral pseudopodia. White lines indicate lamellipodia aligned with the longest cell axis. Cells were scored in two classes: two or more (light gray) versus no or one (dark gray) lateral pseudopod. Errors indicate standard deviation analyzed for >30 cells in three independent experiments. Bar, 10 μm. (E) Scheme suggesting that IGF2BP1 promotes cell polarization and thus stimulates the directionality of cell movement by enhancing PTEN expression. This presumably constrains PIP3-directed activation of RAC1.
In Dictyostelium, PTEN suppresses the formation of lateral pseudopods and thereby promotes cell polarization and directed movement (Wessels et al. 2007). Likewise, the random formation of pseudopods appeared increased upon PTEN knockdown in U2OS cells stably expressing GFP-chZBP1 (Supplemental Movie M4). This was evaluated by quantifying the number of lateral pseudopods. The putative role of IGF2BP1 and PTEN in directing cell polarization was further analyzed by determining the orientation of the MTOC (microtubule organization center) toward a “scratch” in the cell layer (Etienne-Manneville and Hall 2001; Yamana et al. 2006). In cells stably expressing GFP-chZBP1, the knockdown of PTEN led to reduced directionality of cell migration and decreased MTOC orientation, and the number of cells showing two or more lateral pseudopods was increased (Fig. 7B–D). This suggested that IGF2BP1 promoted cell polarization by preventing pseudopod formation due to forced PTEN expression.
PTEN antagonizes PIP3-directed activation of RAC1 and AKT1, which are essentially involved in the control of directed cell migration (Kolsch et al. 2008; Cain and Ridley 2009; Berzat and Hall 2010). Whether IGF2BP1-facilitated control of migration directionality was mediated via these signaling proteins was analyzed by the expression of dominant-negative RAC1 (RAC1DN) or AKT1 (AKT1DN), respectively. RAC1DN prevented the increased formation of lateral pseudopods and the reduction of cell polarization and thus could complement the severe reduction in directed cell migration induced by PTEN knockdown in cells stably expressing GFP-chZBP1 (Fig. 7B–D). This was not observed upon the transient expression of AKT1DN. Hence, by sustaining PTEN expression, IGF2BP1 apparently constrained PIP3-directed RAC1 activation and thereby enforced cell polarization intrinsically. This promoted the directionality of cell migration even in the absence of defined external guidance cues (Fig. 7E).
Discussion
This study reports a novel mechanism that promotes the directed migration of tumor-derived cells by the post-transcriptional fine-tuning of two intracellular signaling networks. The oncofetal RBP IGF2BP1 enhances the velocity of tumor-derived cell migration by inhibiting MAPK4 mRNA translation. This antagonizes MK5-directed phosphorylation of HSP27, leading to reduced G-actin sequestering, and thus ensures proper actin dynamics. Concomitantly, IGF2BP1 promotes the directionality of cell migration by preventing PTEN mRNA degradation. This antagonizes PIP3 signaling and enhances cell polarization by constraining RAC1 activation. Hence, IGF2BP1 promotes the directed migration of tumor-derived cells even in the absence of defined external guidance cues, as previously suggested (Lapidus et al. 2007). We demonstrate that this regulatory role of IGF2BP1 is essentially facilitated by controlling G-actin/F-actin as wells as PIP3/PIP2 equilibriums through the control of MAPK4 and PTEN expression (Supplemental Fig. S10).
IGF2BP1 is a key regulator of spatially restricted ACTB protein synthesis, in particular in neurons (Dahm and Kiebler 2005). This led to the view that the protein promotes growth cone guidance by controlling the availability of actin monomers that feed F-actin polymerization and thus direct the “driving force” of growth cone migration (Condeelis and Singer 2005; Leung et al. 2006; Yao et al. 2006; Sasaki et al. 2010). Here we reveal that IGF2BP1 not only controls the synthesis of “monomeric” actin, but also modulates actin dynamics by antagonizing MAPK4/MK5-directed phosphorylation of HSP27.
MAPK4 and its homolog, ERK3, are classified as atypical MAPKs, both of which activate MK5 (Seternes et al. 2004; Aberg et al. 2006, 2009; Gaestel 2006; Kant et al. 2006; Deleris et al. 2008; Perander et al. 2008). Notably, ERK3 protein is rapidly turned over, whereas MAPK4 is not (Aberg et al. 2006). This suggests that the control of MAPK4 mRNA translation rather than MK5 levels is important in directing downstream signaling. In support of this, we observed MAPK4 up-regulation being more effective than forced MK5 expression. So far, HSP27 and FoxO3a are the only validated MK5 substrates (Gaestel 2006; Kress et al. 2011). HSP27 is phosphorylated by all three MKs (MK2, MK3, and MK5), but only MK5 phosphorylates exclusively at S78 and S82 (Kostenko and Moens 2009). Although the abundance and phosphorylation of HSP27 affect the actin cytoskeleton, little is known about the role of individual phosphorylation sites (Kostenko and Moens 2009). Notably, the mouse and rat homolog of HSP27, HSP25, lacks S78, and thus functional comparisons are limited between rodents and humans (Kostenko and Moens 2009). In vitro, HSP27 efficiently sequesters actin monomers (During et al. 2007). We demonstrate that MK5-directed phosphorylation of HSP27 increases cellular G-actin/F-actin ratios and the congruent association of HSP27 with monomeric ACTB. Hence, the phosphorylation-induced disassembly of HSP27 oligomers presumably enhances the sequestering of actin monomers by HSP27 (Kostenko and Moens 2009; Doshi et al. 2010). In agreement, MAPK4/MK5-induced HSP27 phosphorylation was associated with a loss of stress fibers and led to a significant decrease in cell migration velocity. Thus, IGF2BP1 controls actin dynamics by inhibiting MAPK4 mRNA translation, which antagonizes MK5-directed phosphorylation of HSP27. This is essential to promote the velocity of tumor cell migration, presumably, in particular, at elevated HSP27 expression, frequently observed in primary malignancies (Kostenko and Moens 2009; Calderwood 2010). In support of this, up-regulated expression of HSP27 severely disrupts the actin cytoskeleton (see Supplemental Fig. S7A), whereas this is not observed upon the expression of HSP27-AA. This suggests that with increasing levels of HSP27, phosphorylation at S78 and S82 needs to be restricted to ensure proper actin dynamics and thus maintain or promote the velocity of cell migration. This could be facilitated by the de novo synthesis of IGF2BP1, at least in tumor cells expressing MAPK4 and MK5. However, future studies have to reveal whether IGF2BP1 facilitates these regulatory roles also via the control of additional candidate target mRNAs (see Supplemental Table S1).
The asymmetric distribution of phospholipid signaling with PIP3 at the leading edge versus PIP2 at the trail has a severe impact on the directed movement of cells (Kolsch et al. 2008; Cain and Ridley 2009; Iglesias 2009; Berzat and Hall 2010). This appears to be particularly important in shallow or in the absence of defined guidance cues (Wessels et al. 2007). Under such conditions, PTEN prevents the formation of lateral pseudopods by sustaining an asymmetric PIP3/PIP2 gradient and thus promotes cell polarization intrinsically. In support of this, the IGF2BP1-directed increase in cell polarization and migration directionality is PTEN-dependent. Notably, this is consistent with previous studies that reveal that the forced expression of IGF2BP1 or ZBP1 in cells derived from rat mammary carcinomas promotes the directionality of migration in vitro (Lapidus et al. 2007). We propose that IGF2BP1 constrains PIP3-directed RAC1 activation by enhancing PTEN expression and thereby keeps cells on track. Notably, moderate changes in the levels or activation of RAC1 have profound consequences on cell migration (Pankov et al. 2005), but RAC1 amounts remained largely unaffected by IGF2BP1 (data not shown).
Taken together, our studies indicate that IGF2BP1 promotes both the velocity and directionality of tumor cell migration by controlling the action of at least two intracellular signaling pathways: MK5 and PIP3 signaling. Strikingly, IGF2BP1 enhances the cross-talk of both pathways by antagonizing MK5-mediated as well as AKT-mediated phosphorylation of HSP27. In tumor-derived cells expressing PTEN like U2OS or ES-2, the IGF2BP1-directed inhibition of MK5 appears to prevail in terms of modulating actin dynamics, resulting in enhanced cell migration speed. Concomitantly, IGF2BP1 promotes intrinsic cell polarization and thus the directionality of migration by enforcing the expression of PTEN. The up-regulation of PTEN alone is insufficient to promote the speed or directionality of migration in U2OS cells. This supports the view that PTEN enhances the directionality of migration mainly by enforcing intrinsic cell polarization and preventing the formation of lateral pseudopods. Presumably, this is insufficient to promote migration directionality without an up-regulation of migration speed. However, the loss of PTEN or its activity is a hallmark of tumor progression, and thus down-regulation of PTEN expression was correlated with increased metastatic potential (Chalhoub and Baker 2009). Preliminary studies in glioblastoma-derived U251 cells that lack PTEN and IGF2BP1 confirm that IGF2BP1 antagonizes the phosphorylation of HSP27 upon lentiviral transduction (Supplemental Fig. S11A,B). This correlated with enhanced migration velocity, whereas the directionality of migration remained largely unaffected, presumably due to the lack of PTEN (Supplemental Fig. S11C–F). However, even in U251 cells stably transduced with GFP-chZBP1, the directionality of migration was not enhanced by transiently expressed PTEN. Why PTEN was incapable of promoting the directionality of cell migration in U251 cells remains to be elucidated. Notably, it was shown that directed movement of cells is observed even in the absence of both PI3Ks and PTEN in Dictyostelium (Hoeller and Kay 2007). Hence, alternative regulatory networks can substitute for PTEN-modulated PIP3/PIP2 signaling in the control of cell migration directionality (Kolsch et al. 2008; Cain and Ridley 2009; Chalhoub and Baker 2009). This presumably applies to tumor cells lacking PTEN in which IGF2BP1 apparently promotes the velocity but not the directionality of cell migration, as demonstrated in glioblastoma-derived U373 cells (data not shown) or U251 cells (Supplemental Fig. S11A–F).
In conclusion, our findings provide a novel concept for how tumor cell migration can be modulated at the post-transcriptional level. The IGF2BP1-facilitated control of MAPK4 and PTEN expression regulates and interconnects MK5 with PIP3 signaling. This identifies a pivotal role of MK5 and PTEN in directing tumor cell migration and suggests that IGF2BP1 acts as a governor that fine-tunes speed and the directionality of tumor cell movement in the absence of defined guidance cues. Whether and how IGF2BP1 modulates the invasive potential of tumor-derived cells remains to be determined. We propose that IGF2BP1 promotes the migration of tumor cells along tumor progression, although the subset of involved target mRNAs as well as the outcome of modulated signaling is likely to vary with tumor type and stage.
Materials and methods
Cell culture, transfection, and inhibitors
U2OS, ES-2, U251, and U373 cells were cultured in DMEM supplemented with 10% FBS. Where indicated, cells were transfected with siRNAs or plasmids by RNAiMax or Lipofectamine 2000 (Invitrogen) as previously described (Stohr et al. 2006). For mRNA decay analyses, transcription was blocked by actinomycin D (5 μM). Where indicated, cell stress was induced by sodium arsenate (2.5 mM), and MAPKAPK activity (MK2,3,5) was inhibited by HSP25 kinase inhibitor (10 μM; Calbiochem).
Immunofluorescence, cell migration assays, and microscopy
U2OS cells were grown on glass coverslips, fixed, and processed for indirect immunostaining using indicated antibodies (Supplemental Table S2C) or phalloidin-TRITC labeling (Sigma) as previously described (Stohr et al. 2006). Image acquisition was performed on a Leica SP5 confocal microscope using a 100× TIRF objective or on a TE2000 microscope (Nikon) using a 63× Plan Apo objective. Representative images are shown. For scratch assays, 1 × 105 cells were seeded per well in 24-well plates. Cell layers were wounded 2–3 h after seeding with a rubber policeman. Wound closure was monitored by bright-field microscopy (TS-100, Nikon, 10× objective). Wound closure was determined by quantifying the scratch area using ImageJ and normalization to the initial wound size. For MTOC orientation, MTOCs were labeled by immunostaining for pericentrin. Images were acquired on a Leica SP5 confocal microscope using a 20× magnification. Cells were scored as “polarized” when the MTOC was located within a 90° sector facing directly toward the wound as previously described (Yamana et al. 2006).
For quantification of lateral pseudopodia, cells were seeded on collagen-coated coverslips for 3 h before fixation. Nuclei were labeled with DAPI. Lamellipodia/filopodia outside a 45° angle around the longest cell axis crossing the nucleus were classified as lateral pseudopodia.
For single-cell migration analyses, cells were seeded on collagen-coated glass-bottomed dishes. Single-cell migration was monitored over 5–10 h by time-lapse analyses (one frame per 3 min) based on GFP fluorescence using a Leica TP5 confocal microscope equipped with a Ludin cube life chamber using a 40× (oil) or 20× (air) objective. The velocity of cell migration was determined as mean displacement (microns per minute) of tracked cells using the “manual tracking” plugin (http://rsbweb.nih.gov/ij/plugins/track/track.html) for ImageJ. The directionality of single-cell migration was determined as the ratio of net distance (D) to total track distance (T) based on single-cell tracks.
G-actin/F-actin ratio
To determine cellular G-actin/F-actin ratios, 1 × 104 U2OS cells allowed to adhere overnight were fixed and processed for F-actin labeling by phalloidin-Tritc or for G-actin labeling by DNaseI-Alexa594. G-actin/F-actin ratios were determined by fluorescence spectrometry using DAPI staining for normalization; triplicates of each sample were analyzed.
PI(3,4,5)P3 extraction and lipid–protein overlay
PI(3,4,5)P3 levels were quantified using the PI(3,4,5)P3 Mass Strip kit (Echelon) according to the manufacturer's instructions. In brief, acidic lipids from 5 × 106 U2OS cells were extracted 96 h post-transfection of IGF2BP1-directed, PTEN-directed, or control siRNAs, respectively. Extracts were spotted on the Mass Strips and PI(3,4,5)P3 was determined by lipid–protein overlay analyses. PI(3,4,5)P3 amounts in samples were calculated relative to standards by analyzing overlay assays in a GenGenius Bioimaging system (Syngene).
Identification of IGF2BP1 target transcripts
U2OS cells transfected with indicated siRNAs for 72 h were stressed by arsenate for 2 h. Total RNA was extracted using Trizol and purified using RNeasy MinElute Cleanup kit (Qiagen). RNA integrity and concentration were then examined on an Agilent 2100 Bioanalyzer (Agilent Technologies) using the RNA 6.000 LabChip kit (Agilent Technologies). Changes in RNA abundance were determined by microarray analyses using Affymetrix GeneChips (HG133 plus 2.0) at the microarray core facility of the IZKF (Leipzig, Germany) using the mas5 algorithm for data analyses. Annotated transcripts (Supplemental Table S1) decreased more than eightfold upon IGF2BP1 knockdown and stress were functionally classified using the “functional annotation clustering” tool provided by the NIH database DAVID.
Plasmids and siRNAs/shRNAs
MAPK4, PTEN, and HSP27 cDNAs were cloned by RT–PCR from U2OS cells. PCR products were subcloned in Zero-Blunt plasmid (Invitrogen) or pGEM-T vector (Promega) and sequenced before insertion via EcoRI/XhoI at the 3′ end of a firefly luciferase, Flag tag in pcDNA3.1 (Invitrogen), pEGFP-C2 vector (Clontech), or mRFP-C1 respectively. HSP27 mutants were generated by the insertion of oligonucleotides (Supplemental Table S2A). PTEN nt1932-2243 (NM_000314) was cloned in-frame to the luciferase ORF as previously described (Weidensdorfer et al. 2009). For cloning details and oligonucleotides used, see Supplemental Table S2A. GFP-chZBP1 and the RNAi-insensitive ZBP1 mutant were as previously described (Huttelmaier et al. 2005). GFP-MK5 was a kind gift of O.M. Seternes. GFP-AKT1, GFP-AKT1DN, and GFP-RAC1DN were kind gifts of M. Hatzfeld. siRNAs directed against IGF2BP1 were as previously described (Weidensdorfer et al. 2009). Validated siRNAs directed against PTEN were obtained from Cell Signaling. Sh plasmids were generated using the pSUPER_neo vector system (Oligoengine). For sequence details, see Supplemental Table S2B.
Lentiviruses
Lentiviruses for transduction of cDNAs were generated using the Lenti-X lentiviral expression system (Clontech). A new multiple cloning site (see Supplemental Table S2A) was inserted via AfeI/XbaI in pLVX-puro. Subsequently, EGFP was cloned via XbaI/BamHI, and ZBP1 was inserted in-frame by EcoRI and XhoI. Since transient expression of IGF2BP1/ZBP1 interferes with virus assembly (Zhou et al. 2008), a Tet-responsive CMV promoter was inserted to transiently suppress ZBP1 expression during virus production. For virus production, HEK293T cells were cotransfected by CaPO4 precipitation with pMD2.G (Addgene, no. 12259), psPAX2 (Addgene, no. 12260), pLVX-puro plasmids containing either EGFP alone or EGFP-chZBP1, and pcDNA6/TR (Invitrogen). Lentiviruses were purified 48 h post-transfection by Lentivirus Concentrator (Clontech) according to the manufacturer's instructions.
Luciferase reporter assay
U2OS cells were transfected with siRNAs for 48 h before the transfection of reporter plasmids for 24 h. Luciferase activities were determined by DualGlo (Promega) as previously described (Stohr et al. 2006; Weidensdorfer et al. 2009).
Semiquantitative RT–PCR and qRT–PCR
RNA isolation, reverse transcription, and semiquantitative RT–PCR as well as qRT–PCR were performed as previously described (Stohr et al. 2006). For the oligonucleotide sequences used, see Supplemental Table S2D. Changes in RNA levels were determined by the ΔCt or ΔΔCt method using indicated RNAs for cross-normalization (Stohr et al. 2006). All data were analyzed from at least three independent experiments, and statistical significance was validated by Student`s t-test as indicated.
RNA copurification and coimmunopurification
Immunopurification of IGF2BP1–RNA complexes was essentially performed as previously described (Huttelmaier et al. 2005). In brief, cytoplasmic extracts of U2OS cells were prepared in Tap buffer (10 mM HEPES at pH 7.6, 3 mM MgCL2, 150 mM KCl, 5% glycerol, 0.5% NP-40) supplemented with protease inhibitors, RNAsin, and yeast t-RNA (10 μg/mL). IGF2BP1 was immunopurified using monoclonal antibodies as described previously (Weidensdorfer et al. 2009). IgG beads alone served as controls. After extensive washing with Tap buffer, protein–RNA complexes were eluted by the addition of 0.2% SDS and β-mercaptoethanol. Aliquots of elutes were analyzed by Western blotting, and RNA was isolated by phenol/chloroform extraction. RNA enrichment by IGF2BP1 pull-down was determined by semiquantitative RT-PCR or qRT–PCR using primers listed in Supplemental Table S2D.
For coimmunoprecipitation of ACTB and HSP27, cells were lysed in RIPA-NP40 (50 mM Tris/HCl at pH 7.5, 150 mM NaCl, 0.5% NP40) supplemented with protease and phosphatase inhibitors (Sigma Aldrich, Cocktail 1 and 2). Pull-down of endogenous HSP27 using the polyclonal anti-HSP27 antibody (Millipore) or of GFP-tagged HSP27 mutants using an anti-GFP antibody (Roche) was performed at 4°C. IgG beads alone served as controls. After intense washing, proteins were eluted in SDS sample buffer. Inputs and immunoprecipitations were analyzed by Western blotting with the indicated antibodies.
Western blotting
For Western blotting, total protein extracts were prepared in RIPA-NP40 or BB buffer (20 mM Tris/HCl at pH7.5, 150 mM NaCl, 1.5 mM MgCl2, 8.7% glycerine, 0.05% NP40) and analyzed by an Odyssey infrared scanner (LICOR) as previously described (Weidensdorfer et al. 2009). For the antibodies used, see Supplemental Table S2C.
In vitro transcription and filter-binding analyses
NIR dye labeling of RNA probes and filter-binding assays were performed as recently described (Kohn et al. 2010). The percentage of bound RNA was determined by input-normalized fluorescence intensities plotted against the protein concentration using the Hill equation for fitting.
Acknowledgments
We thank O.M. Seternes for providing GFP-MK5, and M. Hatzfeld for providing GFP-AKT1, GFP-RAC1DN, and GFP-AKT1DN. This work was supported by NIH funding to R.H.S. (NIH AR41480/GM84364) and DFG funding (Hu1547/3-1 and Hu1547/2-1) to S.H.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.177642.111.
References
- Aberg E, Perander M, Johansen B, Julien C, Meloche S, Keyse SM, Seternes OM 2006. Regulation of MAPK-activated protein kinase 5 activity and subcellular localization by the atypical MAPK ERK4/MAPK4. J Biol Chem 281: 35499–35510 [DOI] [PubMed] [Google Scholar]
- Aberg E, Torgersen KM, Johansen B, Keyse SM, Perander M, Seternes OM 2009. Docking of PRAK/MK5 to the atypical MAPKs ERK3 and ERK4 defines a novel MAPK interaction motif. J Biol Chem 284: 19392–19401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An SS, Fabry B, Mellema M, Bursac P, Gerthoffer WT, Kayyali US, Gaestel M, Shore SA, Fredberg JJ 2004. Role of heat shock protein 27 in cytoskeletal remodeling of the airway smooth muscle cell. J Appl Physiol 96: 1701–1713 [DOI] [PubMed] [Google Scholar]
- Berzat A, Hall A 2010. Cellular responses to extracellular guidance cues. EMBO J 29: 2734–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain RJ, Ridley AJ 2009. Phosphoinositide 3-kinases in cell migration. Biol Cell 101: 13–29 [DOI] [PubMed] [Google Scholar]
- Calderwood SK 2010. Heat shock proteins in breast cancer progression—a suitable case for treatment? Int J Hyperthermia 26: 681–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub N, Baker SJ 2009. PTEN and the PI3–kinase pathway in cancer. Annu Rev Pathol 4: 127–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J, Singer RH 2005. How and why does β-actin mRNA target? Biol Cell 97: 97–110 [DOI] [PubMed] [Google Scholar]
- Coulombe P, Meloche S 2007. Atypical mitogen-activated protein kinases: Structure, regulation and functions. Biochim Biophys Acta 1773: 1376–1387 [DOI] [PubMed] [Google Scholar]
- Dahm R, Kiebler M 2005. Cell biology: Silenced RNA on the move. Nature 438: 432–435 [DOI] [PubMed] [Google Scholar]
- Deleris P, Rousseau J, Coulombe P, Rodier G, Tanguay PL, Meloche S 2008. Activation loop phosphorylation of the atypical MAP kinases ERK3 and ERK4 is required for binding, activation and cytoplasmic relocalization of MK5. J Cell Physiol 217: 778–788 [DOI] [PubMed] [Google Scholar]
- Doshi BM, Hightower LE, Lee J 2010. HSPB1, actin filament dynamics, and aging cells. Ann N Y Acad Sci 1197: 76–84 [DOI] [PubMed] [Google Scholar]
- During RL, Gibson BG, Li W, Bishai EA, Sidhu GS, Landry J, Southwick FS 2007. Anthrax lethal toxin paralyzes actin-based motility by blocking Hsp27 phosphorylation. EMBO J 26: 2240–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A 2001. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell 106: 489–498 [DOI] [PubMed] [Google Scholar]
- Findeis-Hosey JJ, Xu H 2010. The use of insulin like-growth factor II messenger RNA binding protein-3 in diagnostic pathology. Hum Pathol 42: 303–314 [DOI] [PubMed] [Google Scholar]
- Gaestel M 2006. MAPKAP kinases—MKs—two's company, three's a crowd. Nat Rev Mol Cell Biol 7: 120–130 [DOI] [PubMed] [Google Scholar]
- Gerits N, Mikalsen T, Kostenko S, Shiryaev A, Johannessen M, Moens U 2007. Modulation of F-actin rearrangement by the cyclic AMP/cAMP-dependent protein kinase (PKA) pathway is mediated by MAPK-activated protein kinase 5 and requires PKA-induced nuclear export of MK5. J Biol Chem 282: 37232–37243 [DOI] [PubMed] [Google Scholar]
- Gertler F, Condeelis J 2011. Metastasis: Tumor cells becoming MENAcing. Trends Cell Biol 21: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller O, Kay RR 2007. Chemotaxis in the absence of PIP3 gradients. Curr Biol 17: 813–817 [DOI] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH 2005. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438: 512–515 [DOI] [PubMed] [Google Scholar]
- Iglesias PA 2009. Spatial regulation of PI3K signaling during chemotaxis. Wiley Interdiscip Rev Syst Biol Med 1: 247–253 [DOI] [PubMed] [Google Scholar]
- Jia Y, Wu SL, Isenberg JS, Dai S, Sipes JM, Field L, Zeng B, Bandle RW, Ridnour LA, Wink DA, et al. 2010. Thiolutin inhibits endothelial cell adhesion by perturbing Hsp27 interactions with components of the actin and intermediate filament cytoskeleton. Cell Stress Chaperones 15: 165–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Schumacher S, Singh MK, Kispert A, Kotlyarov A, Gaestel M 2006. Characterization of the atypical MAPK ERK4 and its activation of the MAPK-activated protein kinase MK5. J Biol Chem 281: 35511–35519 [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH 1994. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J Cell Biol 127: 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobel M, Weidensdorfer D, Reinke C, Lederer M, Schmitt WD, Zeng K, Thomssen C, Hauptmann S, Huttelmaier S 2007. Expression of the RNA-binding protein IMP1 correlates with poor prognosis in ovarian carcinoma. Oncogene 26: 7584–7589 [DOI] [PubMed] [Google Scholar]
- Kohn M, Lederer M, Wachter K, Huttelmaier S 2010. Near-infrared (NIR) dye-labeled RNAs identify binding of ZBP1 to the noncoding Y3-RNA. RNA 16: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolsch V, Charest PG, Firtel RA 2008. The regulation of cell motility and chemotaxis by phospholipid signaling. J Cell Sci 121: 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostenko S, Moens U 2009. Heat shock protein 27 phosphorylation: Kinases, phosphatases, functions and pathology. Cell Mol Life Sci 66: 3289–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostenko S, Johannessen M, Moens U 2009. PKA-induced F-actin rearrangement requires phosphorylation of Hsp27 by the MAPKAP kinase MK5. Cell Signal 21: 712–718 [DOI] [PubMed] [Google Scholar]
- Kress TR, Cannell IG, Brenkman AB, Samans B, Gaestel M, Roepman P, Burgering BM, Bushell M, Rosenwald A, Eilers M 2011. The MK5/PRAK kinase and Myc form a negative feedback loop that is disrupted during colorectal tumorigenesis. Mol Cell 41: 445–457 [DOI] [PubMed] [Google Scholar]
- Lapidus K, Wyckoff J, Mouneimne G, Lorenz M, Soon L, Condeelis JS, Singer RH 2007. ZBP1 enhances cell polarity and reduces chemotaxis. J Cell Sci 120: 3173–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm I, Ross J 2002. Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol Cell Biol 22: 3959–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE 2006. Asymmetrical β-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci 9: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Weinberg RA 2008. Micromanagers of malignancy: Role of microRNAs in regulating metastasis. Trends Genet 24: 448–456 [DOI] [PubMed] [Google Scholar]
- Mounier N, Arrigo AP 2002. Actin cytoskeleton and small heat shock proteins: How do they interact? Cell Stress Chaperones 7: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberman F, Rand K, Maizels Y, Rubinstein AM, Yisraeli JK 2007. VICKZ proteins mediate cell migration via their RNA binding activity. RNA 13: 1558–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankov R, Endo Y, Even-Ram S, Araki M, Clark K, Cukierman E, Matsumoto K, Yamada KM 2005. A Rac switch regulates random versus directionally persistent cell migration. J Cell Biol 170: 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perander M, Aberg E, Johansen B, Dreyer B, Guldvik IJ, Outzen H, Keyse SM, Seternes OM 2008. The Ser(186) phospho-acceptor site within ERK4 is essential for its ability to interact with and activate PRAK/MK5. Biochem J 411: 613–622 [DOI] [PubMed] [Google Scholar]
- Read DE, Gorman AM 2009a. Heat shock protein 27 in neuronal survival and neurite outgrowth. Biochem Biophys Res Commun 382: 6–8 [DOI] [PubMed] [Google Scholar]
- Read DE, Gorman AM 2009b. Involvement of Akt in neurite outgrowth. Cell Mol Life Sci 66: 2975–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH 1997. Characterization of a β-actin mRNA zipcode-binding protein. Mol Cell Biol 17: 2158–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinthone S, Tyagi M, Gerthoffer WT 2008. Small heat shock proteins in smooth muscle. Pharmacol Ther 119: 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Welshhans K, Wen Z, Yao J, Xu M, Goshima Y, Zheng JQ, Bassell GJ 2010. Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local β-actin synthesis and growth cone turning. J Neurosci 30: 9349–9358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seternes OM, Mikalsen T, Johansen B, Michaelsen E, Armstrong CG, Morrice NA, Turgeon B, Meloche S, Moens U, Keyse SM 2004. Activation of MK5/PRAK by the atypical MAP kinase ERK3 defines a novel signal transduction pathway. EMBO J 23: 4780–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Huttelmaier S 2006. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol 175: 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak H, Jang E, Kim SB, Park J, Suk J, Yoon YS, Ahn JK, Lee JH, Joe CO 2007. 14-3-3ε inhibits MK5-mediated cell migration by disrupting F-actin polymerization. Cell Signal 19: 2379–2387 [DOI] [PubMed] [Google Scholar]
- Vainer G, Vainer-Mosse E, Pikarsky A, Shenoy SM, Oberman F, Yeffet A, Singer RH, Pikarsky E, Yisraeli JK 2008. A role for VICKZ proteins in the progression of colorectal carcinomas: Regulating lamellipodia formation. J Pathol 215: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA 2009. MicroRNAs: Crucial multi-tasking components in the complex circuitry of tumor metastasis. Cell Cycle 8: 3506–3512 [DOI] [PubMed] [Google Scholar]
- Vikesaa J, Hansen TV, Jonson L, Borup R, Wewer UM, Christiansen J, Nielsen FC 2006. RNA-binding IMPs promote cell adhesion and invadopodia formation. EMBO J 25: 1456–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidensdorfer D, Stohr N, Baude A, Lederer M, Kohn M, Schierhorn A, Buchmeier S, Wahle E, Huttelmaier S 2009. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA 15: 104–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels D, Lusche DF, Kuhl S, Heid P, Soll DR 2007. PTEN plays a role in the suppression of lateral pseudopod formation during Dictyostelium motility and chemotaxis. J Cell Sci 120: 2517–2531 [DOI] [PubMed] [Google Scholar]
- Yamana N, Arakawa Y, Nishino T, Kurokawa K, Tanji M, Itoh RE, Monypenny J, Ishizaki T, Bito H, Nozaki K, et al. 2006. The Rho–mDia1 pathway regulates cell polarity and focal adhesion turnover in migrating cells through mobilizing Apc and c-Src. Mol Cell Biol 26: 6844–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K, Yisraeli JK 2002. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene 287: 49–54 [DOI] [PubMed] [Google Scholar]
- Yaniv K, Fainsod A, Kalcheim C, Yisraeli JK 2003. The RNA-binding protein Vg1 RBP is required for cell migration during early neural development. Development 130: 5649–5661 [DOI] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ 2006. An essential role for β-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci 9: 1265–1273 [DOI] [PubMed] [Google Scholar]
- Yisraeli JK 2005. VICKZ proteins: A multi-talented family of regulatory RNA-binding proteins. Biol Cell 97: 87–96 [DOI] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ 2001. Neurotrophin-induced transport of a β-actin mRNP complex increases β-actin levels and stimulates growth cone motility. Neuron 31: 261–275 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Rong L, Lu J, Pan Q, Liang C 2008. Insulin-like growth factor II mRNA binding protein 1 associates with Gag protein of human immunodeficiency virus type 1, and its overexpression affects virus assembly. J Virol 82: 5683–5692 [DOI] [PMC free article] [PubMed] [Google Scholar]