Abstract
Despite common belief, accumulation of molecular damage does not play a key role in aging. Still, cancer (an age-related disease) is initiated by molecular damage. Cancer and aging share a lot in common including the activation of the TOR pathway. But the role of molecular damage distinguishes cancer and aging. Furthermore, an analysis of the role of both damage and aging in cancer argues against “a decline, caused by accumulation of molecular damage” as a cause of aging. I also discuss how random molecular damage, via rounds of multiplication and selection, brings about non-random hallmarks of cancer.
Keywords: cancer, target, therapy, leukemia, anticancer drugs
INTRODUCTION
Aging is defined as a decline caused by accumulation of all sorts of damage, in particular, molecular damage. This statement seemed so obvious that it was not questioned. Yet several lines of evidence rule out molecular damage as a cause of aging [1-15]. Yes, of course, molecular damage accumulates over time. But this accumulation is not sufficient to cause organismal death. Eventually it would. But the organism does not live long enough, because another cause terminates life first [8]. This cause is aging, a continuation of developmental growth. Definitely, developmental growth is not driven by accumulation of molecular damage, although molecular damage accumulates. Similarly, aging is not driven by damage.
Growth is stimulated in part by mitogen- and nutrient-sensing (and other) signaling pathways such as mTOR [16-35]. Aging, “an aimless continuation of developmental program”, is driven by the same signaling pathways including mTOR [8, 14, 24]. Aging in turn causes damage: not molecular damage but non-random organ damage (stroke, infarction, renal failure and so on) and death [13]. Seemingly, one objection to this concept is that cancer is caused by molecular damage. And cancer is often a cause of death in mammals. So how may one claim that damage does not drive aging, if it is involved in cancer. Let us discuss this.
Damage in cancer
Damage causes activate oncogenes and de-activate tumor suppressors due to genetic mutations, epigenetic alterations and microRNAs dysregulation [36-57]. Even according to alternative theories, cancer is caused by damage too [58]. So damage is involved in cancer. There are some exceptions, mostly related to embryonic cells. Also, in theory, extra-genetic alterations such as stable activation of oncogenic pathways via positive feedback loops can contribute to malignant phenotype [59]. Finally, positive feedback loops could be established between cancer and normal cells [59-61]. But in general molecular damage is a key factor in cancer origin. In agreement, cancer is associated with genetic instability [59, 62-69].
Not decline but robustness
Due to genetic instability, cancer cells accumulate high levels of unrepaired damage, resulting in genomic mutations and epigenetic alterations as well as aneuploidy [36-49, 70-80]. Despite of accumulation of damage, cancer is neither decline nor ‘wear and tear’. Cancer cells are robust and aggressive. Cancer cells damage organs, thus killing organism. If cancer cells with all damage are so robust, then how possibly aging of normal cells could be “a decline due to accumulation of molecular damage”. In fact, it does not.
Immortality of cancer cells
Cancer is associated with cellular immortality [38, 81-88]. Not only cancer cells can become cell lines but also they can become free-living organisms [89-96]. Such free-living cancer cells spread from one animal to another. Thus, venereal sarcoma in dogs spread as unicellular mammalian organisms for several millennia, once originated from a single cancer cell [89-96]. Thus accumulation of damage is associated with cellular immortality.
Damage is not sufficient to cause cancer
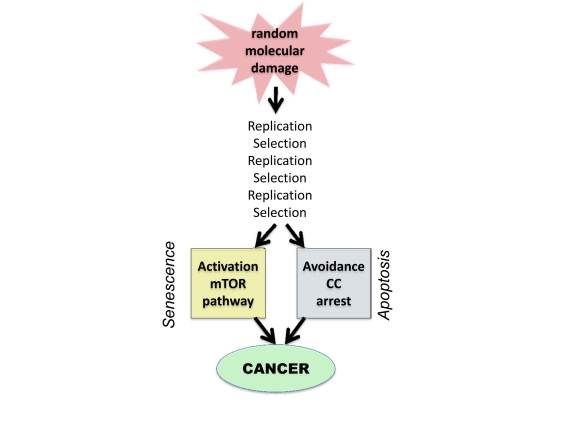
However, molecular damage is not sufficient either to cause cancer or to hurt organism. This damage is multiplied billions of times via cell replication. Also, cells with random mutations undergo non-random selection (Figure 1).
Figure 1. From random damage to cancer.
Random damage undergoes multiple rounds of replication and selection. Aging is one of selection forces that favors cells with oncogenic mutations. Cancer cell is characterized by (a) activation of growth-promoting pathways such as mTOR and (b) loss of cell cycle (CC) control. Isolated activation of mTOR favors senescence, whereas isolated cell cycle progression may trigger apoptosis.
Multiplication and selection
A 1 cm tumor contains 109 (1 billion) cells. Therefore, damage does not passively accumulate but is actively multiplied. Cells undergo clonal selection, analogous to Darwinian selection [70, 97-100]. Importantly, most mutations are so called “passenger” mutations that remain random and useless [72, 79, 80, 101]. But nevertheless they do not decrease cell vitality.
Selective microenvironment
Oncogenic mutations occur randomly. Cancer arises when cellular microenvironment favors oncogenic mutations, creating selective advantage to cells bearing oncogenic mutations. For example, carcinogens not only damage DNA but also cytostatic to normal cells, thus favoring selection of oncogenic mutations that render cells resistant to cytostatic/toxic carcinogens [102, 103]. This is especially apparent with non-damaging carcinogens such as phorbol esters [104]. Cancer therapy can select for additional oncogenic mutations (such as loss of p53), rendering cancer cells not only drug resistant but also increasingly oncogenic [102, 103, 105-108]. Inflammation and chronic infections also favor cancer [109-121]. And the aging microenvironment favors cancer [122-128].
Aging as selective force
Organismal aging is the most important risk factor in common cancers such as prostate, breast, colon, gastric, lung, pancreatic, skin, brain, thyroid (and so on) cancers as well as melanomas and certain leukemias. Calorie restriction [129-137] and rapamycin [138-141], which decelerate aging, also postpone cancer. Why does aging favors cancer? One explanation is that aging stromal cells secrete factors that promote growth of pre-cancer cells [122, 123, 142-144] and aging is associated with pro-inflammation that favors cancer growth [145-147]. The pro-inflammatory NF-kB pathway is involved in both DNA damage response (DDR), cancer and aging [60, 147-156].
One additional explanation is that chronic overactivation of mTOR renders normal cells irresponsive to growth factors [157]. (In fact, mTOR/S6K renders cells resistant to insulin and growth factors [158, 159]). Then, cancer cells, which are growth signal- independent, acquire selective advantage. In theory, by restoring responsiveness of normal cells to mitogenic signals, treatment with rapamycin can eliminate selective advantage for cancer cells. It was predicted that rapamycin can restore responsiveness of aging cells [157]. In fact, mTOR may cause exhaustion of the proliferative potential of stem cells and, in some studies, rapamycin improved the responsiveness of aging stem cells and immune cells [160-163]. As an example, activation of mTOR promoted leukemia-initiated cells, while depleting normal hematopoietic stem cell. Rapamycin not only depleted leukaemia-initiating cells but also restored normal stem cell function [160, 164]. Thus decreased proliferative potential of normal cells is associated with selective advantage to cancer cells.
Non-random activation of the PI3K/mTOR pathway
The PI3K/mTOR pathway is universally involved in cancer [37, 165-180]. It is activated by mutations in PI3K, Ras, Raf, non-receptor and growth factor receptor kinases and autocrine growth factors [165, 177, 181, 182]. Also, inactivation of tumor suppressors such as PTEN, AMPK, TSC2, LKB1, NF1 causes activation of this pathway [160, 169, 183-191]. In addition, the hypertrophic effect is often achieved via activation of downstream mTOR targets, translation factors [178]. Finally, p53, which is lost in cancer, is also a suppressor of the mTOR pathway [192-201]. Therefore, it can suppress conversion of cell cycle arrest to senescence [198-204]. In turn, the GF/PI3K/Akt/mTOR pathway drives cellular mass growth, hypersecrtory phenotype, HIF-1 expression, angiogenic phenotype, high levels of glycolysis and biosyntesis (metabolic switch) and apoptosis avoidance [16-35, 205-208]. In other words, it is involved in most of hallmarks of cancer [38, 88], with a notable exception of loss of cell cycle control. On the other hand, the mTOR pathway is involved in senescent phenotype. Therefore, the second alteration in cancer is deactivation of cell cycle checkpoints. Thus cancer cells can be viewed as cycling senescent cells.
Avoiding cell cycle arrest
In order to proliferate, cell with TOR-activating oncogenes must disable cell cycle control. Inactivation of tumor suppressors such p53, Rb, p16 and activation of c-myc, cyclins D and E, all disable cell cycle control, allowing “pro-senescent” cancer cell to proliferate [209-216]. Still, acute DNA damage, anticancer drugs and induction of p21 or p16 cause cell cycle arrest. Arrested cancer cells rapidly become senescent (geroconversion), revealing their pro-senescent phenotype.
Oncogenic transformation and gerogenic conversion
There are non-mutually exclusive ways to depict oncogenic transformation, as complementary activation/disabling of signaling pathways [88, 217-225]. Here to compare cancer with aging, I view oncogenic transformation as (a) activation of growth-promoting pathways such as mTOR and (b) loss of cell cycle control. Growth promoting pathways can drive either growth or aging, whereas avoidance of cell cycle arrest precludes aging (Fig. 1). In quiescent cells, activation of growth-promoting pathways (such as mTOR) converts quiescence into senescence, a process named gerogenic conversion or geroconversion [226, 227]. In proliferating cells, mTOR is fully activated. Induction of cell cycle arrest, without inhibition of mTOR causes gerogenic conversion too. When cell cycle is arrested, growth-promoting pathways drive hypertrophy and aging instead of growth. The difference between quiescence and senescence was recently discussed in detail [227]. Cellular hyper-functions and feedback signal resistance are manifestations of cellular senescence/aging that lead to age-related diseases [227]. These hallmarks result from excessive activation of signaling pathways not from accumulation of damage.
Why aging is not caused by accumulation of damage
To harbor the active mTOR pathway, cancer cells undergo multiple rounds of selection. In other words, numerous random mutations are selected for non-random activation of mTOR. In contrast it is resting non-dividing cells such as liver, muscle, fat, connective tissue, neurons that undergo aging (geroconversion) in the organism. Not only levels of molecular damage are low in normal cells, but also there is no amplification and selection. So random damage hardly can cause non-random activation of mTOR. Noteworthy, calorie restriction (CR) inhibits mTOR. Even short-term CR suppresses cellular senescence in the organism [228, 229].
Extragenetic activation of mTOR in aging
mTOR pathway is activated by growth factors, hormones, mitogens, pro-inflammatory cytokines and other secretory molecules and nutrients. Cells can overactivate each other, via positive feedback loops. For example in the liver and fat, hyper-active mTOR causes insulin-resistance, which in turn leads to activation mTOR in beta-cells, which produce insulin. Insulin further activates mTOR in the liver and fat.
DNA damage response (DDR) and aging
In proliferating cells, mTOR is fully activated. Acute DNA damage induces DDR and cell cycle arrest. If mTOR is still active, such cells undergo geroconversion. Rapamycin and other inhibitors of the mTOR pathway decelerate geroconversion [198, 200, 206, 226, 230-236]). This is how accelerated senescence is usually induced in proliferating cells (in cell culture). However, in quiescent cells with inactive mTOR, DNA damage does not induce sensecence, whereas activation of mTOR does [226, 237].
In oncogene-induced senescence (OIS), DDR causes cell cycle arrest, leading to senescence [238-245]. Noteworthy, most oncogenes that induce senescence (Ras, Raf, MEK, Akt and so on) activate the mTOR pathway. We can call them TOR-activating oncogenes or gerogenes [14], because they are involved in aging from cells to organisms [14, 246, 247]. Loss of PTEN also activates the mTOR pathway, causing senescence [243]. In OIS, oncogenes induce cell cycle arrest but not necessary DNA damage or even DDR [248, 243, 249]. Furthermore, atypical DDR can occur without DNA damage (pseudo-DDR) [231, 236, 250-256]. DDR pathways and the mTOR pathway are interconnected [257-260]. And it seems that pseudo-DDR and DDR are markers of cellular hyper-activation associated with senescence [145] and can be blocked by rapamycin [231].
Cancer prevention and therapy
Prevention of DNA damage can decrease cancer incidence. For example, non-smoking prevents smoking-induced cancer. Also, cancer can be prevented by decelerating the aging process by calorie restriction and rapamycin. Both calorie restriction and rapamycin delay cancer. Although rapalogs can directly affect cancer cells, rapalogs are only modestly effective as anti-cancer therapy [168, 261, 262], compared with their dramatic preventive effects. In any case, cancer can be prevented without decreasing levels of molecular damage. Furthermore, DNA damaging drugs are cornerstone of cancer therapy. And these drugs are also carcinogens, because anti-cancer and carcinogenic effects are two sides of the same coin [103].
CONCLUSION
Although molecular damage is typically necessary for cancer initiation, this damage limits life span not because of cellular decline but because of cellular robustness. Damage undergoes multiplication and selection. Aging by itself is a selective force that favors cancer probably because aging cells are signal resistant, thus providing selective advantage to cells that by-pass the need in mitogenic signals. In addition to non-random selection for oncogenic mutations, cancer cells accumulate even higher levels of random “passenger” mutations. Despite that cancer cells are robust. It must be expected that a lower rate of DNA damage in normal cells cannot cause cellular decline. Yes, molecular damage accumulates but is not a driving force for aging. Aging would occur in the absence of any molecular damage. On the other hand, yes, molecular damage is involved in something like cancer that can limit lifespan in mammals to some extend. Noteworthy, worms and flies do not die from cancer. Still they undergo PI3K/TOR-dependent aging [263-269].
As already discussed, if quasi-programmed TOR-driven aging would be eliminated, thus extending lifespan, then accumulation of molecular damage would become life-limiting [10]. In any case, in mammals, cellular aging (characterized by cellular overactivation, hyperfunction and secondary signal resistance) can cause diseases, which lead to organ damage. And cancer, an age-related disease, is not an exception: it kills not because cancer cells fail due to decline but because these cells damage organs. Perhaps, cancer is not the only one damage-related disease among aging-dependent conditions. But a subtle interference of molecular damage with TOR-driven aging will be a topic for another article, which will discuss the intricate relationship between non-random organ damage and random molecular damage.
Footnotes
The author of this manuscript has no conflict of interest to declare.
REFERENCES
- Doonan R, McElwee JJ, Matthijssens F, Walker GA, Houthoofd K, Back P, Matscheski A, Vanfleteren JR, Gems D. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 2008;22:3236–3241. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Doonan R. Antioxidant defense and aging in C. elegans: Is the oxidative damage theory of aging wrong? Cell Cyce. 2009;8:1681–1687. doi: 10.4161/cc.8.11.8595. [DOI] [PubMed] [Google Scholar]
- Cabreiro F, Ackerman D, Doonan R, Araiz C, Back P, Papp D, Braeckman BP, Gems D. Increased life span from overexpression of superoxide dismutase in Caenorhabditis elegans is not caused by decreased oxidative damage. Free Radic Biol Med. 2011;51:1575–1582. doi: 10.1016/j.freeradbiomed.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Hekimi S. When a theory of aging ages badly. Cell Mol Life Sci. 2009;67:1–8. doi: 10.1007/s00018-009-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Meng Y, Camp D, Yang W, Jia X, Benard C, Hekimi S. Decreased energy metabolism extends life span in Caenorhabditis elegans without reducing oxidative damage. Genetics. 2010;185:559–571. doi: 10.1534/genetics.110.115378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR, Selman C. The free-radical damage theory: Accumulating evidence against a simple link of oxidative stress to ageing and lifespan. Bioessays. 2011;33:255–259. doi: 10.1002/bies.201000132. [DOI] [PubMed] [Google Scholar]
- Ristow M, Schmeisser S. Extending life span by increasing oxidative stress. Free Radic Biol Med. 2011;51:327–336. doi: 10.1016/j.freeradbiomed.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Aging and immortality: quasi-programmed senescence and its pharmacologic inhibition. Cell Cycle. 2006;5:2087–2102. doi: 10.4161/cc.5.18.3288. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. An anti-aging drug today: from senescence-promoting genes to anti-aging pill. Drug Disc Today. 2007;12:218–224. doi: 10.1016/j.drudis.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Program-like aging and mitochondria: instead of random damage by free radicals. J Cell Biochem. 2007;102:1389–1399. doi: 10.1002/jcb.21602. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Paradoxes of aging. Cell Cycle. 2007;6:2997–3003. doi: 10.4161/cc.6.24.5124. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Aging: ROS or TOR. Cell Cycle. 2008;7:3344–3354. doi: 10.4161/cc.7.21.6965. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. mTOR-driven aging: speeding car without brakes. Cell Cycle. 2009;8:4055–4059. doi: 10.4161/cc.8.24.10310. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Revisiting the antagonistic pleiotropy theory of aging: TOR-driven program and quasi-program. Cell Cycle. 2010;9:3151–3156. doi: 10.4161/cc.9.16.13120. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Why men age faster but reproduce longer than women: mTOR and evolutionary perspectives. Aging. 2010;2:265–273. doi: 10.18632/aging.100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Sarbassov dos D, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Tee AR, Blenis J. mTOR, translational control and human disease. Semin Cell Dev Biol. 2005;16:29–37. doi: 10.1016/j.semcdb.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Dann SG, Selvaraj A, Thomas G. mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med. 2007;13:252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Hands SL, Proud CG, Wyttenbach A. mTOR's role in ageing: protein synthesis or autophagy? Aging. 2009;1:586–597. doi: 10.18632/aging.100070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer HP, Osipov RM, Clements RT, Sellke FW, Bianchi C. Hypercholesterolemia is associated with hyperactive cardiac mTORC1 and mTORC2 signaling. Cell Cycle. 2009;8:1738–1746. doi: 10.4161/cc.8.11.8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV, Hall MN. Growth and aging: a common molecular mechanism. Aging. 2009;1:357–362. doi: 10.18632/aging.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguta M. Control of RNA polymerases I and III by the TOR signaling pathway. Cell Cycle. 2009;8:4023–4024. doi: 10.4161/cc.8.24.10534. [DOI] [PubMed] [Google Scholar]
- Jiang Y. mTOR goes to the nucleus. Cell Cycle. 2009;9:868. [PMC free article] [PubMed] [Google Scholar]
- Dazert E, Hall MN. mTOR signaling in disease. Curr Opin Cell Biol. 2011;23:744–55. doi: 10.1016/j.ceb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Loewith R, Hall MN. Target of Rapamycin (TOR) in Nutrient Signaling and Growth Control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2010;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn CS, Qian SB. mTOR signaling in protein homeostasis: less is more? Cell Cycle. 2011;10:1940–1947. doi: 10.4161/cc.10.12.15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Proud CG. mTOR-mediated regulation of translation factors by amino acids. Biochem Biophys Res Commun. 2004;313:429–436. doi: 10.1016/j.bbrc.2003.07.015. [DOI] [PubMed] [Google Scholar]
- Hall MN. mTOR-what does it do? Transplant Proc. 2008;40:S5–8. doi: 10.1016/j.transproceed.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- McKenna ES, Roberts CW. Epigenetics and cancer without genomic instability. Cell Cycle. 2009;8:23–26. doi: 10.4161/cc.8.1.7290. [DOI] [PubMed] [Google Scholar]
- Timp W, Levchenko A, Feinberg AP. A new link between epigenetic progenitor lesions in cancer and the dynamics of signal transduction. Cell Cycle. 2009;8:383–390. doi: 10.4161/cc.8.3.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle. 2009;8:377–382. doi: 10.4161/cc.8.3.7526. [DOI] [PubMed] [Google Scholar]
- Burdach S, Plehm S, Unland R, Dirksen U, Borkhardt A, Staege MS, Muller-Tidow C, Richter GH. Epigenetic maintenance of stemness and malignancy in peripheral neuroectodermal tumors by EZH2. Cell Cycle. 2009;8:1991–1996. doi: 10.4161/cc.8.13.8929. [DOI] [PubMed] [Google Scholar]
- Chen SS, Sherman MH, Hertlein E, Johnson AJ, Teitell MA, Byrd JC, Plass C. Epigenetic alterations in a murine model for chronic lymphocytic leukemia. Cell Cycle. 2009;8:3663–3667. doi: 10.4161/cc.8.22.9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Kundu ST, Nallur S, Paranjape T, Boeke M, Weidhaas JB, Slack FJ. KRAS alleles: The LCS6 3'UTR variant and KRAS coding sequence mutations in the NCI-60 panel. Cell Cycle. 2012;11 doi: 10.4161/cc.11.2.18794. [DOI] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzi G, Pastorino U, Croce CM. MicroRNAs and lung cancer: from markers to targets. Cell Cycle. 2011;10:2045–2046. doi: 10.4161/cc.10.13.15712. [DOI] [PubMed] [Google Scholar]
- Pekarsky Y, Croce CM. Is miR-29 an oncogene or tumor suppressor in CLL? Oncotarget. 2010;1:224–227. doi: 10.18632/oncotarget.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA. miR-31: a crucial overseer of tumor metastasis and other emerging roles. Cell Cycle. 2010;9:2124–2129. doi: 10.4161/cc.9.11.11843. [DOI] [PubMed] [Google Scholar]
- Mavrakis KJ, Wendel HG. TargetScreen: an unbiased approach to identify functionally important microRNA targets. Cell Cycle. 2010;9:2080–2084. doi: 10.4161/cc.9.11.11807. [DOI] [PubMed] [Google Scholar]
- Duesberg P, Li R. Multistep carcinogenesis: a chain reaction of aneuploidizations. Cell Cycle. 2003;2:202–210. [PubMed] [Google Scholar]
- Blagosklonny MV. Molecular theory of cancer. Cancer Biol Ther. 2005;4:621–627. doi: 10.4161/cbt.4.6.1818. [DOI] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Trimmer C, Lin Z, Whitaker-Menezes D, Chiavarina B, Zhou J, Wang C, Pavlides S, Martinez-Cantarin MP, Capozza F, Witkiewicz AK, Flomenberg N, Howell A, Pestell RG, Caro J, Lisanti MP, et al. Autophagy in cancer associated fibroblasts promotes tumor cell survival: Role of hypoxia, HIF1 induction and NFkappaB activation in the tumor stromal microenvironment. Cell Cycle. 2010;9:3515–3533. doi: 10.4161/cc.9.17.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Martin V, Fueyo J, Lee OH, Xu J, Cortes-Santiago N, Alonso MM, Aldape K, Colman H, Gomez-Manzano C. Tie2/TEK modulates the interaction of glioma and brain tumor stem cells with endothelial cells and promotes an invasive phenotype. Oncotarget. 2010;1:700–709. doi: 10.18632/oncotarget.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Komarova NL, Lengauer C, Vogelstein B, Nowak MA. Dynamics of genetic instability in sporadic and familial colorectal cancer. Cancer Biol Ther. 2002;1:685–692. doi: 10.4161/cbt.321. [DOI] [PubMed] [Google Scholar]
- Li L, Borodyansky L, Yang Y. Genomic instability en route to and from cancer stem cells. Cell Cycle. 2009;8:1000–1002. doi: 10.4161/cc.8.7.8041. [DOI] [PubMed] [Google Scholar]
- Gupta A, Yang Q, Pandita RK, Hunt CR, Xiang T, Misri S, Zeng S, Pagan J, Jeffery J, Puc J, Kumar R, Feng Z, Powell SN, Bhat A, Yaguchi T, Wadhwa R, et al. Cell cycle checkpoint defects contribute to genomic instability in PTEN deficient cells independent of DNA DSB repair. Cell Cycle. 2009;8:2198–2210. doi: 10.4161/cc.8.14.8947. [DOI] [PubMed] [Google Scholar]
- Meyn RE. Linking PTEN with genomic instability and DNA repair. Cell Cycle. 2009;8:2322–2323. [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Tourriere H, Coquelle A, Pasero P. Does interference between replication and transcription contribute to genomic instability in cancer cells? Cell Cycle. 2010;9:1886–1892. doi: 10.4161/cc.9.10.11539. [DOI] [PubMed] [Google Scholar]
- Guirouilh-Barbat JK, Wilhelm T, Lopez BS. AKT1/BRCA1 in the control of homologous recombination and genetic stability: the missing link between hereditary and sporadic breast cancers. Oncotarget. 2010;1:691–699. doi: 10.18632/oncotarget.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CC, Hau PM, Marxer M, Poon RY. The requirement of p53 for maintaining chromosomal stability during tetraploidization. Oncotarget. 2010;1:583–595. doi: 10.18632/oncotarget.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerenwinkel N, Antal T, Dingli D, Traulsen A, Kinzler KW, Velculescu VE, Vogelstein B, Nowak MA. Genetic progression and the waiting time to cancer. PLoS Comput Biol. 2007;3:e225. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, Vogelstein B, Karchin R. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Hot-spot mutations in p110alpha of phosphatidylinositol 3-kinase (pI3K): differential interactions with the regulatory subunit p85 and with RAS. Cell Cycle. 2010;9:596–600. doi: 10.4161/cc.9.3.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AC, van Wely KH. Are aneuploidy and chromosome breakage caused by a CINgle mechanism? Cell Cycle. 2010;9:2275–2280. doi: 10.4161/cc.9.12.11865. [DOI] [PubMed] [Google Scholar]
- McClelland SE, Burrell RA, Swanton C. Chromosomal instability: a composite phenotype that influences sensitivity to chemotherapy. Cell Cycle. 2009;8:3262–3266. doi: 10.4161/cc.8.20.9690. [DOI] [PubMed] [Google Scholar]
- Baker DJ, van Deursen JM. Chromosome missegregation causes colon cancer by APC loss of heterozygosity. Cell Cycle. 2010;9:1711–1716. doi: 10.4161/cc.9.9.11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Feng J, Saldivar JS, Gu D, Bockholt A, Sommer SS. EGFR somatic doublets in lung cancer are frequent and generally arise from a pair of driver mutations uncommonly seen as singlet mutations: one-third of doublets occur at five pairs of amino acids. Oncogene. 2008;27:4336–4343. doi: 10.1038/onc.2008.71. [DOI] [PubMed] [Google Scholar]
- Torkamani A, Schork NJ. Prediction of cancer driver mutations in protein kinases. Cancer Res. 2008;68:1675–1682. doi: 10.1158/0008-5472.CAN-07-5283. [DOI] [PubMed] [Google Scholar]
- Loriaux MM, Levine RL, Tyner JW, Frohling S, Scholl C, Stoffregen EP, Wernig G, Erickson H, Eide CA, Berger R, Bernard OA, Griffin JD, Stone RM, Lee B, Meyerson M, Heinrich MC, et al. High-throughput sequence analysis of the tyrosine kinome in acute myeloid leukemia. Blood. 2008;111:4788–4796. doi: 10.1182/blood-2007-07-101394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohling S, Scholl C, Levine RL, Loriaux M, Boggon TJ, Bernard OA, Berger R, Dohner H, Dohner K, Ebert BL, Teckie S, Golub TR, Jiang J, Schittenhelm MM, Lee BH, Griffin JD, et al. Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell. 2007;12:501–513. doi: 10.1016/j.ccr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. Embo J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CB, Kim NW, Prowse KR, Weinrich SL, Hirsch KS, West MD, Bacchetti S, Hirte HW, Counter CM, Greider CW, et al. Telomerase, cell immortality, and cancer. Cold Spring Harb Symp Quant Biol. 1994;59:307–315. doi: 10.1101/sqb.1994.059.01.035. [DOI] [PubMed] [Google Scholar]
- Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, Sedivy JM, Weinberg RA. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci U S A. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC. Immortalization and transformation of human cells. Mol Cells. 2002;13:351–361. [PubMed] [Google Scholar]
- Lundberg AS, Randell SH, Stewart SA, Elenbaas B, Hartwell KA, Brooks MW, Fleming MD, Olsen JC, Miller SW, Weinberg RA, Hahn WC. Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene. 2002;21:4577–4586. doi: 10.1038/sj.onc.1205550. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Cell immortality and hallmarks of cancer. Cell Cycle. 2003;2:296–299. [PubMed] [Google Scholar]
- Bazarov AV, Hines WC, Mukhopadhyay R, Beliveau A, Melodyev S, Zaslavsky Y, Yaswen P. Telomerase activation by c-Myc in human mammary epithelial cells requires additional genomic changes. Cell Cycle. 2009;8:3373–3378. doi: 10.4161/cc.8.20.9856. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Cohen D. The canine transmissible venereal tumor: a unique result of tumor progression. Adv Cancer Res. 1985;43:75–112. doi: 10.1016/s0065-230x(08)60943-4. [DOI] [PubMed] [Google Scholar]
- Liao KW, Hung SW, Hsiao YW, Bennett M, Chu RM. Canine transmissible venereal tumor cell depletion of B lymphocytes: molecule(s) specifically toxic for B cells. Vet Immunol Immunopathol. 2003;92:149–162. doi: 10.1016/s0165-2427(03)00032-1. [DOI] [PubMed] [Google Scholar]
- Leroi AM, Koufopanou V, Burt A. Cancer selection. Nat Rev Cancer. 2003;3:226–231. doi: 10.1038/nrc1016. [DOI] [PubMed] [Google Scholar]
- Pearse AM, Swift K. Allograft theory: transmission of devil facial-tumour disease. Nature. 2006;439:549. doi: 10.1038/439549a. [DOI] [PubMed] [Google Scholar]
- Siddle HV, Kreiss A, Eldridge MD, Noonan E, Clarke CJ, Pyecroft S, Woods GM, Belov K. Transmission of a fatal clonal tumor by biting occurs due to depleted MHC diversity in a threatened carnivorous marsupial. Proc Natl Acad Sci U S A. 2007;104:16221–16226. doi: 10.1073/pnas.0704580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAloose D, Newton AL. Wildlife cancer: a conservation perspective. Nat Rev Cancer. 2009;9:517–526. doi: 10.1038/nrc2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP. Clonally transmissible cancers in dogs and Tasmanian devils. Oncogene. 2008;27(Suppl 2):S19–30. doi: 10.1038/onc.2009.350. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Tovar C, Hsu A, Bender HS, Kheradpour P, Rebbeck CA, Obendorf D, Conlan C, Bahlo M, Blizzard CA, Pyecroft S, Kreiss A, Kellis M, Stark A, Harkins TT, Marshall Graves JA, et al. The Tasmanian devil transcriptome reveals Schwann cell origins of a clonally transmissible cancer. Science. 2010;327:84–87. doi: 10.1126/science.1180616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schollnberger H, Beerenwinkel N, Hoogenveen R, Vineis P. Cell selection as driving force in lung and colon carcinogenesis. Cancer Res. 2010;70:6797–6803. doi: 10.1158/0008-5472.CAN-09-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–60. [PubMed] [Google Scholar]
- Hempen PM, Zhang L, Bansal RK, Iacobuzio-Donahue CA, Murphy KM, Maitra A, Vogelstein B, Whitehead RH, Markowitz SD, Willson JK, Yeo CJ, Hruban RH, Kern SE. Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers. Cancer Res. 2003;63:994–999. [PubMed] [Google Scholar]
- Zhang Y, Italia MJ, Auger KR, Halsey WS, Van Horn SF, Sathe GM, Magid-Slav M, Brown JR, Holbrook JD. Molecular evolutionary analysis of cancer cell lines. Mol Cancer Ther. 2010;9:279–291. doi: 10.1158/1535-7163.MCT-09-0508. [DOI] [PubMed] [Google Scholar]
- Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, Karchin R, Kinzler KW, Vogelstein B, Nowak MA. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci U S A. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV. Oncogenic resistance to growth-limiting conditions. Nat Rev Cancer. 2002;2:221–225. doi: 10.1038/nrc743. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Carcinogenesis, cancer therapy and chemoprevention. Cell Death Differ. 2005;12:592–602. doi: 10.1038/sj.cdd.4401610. [DOI] [PubMed] [Google Scholar]
- Dotto GP, Parada LF, Weinberg RA. Specific growth response of ras-transformed embryo fibroblasts to tumour promoters. Nature. 1985;318:472–475. doi: 10.1038/318472a0. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Antiangiogenic therapy and tumor progression. Cancer Cell. 2004;5:13–17. doi: 10.1016/s1535-6108(03)00336-2. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Why therapeutic response may not prolong the life of a cancer patient: selection for oncogenic resistance. Cell Cycle. 2005;4:1693–1698. doi: 10.4161/cc.4.12.2259. [DOI] [PubMed] [Google Scholar]
- Fleenor CJ, Marusyk A, DeGregori J. Ionizing radiation and hematopoietic malignancies: altering the adaptive landscape. Cell Cycle. 2010;9:3005–3011. doi: 10.4161/cc.9.15.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz MH, Shen H, Maki CG. Acquisition of p53 mutations in response to the non-genotoxic p53 activator Nutlin-3. Oncogene. 2011;30:4678–4686. doi: 10.1038/onc.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest. 2007;117:1175–1183. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta C, Subhra Kumar B, Larghi P, Rubino L, Mancino A, Sica A. Tumor promotion by tumor-associated macrophages. Adv Exp Med Biol. 2007;604:67–86. doi: 10.1007/978-0-387-69116-9_5. [DOI] [PubMed] [Google Scholar]
- Shan W, Liu J. Inflammation: a hidden path to breaking the spell of ovarian cancer. Cell Cycle. 2009;8:3107–3111. doi: 10.4161/cc.8.19.9590. [DOI] [PubMed] [Google Scholar]
- Silvera D, Schneider RJ. Inflammatory breast cancer cells are constitutively adapted to hypoxia. Cell Cycle. 2009;8:3091–3096. doi: 10.4161/cc.8.19.9637. [DOI] [PubMed] [Google Scholar]
- Gonda TA, Tu S, Wang TC. Chronic inflammation, the tumor microenvironment and carcinogenesis. Cell Cycle. 2009;8:2005–2013. doi: 10.4161/cc.8.13.8985. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhou BP. Inflammation: a driving force speeds cancer metastasis. Cell Cycle. 2009;8:3267–3273. doi: 10.4161/cc.8.20.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradisi A, Mehlen P. Netrin-1, a missing link between chronic inflammation and tumor progression. Cell Cycle. 2010;9:1253–1262. doi: 10.4161/cc.9.7.11072. [DOI] [PubMed] [Google Scholar]
- Barykova YA, Logunov DY, Shmarov MM, Vinarov AZ, Fiev DN, Vinarova NA, Rakovskaya IV, Baker PS, Shyshynova I, Stephenson AJ, Klein EA, Naroditsky BS, Gintsburg AL, Gudkov AV. Association of Mycoplasma hominis infection with prostate cancer. Oncotarget. 2011;2:289–297. doi: 10.18632/oncotarget.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisanti MP, Martinez-Outschoorn UE, Pavlides S, Whitaker-Menezes D, Pestell RG, Howell A, Sotgia F. Accelerated aging in the tumor microenvironment: connecting aging, inflammation and cancer metabolism with personalized medicine. Cell Cycle. 2011;10:2059–2063. doi: 10.4161/cc.10.13.16233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balliet RM, Capparelli C, Guido C, Pestell TG, Martinez-Outschoorn UE, Lin Z, Whitaker-Menezes D, Chiavarina B, Pestell RG, Howell A, Sotgia F, Lisanti MP. Mitochondrial oxidative stress in cancer-associated fibroblasts drives lactate production, promoting breast cancer tumor growth: Understanding the aging and cancer connection. Cell Cycle. 2011;10:4065–4073. doi: 10.4161/cc.10.23.18254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A, Campisi J. Integrating epithelial cancer, aging stroma and cellular senescence. Adv Gerontol. 2003;11:109–116. [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Vicente-Duenas C, Abollo-Jimenez F, Ruiz-Roca L, Alonso-Escudero E, Jimenez R, Cenador MB, Criado FJ, Cobaleda C, Sanchez-Garcia I. The age of the target cell affects B-cell leukaemia malignancy. Aging. 2010;2:908–913. doi: 10.18632/aging.100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CJ, Marusyk A, Zaberezhnyy V, Adane B, DeGregori J. Declining lymphoid progenitor fitness promotes aging-associated leukemogenesis. Proc Natl Acad Sci U S A. 2010;107:21713–21718. doi: 10.1073/pnas.1005486107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CJ, Marusyk A, DeGregori J. Aging-associated changes in hematopoiesis and leukemogenesis: what's the connection? Aging. 2011;3:643–656. doi: 10.18632/aging.100351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Travers JB, Machado C, Somani AK, Spandau DF. Reversing the aging stromal phenotype prevents carcinoma initiation. Aging. 2011;3:407–416. doi: 10.18632/aging.100318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, Andersen JK, Kapahi P, Melov S. Cellular senescence: A link between cancer and age-related degenerative disease? Semin Cancer Biol. 2011;21:354–359. doi: 10.1016/j.semcancer.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hursting SD, Perkins SN, Phang JM. Calorie restriction delays spontaneous tumorigenesis in p53-knockout transgenic mice. Proc Natl Acad Sci U S A. 1994;91:7036–7040. doi: 10.1073/pnas.91.15.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrigan D, Perkins SN, Haines DC, Hursting SD. Adult-onset calorie restriction and fasting delay spontaneous tumorigenesis in p53-deficient mice. Carcinogenesis. 2002;23:817–822. doi: 10.1093/carcin/23.5.817. [DOI] [PubMed] [Google Scholar]
- Hursting SD, Smith SM, Lashinger LM, Harvey AE, Perkins SN. Calories and carcinogenesis: lessons learned from 30 years of calorie restriction research. Carcinogenesis. 2010;31:83–89. doi: 10.1093/carcin/bgp280. [DOI] [PubMed] [Google Scholar]
- Ingram DK, Zhu M, Mamczarz J, Zou S, Lane MA, Roth GS, deCabo R. Calorie restriction mimetics: an emerging research field. Aging Cell. 2006;5:97–108. doi: 10.1111/j.1474-9726.2006.00202.x. [DOI] [PubMed] [Google Scholar]
- Ingram DK, Anson RM, de Cabo R, Mamczarz J, Zhu M, Mattison J, Lane MA, Roth GS. Development of calorie restriction mimetics as a prolongevity strategy. Ann N Y Acad Sci. 2004;1019:412–423. doi: 10.1196/annals.1297.074. [DOI] [PubMed] [Google Scholar]
- Hursting SD, Lavigne JA, Berrigan D, Perkins SN, Barrett JC. Calorie restriction, aging, and cancer prevention: mechanisms of action and applicability to humans. Annu Rev Med. 2003;54:131–152. doi: 10.1146/annurev.med.54.101601.152156. [DOI] [PubMed] [Google Scholar]
- Longo VD, Fontana L. Calorie restriction and cancer prevention: metabolic and molecular mechanisms. Trends Pharmacol Sci. 2010;31:89–98. doi: 10.1016/j.tips.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span–from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV. Calorie restriction: Decelerating mTOR-driven aging from cells to organisms (including humans) Cell Cycle. 2010;9:683–688. doi: 10.4161/cc.9.4.10766. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Prevention of cancer by inhibiting aging. Cancer Biol Ther. 2008;7:1520–1524. doi: 10.4161/cbt.7.10.6663. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandezr E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogenous mice. Nature. 2009;460:392–396. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov VN, Zabezhinski MA, Popovich IG, Piskunova TS, Semenchenko AV, Tyndyk ML, Yurova MN, Antoch MP, Blagosklonny MV. Rapamycin extends maximal lifespan in cancer-prone mice. Am J Pathol. 2010;176:2092–2097. doi: 10.2353/ajpath.2010.091050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov VN, Zabezhinski MA, Popovich IG, Piskunova TS, Semenchenko AV, Tyndyk ML, Yurova MN, Blagosklonny MV. Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle. 2011;10 doi: 10.4161/cc.10.24.18486. [DOI] [PubMed] [Google Scholar]
- Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S, Coppe JP, Krtolica A, Campisi J. Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J Cell Sci. 2005;118:485–496. doi: 10.1242/jcs.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos AR, Coppe JP, Campisi J, Desprez PY. Senescent cells as a source of inflammatory factors for tumor progression. Cancer Metastasis Rev. 2011;29:273–283. doi: 10.1007/s10555-010-9220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudkov AV, Gurova KV, Komarova EA. Inflammation and p53: A Tale of Two Stresses. Genes Cancer. 2011;2:503–516. doi: 10.1177/1947601911409747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- Adler AS, Kawahara TL, Segal E, Chang HY. Reversal of aging by NFkappaB blockade. Cell Cycle. 2008;7:556–559. doi: 10.4161/cc.7.5.5490. [DOI] [PubMed] [Google Scholar]
- Chauncey SS, Boothman DA, Habib AA. The receptor interacting protein 1 mediates a link between NFkappaB and PI3-kinase signaling. Cell Cycle. 2009;8:2671–2672. doi: 10.4161/cc.8.17.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre B, Coqueret O, Perkins ND. Regulation of activity and function of the p52 NF-kappaB subunit following DNA damage. Cell Cycle. 2010;9:4795–4804. doi: 10.4161/cc.9.24.14245. [DOI] [PubMed] [Google Scholar]
- Donato AJ, Pierce GL, Lesniewski LA, Seals DR. Role of NFkappaB in age-related vascular endothelial dysfunction in humans. Aging. 2009;1:678–680. doi: 10.18632/aging.100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko YN, Kuehl WM. A critical role for the NFkB pathway in multiple myeloma. Oncotarget. 2010;1:59–68. doi: 10.18632/oncotarget.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohanna M, Giuliano S, Bonet C, Imbert V, Hofman V, Zangari J, Bille K, Robert C, Bressac-de Paillerets B, Hofman P, Rocchi S, Peyron JF, Lacour JP, Ballotti R, Bertolotto C. Senescent cells develop a PARP-1 and nuclear factor-{kappa}B-associated secretome (PNAS) Genes Dev. 2011;25:1245–1261. doi: 10.1101/gad.625811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan S, Jat PS. Deciphering the role of nuclear factor-kappaB in cellular senescence. Aging. 2011;3:913–919. doi: 10.18632/aging.100390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalev A, Shaposhnikov M. Pharmacological inhibition of NF-kappaB prolongs lifespan of Drosophila melanogaster. Aging. 2011;3:391–394. doi: 10.18632/aging.100314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV. Aging, stem cells, and mammalian target of rapamycin: a prospect of pharmacologic rejuvenation of aging stem cells. Rejuvenation Res. 2008;11:801–808. doi: 10.1089/rej.2008.0722. [DOI] [PubMed] [Google Scholar]
- Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Zhang H, Bajraszewski N, Wu E, Wang H, Moseman AP, Dabora SL, Griffin JD, Kwiatkowski DJ. PDGFRs are critical for PI3K/Akt activation and negatively regulated by mTOR. J Clin Invest. 2007;117:730–738. doi: 10.1172/JCI28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Zheng P. The axis of mTOR-mitochondria-ROS and stemness of the hematopoietic stem cells. Cell Cycle. 2009;8:1158–1160. doi: 10.4161/cc.8.8.8139. [DOI] [PubMed] [Google Scholar]
- Castilho RM, Squarize CH, Chodosh LA, Williams BO, Gutkind JS. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell. 2009;5:279–289. doi: 10.1016/j.stem.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, DePinho RA. mTORC1 signaling governs hematopoietic stem cell quiescence. Cell Cycle. 2009;8:1003–1006. doi: 10.4161/cc.8.7.8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Nakada D, Yilmaz OH, Tothova Z, Joseph NM, Lim MS, Gilliland DG, Morrison SJ. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell. 2010;7:593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Wang TL, Samuels Y, Bardelli A, Cummins JM, DeLong L, Silliman N, Ptak J, Szabo S, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Lengauer C, Velculescu VE. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- Janes MR, Fruman DA. Targeting TOR dependence in cancer. Oncotarget. 2010;1:69–76. doi: 10.18632/oncotarget.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markman B, Dienstmann R, Tabernero J. Targeting the PI3K/Akt/mTOR pathway–beyond rapalogs. Oncotarget. 2010;1:530–543. doi: 10.18632/oncotarget.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28:1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Kittler O, Zhu J, Yang J, Liu G, Hendricks W, Lengauer C, Gabelli SB, Kinzler KW, Vogelstein B, Huso DL, Zhou S. PI3Kalpha inhibitors that inhibit metastasis. Oncotarget. 2010;1:339–348. doi: 10.18632/oncotarget.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dbouk HA, Backer JM. A beta version of life: p110beta takes center stage. Oncotarget. 2010;1:729–733. doi: 10.18632/oncotarget.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli AM, Evangelisti C, Chiarini F, McCubrey JA. The phosphatidylinositol 3-kinase/Akt/mTOR signaling network as a therapeutic target in acute myelogenous leukemia patients. Oncotarget. 2010;1:89–103. doi: 10.18632/oncotarget.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucera C, Lawler J, Hodin R, Parangi S. The BRAFV600E mutation: what is it really orchestrating in thyroid cancer? Oncotarget. 2010;1:751–756. doi: 10.18632/oncotarget.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L. P3Kalpha: a driver of tumor metastasis? Oncotarget. 2010;1:315–316. doi: 10.18632/oncotarget.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Stiegler AL, Boggon TJ, Kobayashi S, Halmos B. EGFR-mutated lung cancer: a paradigm of molecular oncology. Oncotarget. 2010;1:497–514. doi: 10.18632/oncotarget.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian D, Parsyan A, Petroulakis E, Hershey J, Sonenberg N. eIF4B controls survival and proliferation and is regulated by proto-oncogenic signaling pathways. Cell Cycle. 2010;9:4106–4109. doi: 10.4161/cc.9.20.13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia B, Nahle Z, Kenney AM. Double trouble: when sonic hedgehog signaling meets TSC inactivation. Cell Cycle. 2010;9:456–459. doi: 10.4161/cc.9.3.10532. [DOI] [PubMed] [Google Scholar]
- Fujishita T, Aoki M, Taketo MM. The role of mTORC1 pathway in intestinal tumorigenesis. Cell Cycle. 2009;8:3684–3687. doi: 10.4161/cc.8.22.9946. [DOI] [PubMed] [Google Scholar]
- Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci U S A. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La OJ, Murtaugh LC. Notch and Kras in pancreatic cancer: at the crossroads of mutation, differentiation and signaling. Cell Cycle. 2009;8:1860–1864. doi: 10.4161/cc.8.12.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Martin A, Oliveras-Ferraros C, Lopez-Bonet E, Menendez JA. AMPK: Evidence for an energy-sensing cytokinetic tumor suppressor. Cell Cycle. 2009;8:3679–3683. doi: 10.4161/cc.8.22.9905. [DOI] [PubMed] [Google Scholar]
- Peng C, Chen Y, Li D, Li S. Role of Pten in leukemia stem cells. Oncotarget. 2010;1:156–160. doi: 10.18632/oncotarget.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AS, Chapuis N, Lacombe C, Mayeux P, Bouscary D, Tamburini J. LKB1/AMPK/mTOR signaling pathway in hematological malignancies: from metabolism to cancer cell biology. Cell Cycle. 2011;10:2115–2120. doi: 10.4161/cc.10.13.16244. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20:427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hu W, Rajagopal G, Levine AJ. The tumor suppressor p53: cancer and aging. Cell Cycle. 2008;7:842–847. doi: 10.4161/cc.7.7.5657. [DOI] [PubMed] [Google Scholar]
- Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Feng Z, Mak TW, You H, Jin S. Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev. 2006;20:267–275. doi: 10.1101/gad.1363206. [DOI] [PubMed] [Google Scholar]
- Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidenko ZN, Korotchkina LG, Gudkov AV, Blagosklonny MV. Paradoxical suppression of cellular senescence by p53. Proc Natl Acad Sci U S A. 2010;107:9660–9664. doi: 10.1073/pnas.1002298107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotchkina LG, Leontieva OV, Bukreeva EI, Demidenko ZN, Gudkov AV, Blagosklonny MV. The choice between p53-induced senescence and quiescence is determined in part by the mTOR pathway. Aging. 2010;2:344–352. doi: 10.18632/aging.100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontieva O, Gudkov A, Blagosklonny M. Weak p53 permits senescence during cell cycle arrest. Cell Cycle. 2010;9:4323–4327. doi: 10.4161/cc.9.21.13584. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Kepp O, Kroemer G. TP53 and MTOR crosstalk to regulate cellular senescence. Aging. 2010;2:535–537. doi: 10.18632/aging.100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JS, Ryan KM. p53 and senescence: a little goes a long way. Cell Cycle. 2010;9:4050–4051. [PubMed] [Google Scholar]
- Santoro R, Blandino G. p53: The pivot between cell cycle arrest and senescence. Cell Cycle. 2010;9:4262–4263. doi: 10.4161/cc.9.21.13853. [DOI] [PubMed] [Google Scholar]
- Serrano M. Shifting senescence into quiescence by turning up p53. Cell Cycle. 2010;9:4256–4257. doi: 10.4161/cc.9.21.13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Demidenko ZN, Blagosklonny MV. Quantifying pharmacologic suppression of cellular senescence: prevention of cellular hypertrophy versus preservation of proliferative potential. Aging. 2009;1:1008–1016. doi: 10.18632/aging.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidenko ZN, Blagosklonny MV. The purpose of the HIF-1/PHD feedback loop: to limit mTOR-induced HIF-1alpha. Cell Cycle. 2011;10:1557–1562. doi: 10.4161/cc.10.10.15789. [DOI] [PubMed] [Google Scholar]
- Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, Hong S, Berry LS, Reichelt S, Ferreira M, Tavare S, Inoki K, Shimizu S. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332:966–970. doi: 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema RH, Herrera RE, Lam F, Weinberg RA. Growth suppression by p16ink4 requires functional retinoblastoma protein. Proc natl Acad Sci USA. 1995;92:62289–66293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge HJ, Woolthuis CM, de Bont ES, Huls G. Paradoxical down-regulation of p16 mRNA with advancing age in acute myeloid leukemia. Aging (Albany NY) 2009;1:949–953. doi: 10.18632/aging.100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EV, Ravitz MJ, Chen L, Lynch M. Growth controls connect: interactions between c-myc and the tuberous sclerosis complex-mTOR pathway. Cell Cycle. 2009;8:1344–1351. doi: 10.4161/cc.8.9.8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannell IG, Bushell M. Regulation of Myc by miR-34c: A mechanism to prevent genomic instability? Cell Cycle. 2010;9:2726–2730. [PubMed] [Google Scholar]
- Haferkamp S, Tran SL, Becker TM, Scurr LL, Kefford RF, Rizos H. The relative contributions of the p53 and pRb pathways in oncogene-induced melanocyte senescence. Aging. 2009;1:542–556. doi: 10.18632/aging.100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan EJ, Place RF, Basak S, Pookot D, Li LC. miR-449a causes Rb-dependent cell cycle arrest and senescence in prostate cancer cells. Oncotarget. 2010;1:349–358. doi: 10.18632/oncotarget.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akakura S, Nochajski P, Gao L, Sotomayor P, Matsui S, Gelman IH. Rb-dependent cellular senescence, multinucleation and susceptibility to oncogenic transformation through PKC scaffolding by SSeCKS/AKAP12. Cell Cycle. 2010;9:4656–4665. doi: 10.4161/cc.9.23.13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertel A, Dean JL, Rui H, Liu C, Witkiewicz AK, Knudsen KE, Knudsen ES. RB-pathway disruption in breast cancer: differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle. 2010;9:4153–4163. doi: 10.4161/cc.9.20.13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Kauffmann-Zeh A, Rodriguez-Viciana P, Ulrich E, Gilbert C, Coffer P, Downward J, Evan G. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Hueber AO, Evan GI. Traps to catch unwary oncogenes. Trends Genet. 1998;14:364–367. doi: 10.1016/s0168-9525(98)01520-0. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci U S A. 2008;105:5242–5247. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- Chawla R, Procknow JA, Tantravahi RV, Khurana JS, Litvin J, Reddy EP. Cooperativity of Cdk4R24C and Ras in melanoma development. Cell Cycle. 2010;9:3305–3314. doi: 10.4161/cc.9.16.12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- Leontieva OV, Blagosklonny MV. DNA damaging agents and p53 do not cause senescence in quiescent cells, while consecutive re-activation of mTOR is associated with conversion to senescence. Aging (Albany NY) 2010;2:924–935. doi: 10.18632/aging.100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV. Cell cycle arrest is not senescence. Aging. 2011;3:94–101. doi: 10.18632/aging.100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Maddick M, Miwa S, Jurk D, Czapiewski R, Saretzki G, Langie SA, Godschalk RW, Cameron K, von Zglinicki T. Adult-onset, short-term dietary restriction reduces cell senescence in mice. Aging. 2010;2:555–566. doi: 10.18632/aging.100196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland JL. Perspectives on cellular senescence and short term dietary restriction in adults. Aging. 2010;2:542–544. doi: 10.18632/aging.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidenko ZN, Blagosklonny MV. Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell Cycle. 2008;7:3355–3361. doi: 10.4161/cc.7.21.6919. [DOI] [PubMed] [Google Scholar]
- Pospelova TV, Demidenk ZN, Bukreeva EI, Pospelov VA, Gudkov AV, Blagosklonny MV. Pseudo-DNA damage response in senescent cells. Cell Cycle. 2009;8:4112–4118. doi: 10.4161/cc.8.24.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV. Aging-suppressants: cellular senescence (hyperactivation) and its pharmacologic deceleration. Cell Cycle. 2009;8:1883–1887. doi: 10.4161/cc.8.12.8815. [DOI] [PubMed] [Google Scholar]
- Demidenko ZN, Zubova SG, Bukreeva EI, Pospelov VA, Pospelova TV, Blagosklonny MV. Rapamycin decelerates cellular senescence. Cell Cycle. 2009;8:1888–1895. doi: 10.4161/cc.8.12.8606. [DOI] [PubMed] [Google Scholar]
- Demidenko ZN, Shtutman M, Blagosklonny MV. Pharmacologic inhibition of MEK and PI-3K converges on the mTOR/S6 pathway to decelerate cellular senescence. Cell Cycle. 2009;8:1896–1900. doi: 10.4161/cc.8.12.8809. [DOI] [PubMed] [Google Scholar]
- Demidenko ZN, Blagosklonny MV. At concentrations that inhibit mTOR, resveratrol suppresses cellular senescence. Cell Cycle. 2009;8:1901–1904. doi: 10.4161/cc.8.12.8810. [DOI] [PubMed] [Google Scholar]
- Romanov VS, Abramova MV, Svetlikova SB, Bykova TV, Zubova SG, Aksenov ND, Fornace AJ, Jr., Pospelova TV, Pospelov VA. p21(Waf1) is required for cellular senescence but not for cell cycle arrest induced by the HDAC inhibitor sodium butyrate. Cell Cycle. 2010;9:3945–3955. doi: 10.4161/cc.9.19.13160. [DOI] [PubMed] [Google Scholar]
- Wesierska-Gadek J. mTOR and its link to the picture of Dorian Gray - re-activation of mTOR promotes aging. Aging. 2010;2:892–893. doi: 10.18632/aging.100240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Lukas J, Bartkova J. DNA damage response as an anti-cancer barrier: damage threshold and the concept of ‘conditional haploinsufficiency’. Cell Cycle. 2007;6:2344–2347. doi: 10.4161/cc.6.19.4754. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Alimonti A, Nardella C, Chen Z, Clohessy JG, Carracedo A, Trotman LC, Cheng K, Varmeh S, Kozma SC, Thomas G, Rosivatz E, Woscholski R, Cognetti F, Scher HI, Pandolfi PP. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Invest. 2010;120:681–693. doi: 10.1172/JCI40535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27:2801–2809. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- Reddy JP, Peddibhotla S, Bu W, Zhao J, Haricharan S, Du YC, Podsypanina K, Rosen JM, Donehower LA, Li Y. Defining the ATM-mediated barrier to tumorigenesis in somatic mammary cells following ErbB2 activation. Proc Natl Acad Sci U S A. 2010;107:3728–3733. doi: 10.1073/pnas.0910665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkston JM, Garigan D, Hansen M, Kenyon C. Mutations that increase the life span of C. elegans inhibit tumor growth. Science. 2006;313:971–975. doi: 10.1126/science.1121908. [DOI] [PubMed] [Google Scholar]
- Longo VD, Lieber MR, Vijg J. Turning anti-ageing genes against cancer. Nat Rev Mol Cell Biol. 2008;9:903–910. doi: 10.1038/nrm2526. [DOI] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Gutierrez-Martinez P, Soria R, Fernandez-Capetillo O. ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev. 2008;22:297–302. doi: 10.1101/gad.452308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Murga M, Martinez-Pastor B, Ortega-Molina A, Soria R, Collado M, Fernandez-Capetillo O, Serrano M. Limited role of murine ATM in oncogene-induced senescence and p53-dependent tumor suppression. PLoS One. 2009;4:e5475. doi: 10.1371/journal.pone.0005475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus KJ, Hendzel MJ. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol Biol Cell. 2005;16:5013–5025. doi: 10.1091/mbc.E05-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G, Buhmann M, von Zglinicki T. DNA damage foci in mitosis are devoid of 53BP1. Cell Cycle. 2009;8:3379–3383. doi: 10.4161/cc.8.20.9857. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E. DNA lesions and DNA damage response: even long lasting relationships need a “break”. Cell Cycle. 2008;7:3653–3658. doi: 10.4161/cc.7.23.7178. [DOI] [PubMed] [Google Scholar]
- Bencokova Z, Kaufmann MR, Pires IM, Lecane PS, Giaccia AJ, Hammond EM. ATM activation and signaling under hypoxic conditions. Mol Cell Biol. 2009;29:526–537. doi: 10.1128/MCB.01301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T, Hoffbeck AS, Boumendil C, Soutoglou E. DNA damage response in the absence of DNA lesions continued. Cell Cycle. 2009;8:4025–4026. [PubMed] [Google Scholar]
- Bouquet F, Ousset M, Biard D, Fallone F, Dauvillier S, Frit P, Salles B, Muller C. A DNA-dependent stress response involving DNA-PK occurs in hypoxic cells and contributes to cellular adaptation to hypoxia. J Cell Sci. 2011;124:1943–1951. doi: 10.1242/jcs.078030. [DOI] [PubMed] [Google Scholar]
- Alexander A, Walker CL. Differential localization of ATM is correlated with activation of distinct downstream signaling pathways. Cell Cycle. 2010;9:3685–3686. doi: 10.4161/cc.9.18.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Jimenez FJ, Moreno-Manzano V, Mateos-Gregorio P, Royo I, Erceg S, Murguia JR, Sanchez-Puelles JM. FM19G11: A new modulator of HIF that links mTOR activation with the DNA damage checkpoint pathways. Cell Cycle. 2010;9:2803–2813. doi: 10.4161/cc.9.14.12184. [DOI] [PubMed] [Google Scholar]
- Guo Z, Deshpande R, Paull TT. ATM activation in the presence of oxidative stress. Cell Cycle. 2010;9:4805–4811. doi: 10.4161/cc.9.24.14323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1alpha. Mol Cell. 2010;40:509–520. doi: 10.1016/j.molcel.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor B, Gibbons JJ, Abraham RT, Yu K. Targeting mTOR globally in cancer: thinking beyond rapamycin. Cell Cycle. 2009;8:3831–3837. doi: 10.4161/cc.8.23.10070. [DOI] [PubMed] [Google Scholar]
- Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10:868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Babar P, Adamson C, Walker GA, Walker DW, Lithgow GJ. P13-kinase inhibition induces dauer formation, thermotolerance and longevity in C. elegans. Neurobiol Aging. 1999;20:513–519. doi: 10.1016/s0197-4580(99)00094-9. [DOI] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman U, Garcia AM, Busuttil RA, Sorensen D, Lundell M, Kapahi P, Vijg J. Lifespan extension by dietary restriction is not linked to protection against somatic DNA damage in Drosophila melanogaster. Aging Cell. 2009;8:331–338. doi: 10.1111/j.1474-9726.2009.00480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa SD, Kapahi P. Role of TOR signaling in aging and related biological processes in Drosophila melanogaster. Exp Gerontol. 2011;46:382–390. doi: 10.1016/j.exger.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]