Muscle repair is regulated by satellite cells, adult skeletal muscle stem cells that control muscle regeneration by proliferating and fusing with injured myofibers. MyoD is required for muscle regeneration; however, the mechanisms regulating MyoD expression in satellite cells are unclear. In this study, Olson and colleagues have demonstrated that deletion of MASTR and MRTF-A, two members of the Myocardin family of transcription factors, leads to skeletal muscle regeneration defects and down-regulation of MyoD. In addition, MASTR was shown to bind to a MyoD enhancer. Thus, MASTR and MRTF-A cooperate in controlling MyoD expression and muscle regeneration.
Keywords: muscle regeneration, satellite cells, MASTR, MRTF-A, MEF2, MyoD
Abstract
In response to skeletal muscle injury, satellite cells, which function as a myogenic stem cell population, become activated, expand through proliferation, and ultimately fuse with each other and with damaged myofibers to promote muscle regeneration. Here, we show that members of the Myocardin family of transcriptional coactivators, MASTR and MRTF-A, are up-regulated in satellite cells in response to skeletal muscle injury and muscular dystrophy. Global and satellite cell-specific deletion of MASTR in mice impairs skeletal muscle regeneration. This impairment is substantially greater when MRTF-A is also deleted and is due to aberrant differentiation and excessive proliferation of satellite cells. These abnormalities mimic those associated with genetic deletion of MyoD, a master regulator of myogenesis, which is down-regulated in the absence of MASTR and MRTF-A. Consistent with an essential role of MASTR in transcriptional regulation of MyoD expression, MASTR activates a muscle-specific postnatal MyoD enhancer through associations with MEF2 and members of the Myocardin family. Our results provide new insights into the genetic circuitry of muscle regeneration and identify MASTR as a central regulator of this process.
Mammalian skeletal muscle possesses the potential to undergo self-repair in response to injury or disease. The advent of stem cell and regenerative medicine has sparked renewed interest in understanding the molecular and cellular mechanisms of muscle regeneration (Price et al. 2007; Goyenvalle et al. 2011; Tuan 2011). By enhancing muscle regeneration, stem cell-based therapies have potential applications in multiple conditions associated with muscle wasting, including muscular dystrophy, chronic diseases associated with physical inactivity and loss of muscle mass, and aging. Deciphering the molecular mechanisms of muscle regeneration is essential for the development of such therapies.
Muscle repair is a multistep process that includes myofiber degeneration, regeneration, and remodeling (Le Grand and Rudnicki 2007; Ten Broek et al. 2010; Tsivitse 2010). Muscle degeneration is characterized by sarcomere disruption, cell permeabilization, and induction of an acute inflammatory response. The regeneration process is marked by the activation of a myogenic stem cell population, referred to as satellite cells (SCs), which give rise to proliferating myoblasts, followed by myoblast differentiation and fusion into regenerated myofibers. During the final stages of muscle repair, myofibers remodel to produce mature muscle fibers and recover the contractile capacity of the injured muscle.
SCs are the primary population of adult skeletal muscle stem cells, comprising ∼1%–4% of muscle nuclei in normal skeletal muscle (Whalen et al. 1990). Quiescent SCs are characterized by three criteria: a unique localization between the sarcolemma and the basal lamina; a distinct cellular morphology that includes a small cytoplasmic volume, an enlarged nucleus, and condensed chromatin; and expression of the Pax7 transcription factor (Buckingham and Relaix 2007; Biressi and Rando 2010; Kang and Krauss 2010). In response to chemical or mechanical muscle damage, exercise, or overuse, quiescent SCs become activated and migrate to the injury site, where they proliferate and up-regulate MyoD and Myf5 expression. Upon activation, the majority of SCs down-regulate Pax7 expression and undergo terminal muscle cell differentiation, whereas a small proportion of cells down-regulate MyoD expression, maintain Pax7 expression, and revert back to quiescence to sustain the SC pool (Le Grand and Rudnicki 2007; Olguin et al. 2007; Biressi and Rando 2010; Wu et al. 2010).
Expression of the myogenic regulatory factor MyoD specifies the identity of embryonic and adult muscle progenitor cells (Rudnicki et al. 1993; Buckingham 1994; Olson and Klein 1994; Tapscott 2005). MyoD expression, which marks SC activation and differentiation, is required for normal muscle regeneration (Megeney et al. 1996). MyoD knockout mice show severely deficient skeletal muscle regeneration following injury, due to excessive proliferation and defective differentiation of SCs (Sabourin et al. 1999; Yablonka-Reuveni et al. 1999; Cornelison et al. 2000; White et al. 2000; Schuierer et al. 2005). Reminiscent of its expression in activated SCs, differentiating myoblasts up-regulate MyoD and activate transcription of MyoD target genes, resulting in irreversible cell cycle arrest and myogenic differentiation. On the other hand, down-regulation of MyoD in activated SCs causes return to quiescence (Yoshida et al. 1998). Relatively little is known of the upstream activators of MyoD expression. Deciphering the mechanisms that control MyoD expression and function in SCs should provide important insights into the molecular basis of muscle regeneration.
Members of the Myocardin family of transcription factors (Myocardin, MRTF-A, MRTF-B, and MASTR) play important roles in differentiation and remodeling of cardiac, smooth, and skeletal muscle cells (Creemers et al. 2006a; Pipes et al. 2006; Posern and Treisman 2006; Olson and Nordheim 2010). MASTR is a muscle-enriched MEF2 coactivator (Creemers et al. 2006a), whereas Myocardin and MRTF-A/-B are SRF coactivators (Olson and Nordheim 2010). Expression of dominant-negative MRTF-A in mouse skeletal muscle causes muscle hypoplasia (S Li et al. 2005); however, MRTF-A and MRTF-B knockout mice do not show obvious skeletal muscle defects (J Li et al. 2005; Oh et al. 2005; Li et al. 2006; Sun et al. 2006). MRTF-A/-B double-knockout mice revealed that MRTF-A/-B are redundant in the brain and heart, and possibly in other tissues where they are coexpressed (Mokalled et al. 2010; EN Olson and MH Mokalled, unpubl.). The in vivo functions of MASTR and MRTF-A/-B in mammalian skeletal muscle development, maintenance, or regeneration remain unknown.
Here, we show that MASTR and MRTF-A are up-regulated in SCs in response to injury. Global and SC-specific deletion of MASTR in mice causes impaired muscle regeneration in response to chemical injury and muscular dystrophy. This regeneration defect is exacerbated by MRTF-A deletion and is due to aberrant differentiation and excessive proliferation of SCs. These abnormalities resemble those of mice lacking MyoD, which is down-regulated in the absence of MASTR and MRTF-A. Consistent with a regulatory role of MASTR in the control of MyoD expression, MASTR activates a muscle-specific MyoD enhancer together with MEF2 and MRTF-A. These findings reveal a previously unrecognized regulatory network within myogenic stem cells in which MASTR functions as a primary upstream activator of skeletal muscle regeneration.
Results
Up-regulation of MASTR in response to muscle injury
Members of the Myocardin family of transcription factors have been implicated in pathological remodeling of cardiac and vascular smooth muscle (Parmacek 2007; Olson and Nordheim 2010; Small et al. 2010), but little is known of their potential roles in skeletal muscle disease. To investigate the involvement of this family of coactivators in the response of skeletal muscle to injury, we surveyed the expression of MASTR, MRTF-A, and MRTF-B in response to a series of skeletal muscle injuries that induce regeneration.
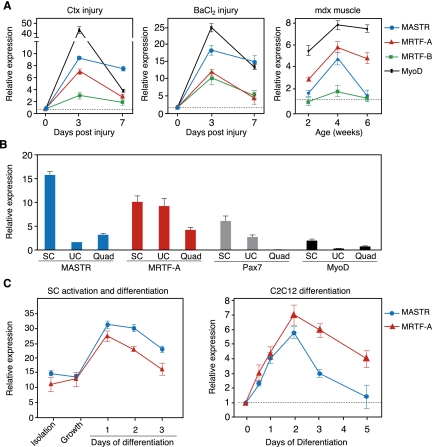
Injection of skeletal muscle with cardiotoxin (Ctx) causes myofiber degeneration followed by regeneration and repair, as activated SCs proliferate, differentiate, and fuse with injured myofibers (Yan et al. 2003). As shown in Figure 1A, MASTR was up-regulated in the adult tibialis anterior (TA) muscle at days 3 and 7 following Ctx injury. MRTF-A was also up-regulated in response to Ctx injury, whereas MRTF-B showed relatively little change in expression. The pattern of MASTR and MRTF-A expression during muscle regeneration resembled that of MyoD. Injection of the TA muscle with barium chloride (BaCl2) also causes muscle injury followed by regeneration and was accompanied by similar changes in expression of MASTR, MRTF-A, and MyoD (Fig. 1A). In addition, MASTR and MRTF-A were up-regulated in parallel with MyoD in the TA muscle of mdx mice, concomitant with muscle degeneration and regeneration (Fig. 1A).
Figure 1.
Up-regulation of MASTR and MRTF-A during muscle regeneration. (A) Shown is the expression of MASTR, MRTF-A, and MRTF-B in the TA muscle following Ctx and barium chloride (BaCl2) injuries and in mdx mice. qPCR shows MASTR and MRTF-A up-regulation 3 d after Ctx and BaCl2 injection. MASTR and MRTF-A are also up-regulated in the TA muscle of 2-, 4-, and 6-wk-old mdx mice compared with wild-type (WT) mice. For Ctx and BaCl2 injury, relative expression at days 3 and 7 post-injury is normalized to GAPDH levels and to baseline level at day 0. For mdx mice, relative expression is normalized to GAPDH levels and to wild-type expression for each time point. MyoD expression is used as control for muscle regeneration and parallels MASTR and MRTF-A expression patterns. (B) Expression of MASTR and MRTF-A in sorted SCs. Sorted SCs (SC), mononuclear unsorted cells (UCs), and quadriceps mRNA samples (Quad) were used for qPCR. Relative expression is normalized to the levels of ribosomal 18S RNA. The levels of Pax7 and MyoD are shown as controls. (C) Up-regulation of MASTR and MRTF-A expression during SC and C2C12 differentiation. Sorted SCs, activated SCs maintained under growth conditions, and differentiating SCs at days 1, 2, or 3 of differentiation were used for qPCR. MASTR and MRTF-A are expressed in sorted and activated SCs, and are up-regulated at days 1 and 2 of differentiation. Similarly, qPCR on differentiating C2C12 cultures shows approximately sixfold to sevenfold up-regulation of MASTR and MRTF-A by day 2 of differentiation.
Expression of MASTR and MRTF-A in SCs
To determine whether MASTR and MRTF-A are expressed in SCs in response to muscle injury, SCs were isolated from adult skeletal muscle and sorted using positive selection for CD34 and negative selection of Sca1, CD31, and CD45 (Supplemental Fig. S1A,B; Beauchamp et al. 2000; Sherwood et al. 2004; Montarras et al. 2005; Sacco et al. 2008). MASTR expression was enriched in sorted SCs compared with either unsorted cells (UCs) or whole quadriceps (Quad), whereas MRTF-A was expressed at comparable levels in SCs and UCs (Fig. 1B). Pax7 and MyoD expression was also enriched in sorted SCs (Fig. 1B; Supplemental Fig. S1C), confirming the identity of this cell population. MASTR and MRTF-A expression was unchanged when SCs were activated to proliferate by exposure to growth medium and was up-regulated when SCs were induced to differentiate into myotubes in the presence of differentiation medium (Fig. 1C; Musaro and Barberi 2010). Similarly, MASTR and MRTF-A expression was up-regulated during differentiation of C2C12 myoblasts into myotubes (Fig. 1C). These findings suggested the potential involvement of MASTR and MRTF-A in SC-mediated responses to muscle injury and differentiation.
MASTR is required for skeletal muscle regeneration
To investigate the function of MASTR in vivo, we generated a conditional MASTR knockout (MKO) allele by inserting loxP sites for Cre-mediated deletion in introns 1 and 7 of the protein-coding region of the gene using homologous recombination. Expression of Cre recombinase resulted in deletion of exons 2–7 of the MASTR locus, thereby eliminating the MEF2-binding and SAP domains and generating what is expected to be a null allele (Supplemental Fig. S2A). Correct targeting and germline transmission of the MASTR allele was confirmed by Southern blot and PCR of genomic DNA (Supplemental Fig. S2B). By breeding MASTRFloxed mice to CAG-Cre transgenic mice, we obtained global MKO mice (Supplemental Fig. S2C), which were viable, fertile, and without obvious phenotypic defects.
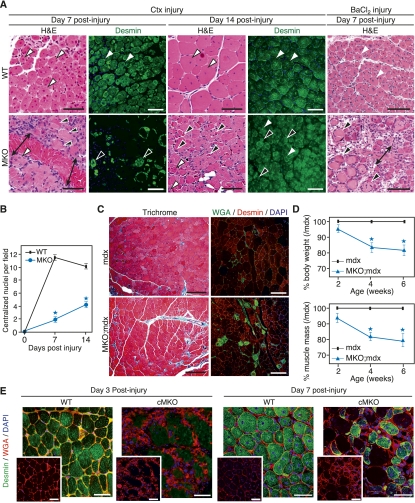
We subjected MKO mice to Ctx injury and evaluated their regeneration potential. Newly regenerated myofibers are readily identifiable by the presence of centralized nuclei, which distinguish them from pre-existing myofibers, in which nuclei are positioned at the cell periphery. By H&E staining, wild-type TA muscles were mostly composed of regenerating myofibers 7 d post-injury (Fig. 2A). MKO TA muscles at 7 d post-injury showed no centralized nuclei and instead were composed of degenerating necrotic fibers, fibrotic tissue, and inflammatory cells (Fig. 2A). By day 14 post-injury, regenerated wild-type myofibers remained centrally nucleated and became homogeneous in size, indicative of their maturation (Fig. 2A). In contrast, MKO myofibers at day 14 post-injury remained heterogeneous in size and were fewer in number compared with wild-type muscle (Fig. 2A).
Figure 2.
Aberrant muscle regeneration of MASTR-null mice. (A) Regeneration of MKO muscle following chemical injury. TA muscle from wild-type (WT) and MKO mice was injected with Ctx and BaCl2 and assayed for regeneration by H&E and by Desmin immunohistochemistry at days 7 and 14 post-injury. Regenerating and degenerating fibers are indicated by white and black arrowheads, respectively. At day 7 post-Ctx or post-BaCl2 injury, wild-type muscle contains regenerating myofibers that are centrally nucleated and heterogeneous in size. MKO muscle shows fewer regenerating fibers and is mostly composed of necrotic fibers and regions of fibrotic tissue and inflammatory cells (black arrows). By day 14 post-Ctx injury, regenerating wild-type fibers are mature, centrally nucleated, and homogeneous in size, whereas regenerating MKO fibers are heterogeneous in size and decreased in number. Bar, 65 μm. (B) Quantitation of regeneration shows a significant decrease in the number of centralized nuclei in MKO muscle compared with wild-type muscle at days 7 and 14 post-injury. Analysis was performed on five animals for each genotype and time point and on five sections from each animal; (*) P < 0.01. (C) Enhanced mdx regeneration defect by MASTR deletion. Trichrome staining and immunohistochemistry for Desmin and WGA show enhanced muscle damage and fibrosis in 4-wk-old MKO;mdx mice compared with mdx mice. Bar, 125 μm. (D) MKO;mdx mice have decreased body mass and muscle mass compared with littermate mdx mice; (*) P < 0.01. (E) Impaired regeneration of cMKO mice following Ctx injury. Staining for Desmin and WGA shows defective TA muscle regeneration of cMKO mice at days 3 and 7 post-Ctx injury. Bar, 45 μm.
Wild-type muscle injured with Ctx initiates expression of Desmin, a marker of skeletal muscle differentiation, 7 d post-injury and shows robust Desmin expression in mature myofibers by day 14 post-injury (Fig. 2A). Desmin-positive myofibers were absent in MKO TA muscle 7 d post-injury and were still fewer in number and immature at day 14 post-injury (Fig. 2A). In addition, the number of centralized nuclei in MKO mice at days 7 and 14 post-injury was significantly decreased compared with wild-type muscle (Fig. 2B). Muscle regeneration in response to injection of BaCl2 was also compromised in MKO mice (Fig. 2A). Thus, MASTR is required for efficient regeneration in two independent models of muscle injury.
To further investigate the requirement of MASTR for skeletal muscle regeneration, we crossed the MKO mice to mdx mice, which harbor a null mutation in the Dystrophin gene. Mdx mice undergo extensive muscle degeneration and regeneration at ∼3 wk of age, until they exhaust their regeneration capacity by ∼7 wk of age (McGeachie et al. 1993; Banks and Chamberlain 2008). Trichrome and wheat germ agglutinin (WGA) stainings revealed severe muscle damage and increased fibrosis in MKO;mdx TA muscle compared with mdx muscle (Fig. 2C). MKO;mdx mice also showed a 20% reduction in body weight and muscle weight at 4 and 6 wk of age compared with mdx mice (Fig. 2D). We conclude that MASTR regulates skeletal muscle regeneration after chemical injury and in a muscular dystrophy disease model.
Aberrant muscle regeneration resulting from SC-specific deletion of MASTR
MASTR expression in proliferating and differentiating SCs and the requirement for MASTR in skeletal muscle regeneration suggest that MASTR regulates SC-mediated skeletal muscle regeneration. However, because MASTR is expressed in mature muscle fibers in addition to SCs, global gene deletion could not definitely distinguish its function in one population versus the other. Therefore, to directly test the role of MASTR in SCs, we crossed the MASTRFloxed mice to Pax7-Cre-ERT2 mice, in which a Cre recombinase-estrogen receptor expression cassette was knocked into the Pax7 locus, allowing for tamoxifen-inducible, SC-specific expression of Cre (Lepper et al. 2009). We subjected the MASTRFloxed/Floxed;Pax7-Cre-ERT2 mice, hereafter referred to as conditional MKO (cMKO) mice, to tamoxifen treatment followed by Ctx injury. At days 3 and 7 post-injury, control TA muscle showed significant recovery, with total replacement of damaged myofibers with newly regenerated, Desmin-positive, centrally nucleated myofibers (Fig. 2E). However, as observed with the global deletion of MASTR, muscle regeneration was defective in cMKO mice, with a significant decrease in the number of regenerating myofibers, persistence of necrotic fibers, and greatly reduced Desmin expression (Fig. 2F). These findings demonstrate that MASTR performs a SC-autonomous role in regulating skeletal muscle regeneration after injury.
MASTR and MRTF-A cooperatively regulate muscle mass and regeneration
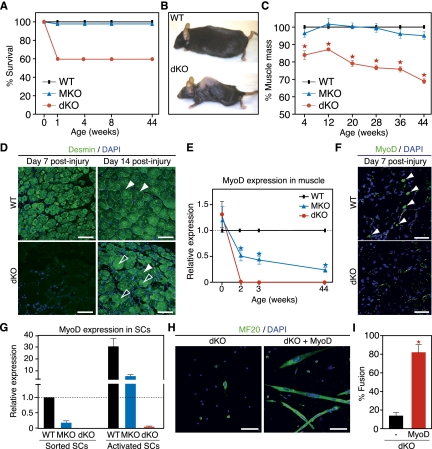
To determine whether MRTF-A and MASTR might act in a common pathway during skeletal muscle regeneration, we generated MASTR/MRTF-A double-knockout (dKO) mice by crossing MKO mice to the previously generated MRTF-A knockout (AKO) mouse line (Li et al. 2006). AKO mice showed no abnormalities in muscle regeneration compared with wild-type mice (Supplemental Fig. S3A). Approximately 40% of the dKO mice showed perinatal lethality (Fig. 3A), which may reflect an essential function for MASTR and MRTF-A during neuronal development. dKO mice that survived beyond postnatal day 1 (P1) were viable and fertile, but showed a significant decrease in body mass and muscle mass, as shown by nuclear magnetic resonance (NMR) analysis of lean mass content (Fig. 3B,C). Muscle mass was not significantly changed in MKO or AKO mice (Li et al. 2006; Sun et al. 2006). By 44 wk of age, dKO mice showed dramatic muscle wasting (Fig. 3B) but normal TA muscle histology, with only a mild decrease in myofiber size compared with either wild-type or MKO TA muscle (Supplemental Fig. S4A). In addition, the regeneration defect of MKO mice was significantly enhanced in dKO mice (Fig. 3D). For example, dKO TA muscle completely lacked regenerating myofibers 7 d post-injury and showed more severe myofiber dropout following injury than MKO mice.
Figure 3.
Cooperative regulation of MyoD expression, muscle mass, and muscle regeneration by MASTR and MRTF-A. (A) Partial lethality of dKO mice at birth. Survival curve shows ∼40% perinatal lethality of dKO mice compared with wild-type (WT) or MKO mice. Mice that survived the first week of postnatal life were viable and fertile. (B) Loss of muscle mass and kyphosis of dKO mice by 44 wk of age. (C) Decreased muscle mass in dKO mice. Percent lean mass was measured by NMR from wild-type, MKO, and dKO mice. dKO mice are significantly leaner than wild-type or MKO littermates. Lean mass content decreased with age and reached ∼70% in 44-wk-old dKO mice. MKO mice do not show a significant decrease in muscle mass at baseline; (*) P < 0.01. (D) Defective regeneration of dKO muscle following Ctx injury. Regeneration of the TA muscle was examined by Desmin immunohistochemistry at days 7 and 14 after Ctx administration. Regenerating and degenerating fibers are indicated by white and black arrowheads, respectively. Bar, 65 μm. (E) Down-regulation of MyoD mRNA levels in MKO and dKO muscle. Hindlimb neonatal muscle and TA muscle from 2-, 3-, and 44-wk-old mice were used for qPCR. MyoD levels were normalized to GAPDH and wild-type MyoD levels for each time point. MyoD expression was significantly diminished in MKO muscle and was undetectable in dKO muscle, starting from 2 wk of age; (*) P < 0.01. (F) Down-regulation of MyoD protein levels in dKO mice. MyoD immunohistochemistry on injured TA muscle from wild-type and dKO mice reveals a dramatic decrease in the number of MyoD-positive cells in dKO muscle at day 7 post-injury. (G) Down-regulation of MyoD mRNA levels in MKO and dKO SCs. Sorted and activated SCs from wild-type, MKO, and dKO mice were used for qPCR. Relative expression was normalized to MyoD levels in wild-type sorted SCs. MyoD levels were down-regulated in MKO SCs and were undetectable in dKO SCs. (H) Rescue of the dKO SC differentiation defect by MyoD expression. dKO SCs were infected with control or MyoD-expressing retrovirus, allowed to differentiate for 4 d, and stained with anti-myosin antibody to assay differentiation. MyoD overexpression rescues the dKO differentiation defect. Bar, 90 μm. (I) Quantification of the percent myotube fusion confirms that MyoD overexpression rescues the dKO SC differentiation defect; (*) P < 0.01.
Regulation of MyoD expression by MASTR and MRTF-A
The regenerative defects in MKO and dKO mice are remarkably similar to those of MyoD-null mice (Megeney et al. 1996). To determine whether MyoD down-regulation might account for the observed abnormalities, we analyzed MyoD expression in wild-type and mutant muscle. As shown in Figure 3E, MyoD expression was comparable between wild-type and dKO mice at birth, but was strikingly undetectable in dKO mice at P14. MyoD expression was also diminished in MKO mice, but unchanged in AKO mice (Fig. 3E; Supplemental Fig. S3B). By immunohistochemistry, MyoD protein was undetectable in the TA muscle of dKO mice 7 d post-injury (Fig. 3F).
MyoD expression was also significantly down-regulated in sorted MKO SCs compared with wild-type SCs (Fig. 3G). Upon activation in culture, wild-type SCs induce MyoD expression, whereas activated MKO SCs showed blunted MyoD induction (Fig. 3G). In addition, MyoD levels were undetectable in sorted or activated dKO SCs. MyoD expression is therefore sensitive to MASTR levels under multiple contexts, and MASTR plus MRTF-A appear essential for normal postnatal expression of MyoD.
To determine whether the block to MyoD expression was sufficient to account for the failure in differentiation of dKO SCs, we performed an in vitro MyoD rescue experiment (Fig. 3H). Forced expression of MyoD by retroviral delivery rescued the differentiation defect of dKO SCs, as shown by increased myotube formation in MyoD-infected dKO SCs compared with control-infected dKO SCs, suggesting that the dKO phenotype is due to MyoD down-regulation (Fig. 3I).
MASTR restricts proliferation and promotes differentiation of SCs
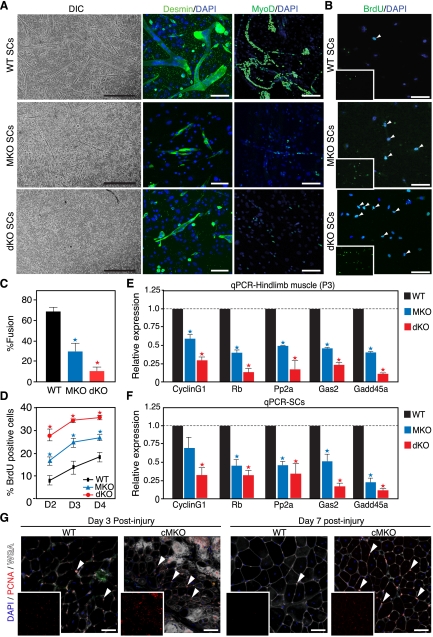
To understand the temporal requirement for MASTR and MRTF-A during muscle regeneration, we monitored the growth and differentiation of MKO and dKO SCs under growth and differentiation culture conditions. Under differentiation conditions, MKO SCs formed fewer myotubes compared with wild-type cultures, and dKO SCs showed even fewer myotubes than MKO SCs (Fig. 4A,C). When equal numbers of wild-type and MKO cells were plated at subconfluency under growth conditions, MKO and dKO SCs achieved confluency faster than wild-type SCs, suggesting a hyperproliferative phenotype (data not shown). To confirm that MKO and dKO SCs are hyperproliferative under growth conditions, we assayed cell proliferation with a 1-h BrdU pulse and found that mutant SCs showed a significant increase in BrdU incorporation compared with wild-type cells (Fig. 4B,D). Moreover, consistent with our in vitro proliferation assays, the number of proliferating PCNA-positive cells was increased in injured MKO muscle at days 3 and 7 post-injury (Fig. 4G). These results confirm that the MKO phenotype was enhanced by deletion of MRTF-A and suggest that MASTR and MRTF-A regulate muscle regeneration by restricting proliferation and promoting differentiation of SCs.
Figure 4.
Increased proliferation and defective differentiation of MKO and dKO SCs. (A) Defective differentiation of MKO and dKO SCs in vitro. Bright-field microscope images show impaired differentiation of MKO SCs and enhanced differentiation defect of dKO SCs, compared with wild-type (WT) cells. Desmin immunohistochemistry confirms aberrant differentiation of MKO and dKO SCs. Extensive multinucleated myotube networks are observed in wild-type but not mutant SC cultures. MyoD immunohistochemistry shows that the number of MyoD-positive cells is decreased in MKO SC cultures compared with wild-type cells. Bars: for DIC, 200 μm; for Desmin, 65 μm; for MyoD, 90 μm. (B) Increased proliferation of MKO and dKO cells. Wild-type, MKO, and dKO SCs were maintained under growth conditions and pulsed with BrdU for 1 h. Anti-BrdU immunocytochemistry shows that the number of BrdU-positive cells (white arrowheads) is increased in MKO and dKO SCs at days 2, 3, and 4 of culture. Bar, 70 μm. (C) Quantitation of MKO and dKO SC differentiation. Percent myotube fusion, calculated from A, confirms the differentiation defect of MKO and dKO SCs; (*) P < 0.01. (D) Quantitation of MKO and dKO SC proliferation. Percent BrdU-positive cells, calculated from B, confirms the increase in proliferation of MKO SCs compared with wild-type cells; (*) P < 0.01. (E,F) Down-regulation of genes involved in cell cycle arrest following MASTR deletion. Wild-type, MKO, and dKO TA muscle (E) and SC cultures (F) were used for qPCR. Expression of CyclinG1, Retinoblastoma (Rb), protein phosphatase 2 (Pp2a), growth arrest-specific 2 (Gas2), and growth arrest and DNA damage-inducible 45a (Gadd45a) mRNA is significantly diminished following MASTR/MRTF-A deletion; (*) P < 0.01. (G) Increased cell proliferation in the cMKO muscle following Ctx injury. PCNA immunohistochemistry shows an increase in the number of PCNA-positive cells (arrowheads) in the cMKO TA muscle at days 3 and 7 post-injury.
To further explore the molecular basis of the MKO and dKO regeneration phenotype, we performed microarray analysis on activated wild-type and MKO SCs. Gene ontology cluster analysis of down-regulated transcripts uncovered a significant enrichment of genes involved in cell cycle regulation, mRNA processing, and actin cytoskeletal dynamics (Supplemental Fig. S5; Supplemental Table S1). Microarray analysis also revealed down-regulation of MyoD expression in MKO SCs. Strikingly, the set of down-regulated transcripts in MKO SCs parallels that of MyoD-null cells (Supplemental Fig. S5; Seale et al. 2004; Ishibashi et al. 2005; Asakura et al. 2007), suggesting that MASTR and MyoD act in a common molecular pathway to control SC functions.
Since mutant SCs showed hyperproliferation, we validated the expression of cell cycle arrest genes down-regulated in both our MKO SC microarray and the published MyoD microarray by quantitative PCR (qPCR) (Hosoyama et al. 2011; Seale et al. 2004; Tomczak et al. 2004; Ishibashi et al. 2005; Asakura et al. 2007). Indeed, expression of CyclinG1, Retinoblastoma (Rb), protein phosphatase 2 (Pp2a), growth arrest-specific 2 (Gas2), and growth arrest and DNA damage-inducible 45a (Gadd45a) was significantly reduced in MKO and dKO whole muscle and SCs compared with wild-type controls (Fig. 4E,F). The similarities between the MKO and MyoD knockout mouse phenotypes and gene expression profiles, and the down-regulation of MyoD in our mutant mice, suggest that MASTR and MRTF-A regulate MyoD expression to initiate cell cycle arrest and promote differentiation of SCs.
MEF2A and MEF2C are expressed in SCs and are required for SC differentiation
MASTR is a potent coactivator of MEF2 transcriptional activity (Creemers et al. 2006b); however, the expression and potential function of MEF2 transcription factors in SCs remain uncharacterized. To investigate the mechanism by which MASTR regulates MyoD expression, we first assayed the expression of MEF2 family members in sorted, proliferating, and differentiating SCs by qPCR. MEF2A expression was detected at low levels in sorted and proliferating SCs and peaked at day 2 of differentiation; enriched MEF2C expression was detected in sorted, proliferating, and differentiating SCs (Fig. 5A). These results confirmed previous microarray results (data not shown) and showed that MEF2A and MEF2C, but not MEF2B or MEF2D, are enriched and up-regulated during SC activation and differentiation in vitro (Fig. 5A). MEF2A/C are therefore expressed in a manner similar to MASTR in proliferating and differentiating SCs. We then colabeled MEF2 and MyoD in the mouse TA muscle at days 7 and 14 post-Ctx injury using a MEF2 antibody that detects MEF2A, MEF2C, and MEF2D (Haberland et al. 2007). Interestingly, MEF2 was expressed in almost all MyoD-expressing SCs at days 7 and 14 post-injury (Fig. 5B). By day 14 post-injury, MEF2 expression was maintained in a subset of interstitial cells that were negative for MyoD and in centrally nucleated, regenerating myofibers (Fig. 5B). These expression studies suggest that MEF2A/C could cooperate with MASTR to regulate muscle regeneration.
Figure 5.
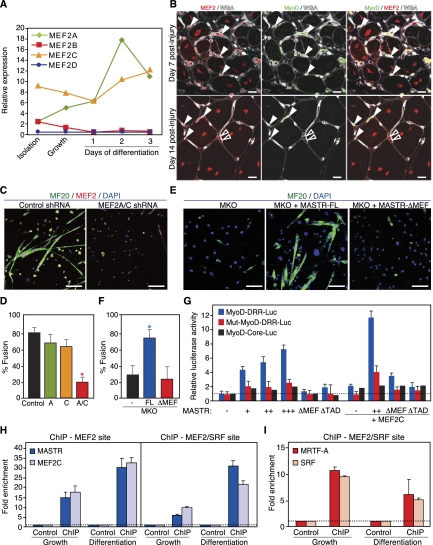
Activation of the MyoD-DRR enhancer by MASTR, MEF2, and MRTF-A. (A) Regulation of the MEF2 family during SC activation and proliferation. Shown is the expression of MEF2A, MEF2B, MEF2C, and MEF2D in SCs at isolation, under growth conditions, and at days 1, 2, and 3 of differentiation by qPCR. Relative expression was normalized to 18S RNA levels. (B) Expression of MEF2 in activated MyoD-positive SCs. Immunohistochemistry was performed on TA muscle at days 7 and 14 post-Ctx injury using anti-MEF2 antibody that detects MEF2A, MEF2C, and MEF2D isoforms. MyoD and WGA staining shows colocalization of MEF2-positive cells to activated SCs. White arrowheads indicate cells that are double positive for MEF2 and MyoD, whereas black arrowheads represent cells that are positive for MEF2 and negative for MyoD. Bar, 10 μm. (C) Defective SC differentiation following knockdown of MEF2A and MEF2C in vitro. SCs infected with a combination of MEF2A and MEF2C shRNA viruses show delayed differentiation and myotube formation compared with control cells infected with an empty shRNA virus. Bar, 65 μm. (D) Quantitation of the percent myotube fusion confirms that MEF2A/C knockdown inhibits SC differentiation. Knockdown of either MEF2A or MEF2C alone does not cause a significant differentiation defect; (*) P < 0.01. (E) Rescue of the MKO SC differentiation defect by MASTR-FL, but not by MASTR-ΔMEF. MKO SCs were transfected with control, MASTR-FL-expressing, or MASTR-ΔMEF-expressing vectors; allowed to differentiate for 4 d under low-serum culture conditions; and stained with anti-Myosin antibody to assay differentiation. Unlike MASTR-FL overexpression, which results in myotube formation in MKO SCs, MASTR-ΔMEF fails to rescue the MKO SC differentiation defect. Bar, 90 μm. (F) Quantitation of the percent myotube fusion confirms that overexpression of MASTR-FL, but not MASTR-ΔMEF, rescues the MKO SC differentiation defect; (*) P < 0.01. (G) Activation of the MyoD-DRR enhancer by MASTR and MEF2. C2C12 cells were transfected with the MyoD-DRR-Luciferase vector and expression vectors encoding various combinations of MASTR and MEF2C and assayed for luciferase activity. The MyoD-core-Luciferase vector was used as a negative control. The Mut-MyoD-DRR-Luciferase vector, in which the MEF2 and MEF2/SRF-binding sites were mutated, was also used as a control. MASTR alone or with MEF2C activates the MyoD-DRR but not the MyoD-core vectors. MASTR mutants that lack either the MEF2-binding (ΔMEF) or the transactivation (ΔTAD) domains do not induce MyoD-DRR activation. Mutation of the MEF2- and MEF2/SRF-binding sites on the DRR enhancer blocks reporter activation. (H) Binding of MASTR and MEF2 to the MyoD-DRR enhancer. Proliferating and differentiating C2C12 cells were transfected with MASTR and MEF2C and used for ChIP assay. Enrichment of MASTR and MEF2C at the MyoD DRR enhancer was quantified by qPCR and normalized to enrichment in C2C12 cells transfected with empty vectors. MASTR and MEF2C are enriched at the MEF2- and MEF2/SRF-binding sites of the DRR enhancer. (I) Binding of MRTF-A and SRF to the MyoD-DRR enhancer. Proliferating and differentiating C2C12 cells were transfected with MRTF-A and SRF and used for ChIP assay. Enrichment of MRTF-A and SRF at the MyoD-DRR enhancer was quantified by qPCR and normalized to enrichment in C2C12 cells transfected with empty vectors. Comparable enrichment was obtained for MRTF-A and SRF under both proliferation and differentiation conditions.
To examine the function of MEF2 in SCs, we knocked down MEF2A and MEF2C in cultured SCs using shRNA lentivirus and assessed SC differentiation by myosin (MF20) immunohistochemistry. Differentiation and fusion were defective in SCs infected with MEF2A/C-shRNA compared with control shRNA-infected cells (Fig. 5C,D). However, knockdown of MEF2A or MEF2C alone did not impair SC differentiation, suggesting a redundant role for the MEF2 family members in differentiating SCs (Fig. 5D). These results demonstrate that MEF2A and MEF2C are expressed in SCs and are required for SC differentiation and suggest that MASTR and MEF2A/C cooperatively regulate MyoD expression and muscle regeneration.
To investigate the requirement for MASTR/MEF2 interaction in SC differentiation, we attempted to rescue the MKO phenotype by overexpressing full-length MASTR (FL) and a MASTR mutant lacking the MEF2-binding domain (ΔMEF) in MKO SCs. MASTR-FL restored MKO SC differentiation, whereas MASTR-ΔMEF failed to rescue the MKO differentiation defect (Fig. 5E,F). These findings suggest that the MKO phenotype is mediated by the association of MASTR with MEF2.
MASTR and MEF2 bind and activate the MyoD DRR enhancer
MyoD transcription is controlled by an embryonic core enhancer and an adult DRR enhancer. Activation of the DRR enhancer at P14 is concomitant with inactivation of the core enhancer and correlates with the time at which MyoD expression decreases in MKO and dKO muscle (Fig. 3E; Kucharczuk et al. 1999; Chen et al. 2001, 2002). Interestingly, the DRR enhancer harbors one canonical MEF2-binding site and a MEF2/SRF hybrid site (L'Honore et al. 2007), suggesting that MASTR and MEF2 coregulate the DRR enhancer to induce MyoD expression. To test this hypothesis, we performed luciferase assays in C2C12 myoblasts using MyoD-DRR and MyoD-core reporter constructs. MASTR alone or with MEF2 activated the DRR but not the core enhancer in a dose-dependent fashion (Fig. 5G). MASTR mutants lacking either the MEF2-binding domain or the transactivation domain failed to activate the MyoD-DRR reporter, suggesting that the partnership between MASTR and MEF2 is required for MyoD activation. Moreover, mutating the MEF2-binding site and the MEF2/SRF hybrid sites blocked reporter activation, suggesting that MASTR and MEF2 directly regulate DRR activation (Fig. 5G). Chromatin immunoprecipitation (ChIP) in proliferating C2C12 myoblasts confirmed that MASTR and MEF2 bind the MEF2 and MEF2/SRF hybrid sites of the native MyoD DRR enhancer (Fig. 5H). Strikingly, binding of MASTR and MEF2 to the MyoD DRR enhancer significantly increased in differentiating C2C12 cells (Fig. 5H).
Since MASTR and MRTF-A exert their functions through association with MEF2 and SRF, respectively, these findings suggested that the MASTR/MEF2 and MRTF/SRF pathways converge to direct skeletal muscle regeneration by restricting proliferation and promoting differentiation of SCs. Indeed, ChIP assays showed binding of MRTF-A and SRF to the MEF2/SRF hybrid site of the DRR enhancer (Fig. 5I). We conclude that the MASTR/MEF2 complex is recruited to the MyoD DRR enhancer with MRTF-A and SRF and activates MyoD expression in differentiating SCs.
Discussion
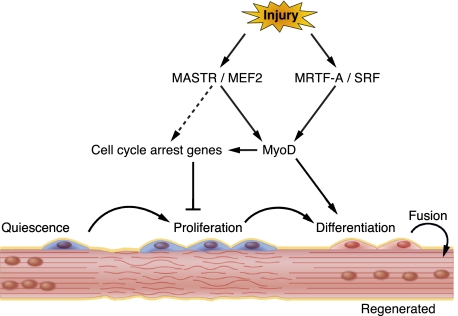
This study identifies members of the Myocardin and MEF2 transcription factor families as key components of the gene regulatory network that drives MyoD expression and muscle regeneration in adult mice. MASTR and MRTF-A are up-regulated in SCs in response to injury and muscular dystrophy. Global and SC-specific loss-of-function studies in mice demonstrate that MASTR and MRTF-A regulate MyoD expression, direct SCs to exit the cell cycle and differentiate, and promote muscle repair following injury. MASTR activates a muscle-specific postnatal MyoD enhancer through association with MEF2 and MRTFs. A model to account for these findings is shown in Figure 6.
Figure 6.
Schematic model for MASTR, MEF2, and MRTF-A function during SC differentiation. MASTR coactivates MEF2 and cooperates with MRTF-A to regulate MyoD expression, SC differentiation, and skeletal muscle regeneration in response to injury.
Essential functions for MRTFs during skeletal muscle regeneration
MASTR was originally identified as a muscle-enriched MEF2 coactivator and was reported to activate the myogenic program in Xenopus (Creemers et al. 2006b; Meadows et al. 2008); however, its function in mammalian muscle development or regeneration was not characterized. Mice lacking either MRTF-A or MRTF-B do not show obvious skeletal muscle defects (J Li et al. 2005; Oh et al. 2005; Li et al. 2006; Sun et al. 2006); however, MASTR/MRTF-A deletion causes age-dependent muscle wasting, defective muscle regeneration, and failure to activate postnatal MyoD expression. Unlike MKO mice, which show a significant delay in their regenerative capacity, AKO mice regenerate normally after injury, suggesting that MASTR and MRTF-A perform cooperative and nonredundant functions during skeletal muscle regeneration. The lack of a regeneration defect in AKO mice could be explained by redundancy of MRTF-A and MRTF-B in SCs. Indeed, MRTF-A and MRTF-B share the highest homology among MRTF family members and perform interchangeable functions in multiple cell and tissue contexts (Cen et al. 2003; Mokalled et al. 2010). In this regard, brain- and heart-specific deletions of both MRTFs result in multiple developmental neuronal and cardiac defects and lethality before adulthood, while the brain and heart develop normally in MRTF-A or MRTF-B single-knockout mice (Mokalled et al. 2010; EN Olson and MH Mokalled, unpubl.). Although MRTF-B is expressed at low levels in regenerating and dystrophic muscle, this level of expression may be sufficient to compensate for the absence of MRTF-A. This study is the first to demonstrate a function for the Myocardin family of transcription factors in SC differentiation and muscle regeneration.
Regulation of the MyoD DRR enhancer by MRTFs and MEF2
Our study adds the Myocardin and MEF2 families of transcription factors to the multiple factors that drive postnatal MyoD expression in SCs to regulate adult muscle regeneration. Similar to our mutant mice, MyoD-null SCs proliferate excessively, fail to differentiate, and exhibit a molecular signature reflective of a proliferative state (Megeney et al. 1996; Sabourin et al. 1999; White et al. 2000). In mice as well as in humans, myogenic expression of MyoD is linked to the activity of two muscle-specific enhancers: a 300-base-pair (bp) core and a 750-bp DRR enhancer (Goldhamer et al. 1992, 1995; Tapscott et al. 1992; Asakura et al. 1995; Chen and Goldhamer 1999; Chen et al. 2001, 2002). The core enhancer controls MyoD expression during development but is inactive after birth (Faerman et al. 1995; Goldhamer et al. 1995; Chen and Goldhamer 1999; Chen et al. 2001), whereas the DRR enhancer is dispensable during development but is essential in mature muscle and activated SCs (Chen et al. 2002; L'Honore et al. 2003). Interestingly, the level of MyoD expression is comparable between wild-type and dKO muscle at birth, but undetectable at 2 wk of age, suggesting that MASTR and MRTF-A control postnatal MyoD expression in SCs via the DRR enhancer. The striking temporal regulation of MyoD in MKO and dKO mice parallels that of MyoD activation in postnatal muscle and supports our finding that MRTFs regulate MyoD DRR activation in SCs. Our findings demonstrate that MASTR binds and activates the MyoD DRR enhancer through association with MEF2 and MRTF-A.
Novel functions for MEF2 transcription factors in muscle regeneration
MEF2 has been implicated in multiple aspects of muscle development (Potthoff and Olson 2007; Olson and Nordheim 2010), yet the functions of MEF2 in either SC differentiation or muscle regeneration are unknown. Although Drosophila MEF2 was shown to activate the MyoD ortholog nautilus, these findings were not extended to mammalian skeletal muscle (Sandmann et al. 2006). Here, we demonstrate that MASTR coactivates MEF2 to drive SC differentiation by showing that (1) MEF2A/C are up-regulated with MASTR in differentiating SCs and are required for SC differentiation, (2) the MASTR/MEF2C complex binds to and activates the adult MyoD DRR enhancer in myoblasts, and (3) the MASTR-ΔMEF mutant fails to rescue the MKO SC phenotype. Interestingly, MEF2A and MEF2C are dysregulated in patients with myotonic dystrophy (Bachinski et al. 2010), suggesting their involvement in skeletal muscle regeneration in humans.
Unlike MASTR, which coactivates MEF2, MRTF-A is an SRF activator. SRF binds to the MEF2/SRF hybrid site of the MyoD DRR enhancer to regulate MyoD expression during muscle regeneration (L'Honore et al. 2003). In contrast to transgenic mice overexpressing a DRR-β-galactosidase reporter, which activate β-galactosidase expression following muscle injury, mutation of the MEF2/SRF site fails to activate DRR reporter expression in injured mice (L'Honore et al. 2003). Our findings demonstrate that MEF2 and SRF bind to the MEF2/SRF hybrid site and confirm the relevance of this site for activating MyoD expression during skeletal muscle regeneration. In addition, SRF selectively regulates the levels of MyoD in myoblasts without affecting the levels of Myf5 (Gauthier-Rouviere et al. 1996; Soulez et al. 1996; Carnac et al. 1998). Interestingly, Myf5 expression was also unchanged in our dKO mice (Supplemental Fig. S4B), suggesting that the regulatory functions of MASTR and MRTF-A are specific to MyoD, but not to all muscle regulatory factors.
By unveiling novel functions for the Myocardin family of transcription factors in SC differentiation and muscle regeneration, this study suggests that MRTFs could act as direct sensors of the SC niche and may present a potential therapeutic target for muscle regenerative therapy. Dynamic signaling from the SC niche dictates SC activation in settings of homeostasis or injury, respectively (Gopinath and Rando 2008; Kuang et al. 2008). MRTFs couple extracellular signaling to gene transcription and could potentially sense extracellular alterations of the SC microenvironment. For example, MRTF-A and MRTF-B are sensitive to extracellular Rho signaling, which drives their nuclear translocation and consequent transcriptional activation (Miralles et al. 2003; Posern and Treisman 2006; Olson and Nordheim 2010). Interestingly, inhibition of RhoA in myoblast cultures, whether by dominant-negative expression or antagonist treatment, specifically alters MyoD but not Myf5 expression (Carnac et al. 1998). This effect on MyoD expression is specific to RhoA, but not to Rac or Cdc42, and requires functional SRF (Carnac et al. 1998). Moreover, concomitant with age-dependent changes within the SC systemic microenvironment (Hall et al. 2010; Conboy et al. 2005; Conboy and Rando 2005), MRTF expression levels were reported to decrease with aging (Sakuma et al. 2008), suggesting dynamic cross-talk between MRTFs and the SC niche. Because MRTFs link extracellular signaling to changes in gene regulation and are required for MyoD expression and SC differentiation, it will be of interest to examine whether perturbation of the SC niche affects MRTF signaling and whether manipulation of MRTF signaling affects muscle regeneration.
Materials and methods
Mouse lines
The MKO allele was generated using homologous recombination in embryonic stem (ES) cells. The pGKNEO-F2L2DTA vector, which contains a neomycin resistance gene flanked by FRT and loxP sites and a diphtheria toxin gene cassette, was used for MASTR targeting. The 5′, knockout, and 3′ arms of the targeting construct were generated by high-fidelity PCR amplification (Roche Expand High-Fidelity Long Template). The targeting vector was linearized with PvuI and electroporated into 129SvEv-derived ES cells. Isolated ES cell clones were analyzed for incorporation of the 5′ and 3′ loxP sites by Southern blotting using 5′ and 3′ probes. Clones with the targeted MASTR allele were injected into 3.5-d C57BL/6 blastocysts, and the resulting chimeras were crossed to C57BL/6 females to achieve germline transmission of the targeted (MASTRneo-loxP) allele. The MASTRneo-loxP mice were crossed to Flpe transgenic mice and then to CAG-Cre transgenic mice to obtain the MKO allele. MKO mice were then crossed to AKO mice (Li et al. 2006) to generate dKO mice. MKO mice were also crossed to mdx mice to generate MKO;mdx mice. MASTRFloxed mice were crossed to Pax7-Cre-ERT2 mice (Lepper et al. 2009) to generate the SC-specific cMKO allele.
SC isolation
Mononuclear cells were isolated from adult mouse hindlimb muscles following muscle digestion and myofiber dissociation (Sherwood et al. 2004). Eight-week-old to 12-wk-old mice were used for all experiments. CD34+/Sca1−/CD31−/CD45− SCs were then sorted from mononuclear cells with a yield of ∼4%. Gating of the SC population was determined using a negative control sample stained with phycoerythrin (PE)-conjugated antibodies for Sca1/CD31/CD45, and a positive control sample stained with Alexa-488-conjugated anti-CD34 antibody. SCs were then maintained under growth conditions (Ham F20 with 20% fetal calf serum, supplemented with 5 ng/μL bFGF) or differentiation conditions (DMEM1 with 2% horse serum).
BrdU labeling
SC cultures were maintained under growth conditions. Wild-type, MKO, and dKO SCs were labeled with BrdU at a final concentration of 10 μM for 1 h at days 2, 3, and 4 of culture. BrdU immunohistochemistry was then used to assay BrdU incorporation.
Chemical muscle injury
Chemical injury experiments were performed using Ctx and BaCl2 (Yan et al. 2003; Cornelison et al. 2004). Briefly, mice were anesthetized with 5% Avertin. Left TA muscles were injected with 100 μL of 10 μM Ctx or 50 μL of 1.2% BaCl2 in sterile saline. Mice were allowed to recover for 3, 7, or 14 d post-injury. Injected and contralateral TA muscles were harvested for sectioning and staining.
Histology and immunohistochemistry
Mice were anesthetized and transcardially perfused with PBS, followed by 4% paraformaldehyde prior to muscle dissection. TA muscles were then post-fixed in 4% paraformaldehyde for 48 h, embedded in paraffin, and sectioned. Sections were stained with H&E or WGA using standard procedures. Immunohistochemistry using monoclonal anti-BrdU (Roche), monoclonal anti-MyoD (BD Biosciences), monoclonal anti-Desmin (DAKO), monoclonal anti-MF20 (Hybridoma Bank), polyclonal anti-PCNA (Santa Cruz Biotechnology), and polyclonal anti-MEF2-C21 (Santa Cruz Biotechnology) antibodies was performed using standard protocols.
Quantitative real-time PCR
Total RNA was purified from the indicated tissues or cells using TRIzol reagent according to the manufacturer's instructions (Invitrogen). For RT–PCR, total RNA was used as a template for RT using random hexamer primers. qPCR was performed using TaqMan probes (ABI).
Luciferase assays
The MyoD-Core and MyoD-DRR luciferase constructs were cloned (Tapscott et al. 1992). C2C12 cells, plated in 24-well plates (5 × 104 cells per well), were transfected with 150 ng of luciferase construct, 20 ng of pCMV-LacZ, and a total of 250 ng of expression plasmids encoding MASTR, MEF2C, or an empty control vector. FuGENE 6 reagent (1.4 μL) (Roche) was complexed with the DNA and added to freshly plated C2C12 cells for 12 h. Cells were harvested 48 h after transfection in 150 μL of passive lysate buffer (Promega). Twenty microliters of cell lysate was used for luciferase or β-galactosidase assays. Luciferase activity was normalized to β-galactosidase levels and to luciferase activity in C2C12 cells transfected with a mix of reporter genes and empty vector.
ChIP
ChIP assays were performed using EZ ChIP (Millipore) following the manufacturer's instructions. Briefly, C2C12 cells were transfected with either a negative control empty vector or with Flag-MASTR, myc-MEF2C, myc-MRTF-A, or Flag-SRF vectors as previously described. Medium was changed 12 h after transfection into either growth or differentiation medium and collected 24 h later for ChIP. For each immunoprecipitation, chromatin from 3 × 106 cells was cross-linked with 1% formaldehyde and sonicated into 400- to 1000-bp fragments. Ten percent of the chromatin from the control, MASTR, MEF2C, MRTF-A, and SRF samples was used to purify the input DNA. The other 90% of the chromatin was immunoprecipitated with anti-Flag or anti-myc agarose affinity beads (Sigma). DNA was purified from the ChIP and input samples and was analyzed by qPCR using primers spanning the MEF2-binding or MEF2/SRF hybrid site of the MyoD-DRR enhancer. Fold enrichment for each of the control, MASTR, MEF2C, and MRTF-A ChIP samples was normalized first to the enrichment in the corresponding input sample and then to the enrichment in the control sample transfected with empty vector.
Statistical analysis
Results are expressed as means ± SEM. Unpaired two-tailed Student's t-test with Welch correction was performed to determine statistical significance between groups. P-values of <0.01 were considered significant.
Acknowledgments
We are grateful to Chen-Ming Fan (Carnegie Institute, Maryland) for providing the Pax7-Cre-ERT2 mice, Jose Cabrera for graphics, and Sasha Qi and Lil Sutherland for technical help. This work was supported by grants from the NIH and the Robert A. Welch foundation to E.N.O.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.179663.111.
References
- Asakura A, Lyons GE, Tapscott SJ 1995. The regulation of MyoD gene expression: Conserved elements mediate expression in embryonic axial muscle. Dev Biol 171: 386–398 [DOI] [PubMed] [Google Scholar]
- Asakura A, Hirai H, Kablar B, Morita S, Ishibashi J, Piras BA, Christ AJ, Verma M, Vineretsky KA, Rudnicki MA 2007. Increased survival of muscle stem cells lacking the MyoD gene after transplantation into regenerating skeletal muscle. Proc Natl Acad Sci 104: 16552–16557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachinski LL, Sirito M, Bohme M, Baggerly KA, Udd B, Krahe R 2010. Altered MEF2 isoforms in myotonic dystrophy and other neuromuscular disorders. Muscle Nerve 42: 856–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks GB, Chamberlain JS 2008. The value of mammalian models for duchenne muscular dystrophy in developing therapeutic strategies. Curr Top Dev Biol 84: 431–453 [DOI] [PubMed] [Google Scholar]
- Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A, Buckingham ME, Partridge TA, Zammit PS 2000. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biol 151: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biressi S, Rando TA 2010. Heterogeneity in the muscle satellite cell population. Semin Cell Dev Biol 21: 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham, M 1994. Muscle differentiation. Which myogenic factors make muscle? Curr Biol 4: 61–63 [DOI] [PubMed] [Google Scholar]
- Buckingham M, Relaix F 2007. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol 23: 645–673 [DOI] [PubMed] [Google Scholar]
- Carnac G, Primig M, Kitzmann M, Chafey P, Tuil D, Lamb N, Fernandez A 1998. RhoA GTPase and serum response factor control selectively the expression of MyoD without affecting Myf5 in mouse myoblasts. Mol Biol Cell 9: 1891–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen B, Selvaraj A, Burgess RC, Hitzler JK, Ma Z, Morris SW, Prywes R 2003. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol Cell Biol 23: 6597–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Goldhamer DJ 1999. Transcriptional mechanisms regulating MyoD expression in the mouse. Cell Tissue Res 296: 213–219 [DOI] [PubMed] [Google Scholar]
- Chen JC, Love CM, Goldhamer DJ 2001. Two upstream enhancers collaborate to regulate the spatial patterning and timing of MyoD transcription during mouse development. Dev Dyn 221: 274–288 [DOI] [PubMed] [Google Scholar]
- Chen JC, Ramachandran R, Goldhamer DJ 2002. Essential and redundant functions of the MyoD distal regulatory region revealed by targeted mutagenesis. Dev Biol 245: 213–223 [DOI] [PubMed] [Google Scholar]
- Conboy IM, Rando TA 2005. Aging, stem cells and tissue regeneration: Lessons from muscle. Cell Cycle 4: 407–410 [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA 2005. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433: 760–764 [DOI] [PubMed] [Google Scholar]
- Cornelison DD, Olwin BB, Rudnicki MA, Wold BJ 2000. MyoD−/− satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev Biol 224: 122–137 [DOI] [PubMed] [Google Scholar]
- Cornelison DD, Wilcox-Adelman SA, Goetinck PF, Rauvala H, Rapraeger AC, Olwin BB 2004. Essential and separable roles for Syndecan-3 and Syndecan-4 in skeletal muscle development and regeneration. Genes Dev 18: 2231–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creemers EE, Sutherland LB, McAnally J, Richardson JA, Olson EN 2006a. Myocardin is a direct transcriptional target of Mef2, Tead and Foxo proteins during cardiovascular development. Development 133: 4245–4256 [DOI] [PubMed] [Google Scholar]
- Creemers EE, Sutherland LB, Oh J, Barbosa AC, Olson EN 2006b. Coactivation of MEF2 by the SAP domain proteins myocardin and MASTR. Mol Cell 23: 83–96 [DOI] [PubMed] [Google Scholar]
- Faerman A, Goldhamer DJ, Puzis R, Emerson CP Jr, Shani M 1995. The distal human myoD enhancer sequences direct unique muscle-specific patterns of lacZ expression during mouse development. Dev Biol 171: 27–38 [DOI] [PubMed] [Google Scholar]
- Gauthier-Rouviere C, Vandromme M, Tuil D, Lautredou N, Morris M, Soulez M, Kahn A, Fernandez A, Lamb N 1996. Expression and activity of serum response factor is required for expression of the muscle-determining factor MyoD in both dividing and differentiating mouse C2C12 myoblasts. Mol Biol Cell 7: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldhamer DJ, Faerman A, Shani M, Emerson CP Jr 1992. Regulatory elements that control the lineage-specific expression of myoD. Science 256: 538–542 [DOI] [PubMed] [Google Scholar]
- Goldhamer DJ, Brunk BP, Faerman A, King A, Shani M, Emerson CP Jr 1995. Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development 121: 637–649 [DOI] [PubMed] [Google Scholar]
- Gopinath SD, Rando TA 2008. Stem cell review series: Aging of the skeletal muscle stem cell niche. Aging Cell 7: 590–598 [DOI] [PubMed] [Google Scholar]
- Goyenvalle A, Seto JT, Davies KE, Chamberlain J 2011. Therapeutic approaches to muscular dystrophy. Hum Mol Genet 20: R69–R78 doi: 10.1093/hmg/ddr105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Arnold MA, McAnally J, Phan D, Kim Y, Olson EN 2007. Regulation of HDAC9 gene expression by MEF2 establishes a negative-feedback loop in the transcriptional circuitry of muscle differentiation. Mol Cell Biol 27: 518–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JK, Banks GB, Chamberlain JS, Olwin BB 2010. Prevention of muscle aging by myofiber-associated satellite cell transplantation. Sci Transl Med 2: 57ra83 doi: 10.1126/scitranslmed.3001081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoyama T, Nishijo K, Prajapati SI, Li G, Keller C 2011. Rb1 gene inactivation expands satellite cell and postnatal myoblast pools. J Biol Chem 286: 19556–19564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi J, Perry RL, Asakura A, Rudnicki MA 2005. MyoD induces myogenic differentiation through cooperation of its NH2- and COOH-terminal regions. J Cell Biol 171: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Krauss RS 2010. Muscle stem cells in developmental and regenerative myogenesis. Curr Opin Clin Nutr Metab Care 13: 243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Gillespie MA, Rudnicki MA 2008. Niche regulation of muscle satellite cell self-renewal and differentiation. Cell Stem Cell 2: 22–31 [DOI] [PubMed] [Google Scholar]
- Kucharczuk KL, Love CM, Dougherty NM, Goldhamer DJ 1999. Fine-scale transgenic mapping of the MyoD core enhancer: MyoD is regulated by distinct but overlapping mechanisms in myotomal and non-myotomal muscle lineages. Development 126: 1957–1965 [DOI] [PubMed] [Google Scholar]
- Le Grand F, Rudnicki M 2007. Satellite and stem cells in muscle growth and repair. Development 134: 3953–3957 [DOI] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan CM 2009. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 460: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Honore A, Lamb NJ, Vandromme M, Turowski P, Carnac G, Fernandez A 2003. MyoD distal regulatory region contains an SRF binding CArG element required for MyoD expression in skeletal myoblasts and during muscle regeneration. Mol Biol Cell 14: 2151–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Honore A, Rana V, Arsic N, Franckhauser C, Lamb NJ, Fernandez A 2007. Identification of a new hybrid serum response factor and myocyte enhancer factor 2-binding element in MyoD enhancer required for MyoD expression during myogenesis. Mol Biol Cell 18: 1992–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhu X, Chen M, Cheng L, Zhou D, Lu MM, Du K, Epstein JA, Parmacek MS 2005. Myocardin-related transcription factor B is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc Natl Acad Sci 102: 8916–8921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Czubryt MP, McAnally J, Bassel-Duby R, Richardson JA, Wiebel FF, Nordheim A, Olson EN 2005. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc Natl Acad Sci 102: 1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chang S, Qi X, Richardson JA, Olson EN 2006. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol Cell Biol 26: 5797–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachie JK, Grounds MD, Partridge TA, Morgan JE 1993. Age-related changes in replication of myogenic cells in mdx mice: Quantitative autoradiographic studies. J Neurol Sci 119: 169–179 [DOI] [PubMed] [Google Scholar]
- Meadows SM, Warkman AS, Salanga MC, Small EM, Krieg PA 2008. The myocardin-related transcription factor, MASTR, cooperates with MyoD to activate skeletal muscle gene expression. Proc Natl Acad Sci 105: 1545–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA 1996. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev 10: 1173–1183 [DOI] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113: 329–342 [DOI] [PubMed] [Google Scholar]
- Mokalled MH, Johnson A, Kim Y, Oh J, Olson EN 2010. Myocardin-related transcription factors regulate the Cdk5/Pctaire1 kinase cascade to control neurite outgrowth, neuronal migration and brain development. Development 137: 2365–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M 2005. Direct isolation of satellite cells for skeletal muscle regeneration. Science 309: 2064–2067 [DOI] [PubMed] [Google Scholar]
- Musaro A, Barberi L 2010. Isolation and culture of mouse satellite cells. Methods Mol Biol 633: 101–111 [DOI] [PubMed] [Google Scholar]
- Oh J, Richardson JA, Olson EN 2005. Requirement of myocardin-related transcription factor-B for remodeling of branchial arch arteries and smooth muscle differentiation. Proc Natl Acad Sci 102: 15122–15127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olguin HC, Yang Z, Tapscott SJ, Olwin BB 2007. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J Cell Biol 177: 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN, Klein WH 1994. bHLH factors in muscle development: Dead lines and commitments, what to leave in and what to leave out. Genes Dev 8: 1–8 [DOI] [PubMed] [Google Scholar]
- Olson EN, Nordheim A 2010. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol 11: 353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmacek MS 2007. Myocardin-related transcription factors: Critical coactivators regulating cardiovascular development and adaptation. Circ Res 100: 633–644 [DOI] [PubMed] [Google Scholar]
- Pipes GC, Creemers EE, Olson EN 2006. The myocardin family of transcriptional coactivators: Versatile regulators of cell growth, migration, and myogenesis. Genes Dev 20: 1545–1556 [DOI] [PubMed] [Google Scholar]
- Posern G, Treisman R 2006. Actin' together: Serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol 16: 588–596 [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN 2007. MEF2: A central regulator of diverse developmental programs. Development 134: 4131–4140 [DOI] [PubMed] [Google Scholar]
- Price FD, Kuroda K, Rudnicki MA 2007. Stem cell based therapies to treat muscular dystrophy. Biochim Biophys Acta 1772: 272–283 [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75: 1351–1359 [DOI] [PubMed] [Google Scholar]
- Sabourin LA, Girgis-Gabardo A, Seale P, Asakura A, Rudnicki MA 1999. Reduced differentiation potential of primary MyoD−/− myogenic cells derived from adult skeletal muscle. J Cell Biol 144: 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM 2008. Self-renewal and expansion of single transplanted muscle stem cells. Nature 456: 502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma K, Akiho M, Nakashima H, Akima H, Yasuhara M 2008. Age-related reductions in expression of serum response factor and myocardin-related transcription factor A in mouse skeletal muscles. Biochim Biophys Acta 1782: 453–461 [DOI] [PubMed] [Google Scholar]
- Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EE 2006. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell 10: 797–807 [DOI] [PubMed] [Google Scholar]
- Schuierer MM, Mann CJ, Bildsoe H, Huxley C, Hughes SM 2005. Analyses of the differentiation potential of satellite cells from myoD−/−, mdx, and PMP22 C22 mice. Bmc Musculoskel Dis 6: 15 doi: 10.1186/1471-2474-6-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Ishibashi J, Holterman C, Rudnicki MA 2004. Muscle satellite cell-specific genes identified by genetic profiling of MyoD-deficient myogenic cell. Dev Biol 275: 287–300 [DOI] [PubMed] [Google Scholar]
- Sherwood RI, Christensen JL, Conboy IM, Conboy MJ, Rando TA, Weissman IL, Wagers AJ 2004. Isolation of adult mouse myogenic progenitors: Functional heterogeneity of cells within and engrafting skeletal muscle. Cell 119: 543–554 [DOI] [PubMed] [Google Scholar]
- Small EM, Thatcher JE, Sutherland LB, Kinoshita H, Gerard RD, Richardson JA, Dimaio JM, Sadek H, Kuwahara K, Olson EN 2010. Myocardin-related transcription factor-a controls myofibroblast activation and fibrosis in response to myocardial infarction. Circ Res 107: 294–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulez M, Rouviere CG, Chafey P, Hentzen D, Vandromme M, Lautredou N, Lamb N, Kahn A, Tuil D 1996. Growth and differentiation of C2 myogenic cells are dependent on serum response factor. Mol Cell Biol 16: 6065–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Boyd K, Xu W, Ma J, Jackson CW, Fu A, Shillingford JM, Robinson GW, Hennighausen L, Hitzler JK, et al. 2006. Acute myeloid leukemia-associated Mkl1 (Mrtf-a) is a key regulator of mammary gland function. Mol Cell Biol 26: 5809–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132: 2685–2695 [DOI] [PubMed] [Google Scholar]
- Tapscott SJ, Lassar AB, Weintraub H 1992. A novel myoblast enhancer element mediates MyoD transcription. Mol Cell Biol 12: 4994–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Broek RW, Grefte S, Von den Hoff JW 2010. Regulatory factors and cell populations involved in skeletal muscle regeneration. J Cell Physiol 224: 7–16 [DOI] [PubMed] [Google Scholar]
- Tomczak KK, Marinescu VD, Ramoni MF, Sanoudou D, Montanaro F, Han M, Kunkel LM, Kohane IS, Beggs AH 2004. Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J 18: 403–405 [DOI] [PubMed] [Google Scholar]
- Tsivitse S 2010. Notch and Wnt signaling, physiological stimuli and postnatal myogenesis. Int J Biol Sci 6: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan RS 2011. Role of adult stem/progenitor cells in osseointegration and implant loosening. Int J Oral Maxillofac Implants 26: 50–62 [PubMed] [Google Scholar]
- Whalen RG, Harris JB, Butler-Browne GS, Sesodia S 1990. Expression of myosin isoforms during notexin-induced regeneration of rat soleus muscles. Dev Biol 141: 24–40 [DOI] [PubMed] [Google Scholar]
- White JD, Scaffidi A, Davies M, McGeachie J, Rudnicki MA, Grounds MD 2000. Myotube formation is delayed but not prevented in MyoD-deficient skeletal muscle: Studies in regenerating whole muscle grafts of adult mice. J Histochem Cytochem 48: 1531–1544 [DOI] [PubMed] [Google Scholar]
- Wu X, Wang S, Chen B, An X 2010. Muscle-derived stem cells: Isolation, characterization, differentiation, and application in cell and gene therapy. Cell Tissue Res 340: 549–567 [DOI] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z, Rudnicki MA, Rivera AJ, Primig M, Anderson JE, Natanson P 1999. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev Biol 210: 440–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Choi S, Liu X, Zhang M, Schageman JJ, Lee SY, Hart R, Lin L, Thurmond FA, Williams RS 2003. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J Biol Chem 278: 8826–8836 [DOI] [PubMed] [Google Scholar]
- Yoshida N, Yoshida S, Koishi K, Masuda K, Nabeshima Y 1998. Cell heterogeneity upon myogenic differentiation: Down-regulation of MyoD and Myf-5 generates ‘reserve cells.’ J Cell Sci 111: 769–779 [DOI] [PubMed] [Google Scholar]