Abstract
Oocytes from Xenopus laevis were injected with purified amber (UAG), ochre (UAA), and opal (UGA) suppressor tRNAs from yeasts. The radioactively labeled proteins translated from the endogenous mRNAs were then separated on two-dimensional gels. All three termination codons are used in a single cell, the Xenopus laevis oocyte. But a surprisingly low number of readthrough polypeptides were observed from the 600 mRNAs studied in comparison to uninjected oocytes. The experimental data are compared with the conclusions obtained from the compilation of all available termination sequences on eukaryotic and prokaryotic mRNAs. This comparison indicates that the apparent resistance of natural termination codons against readthrough, as observed by the microinjection experiments, cannot be explained by tandem or very close second stop codons. Instead it suggests that specific context sequences around the termination codons may play a role in the efficiency of translation termination.
Full text
PDF




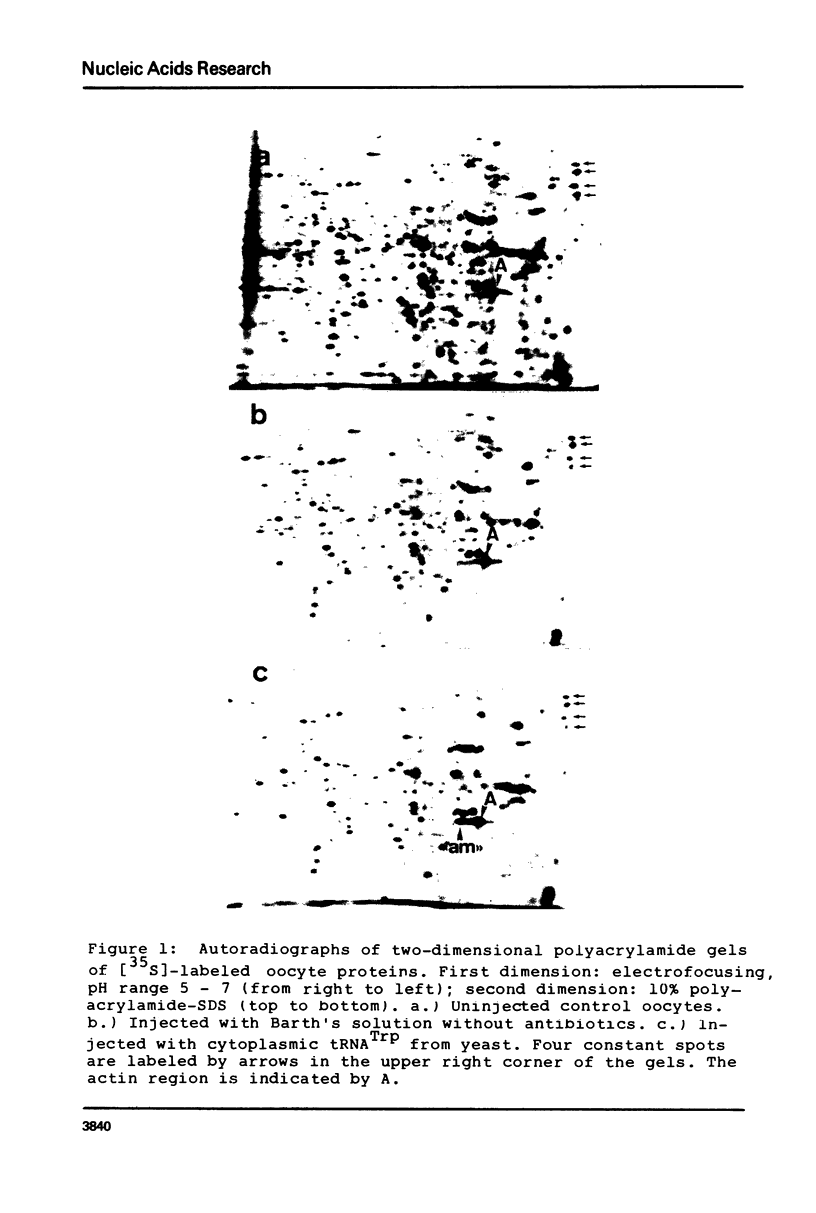
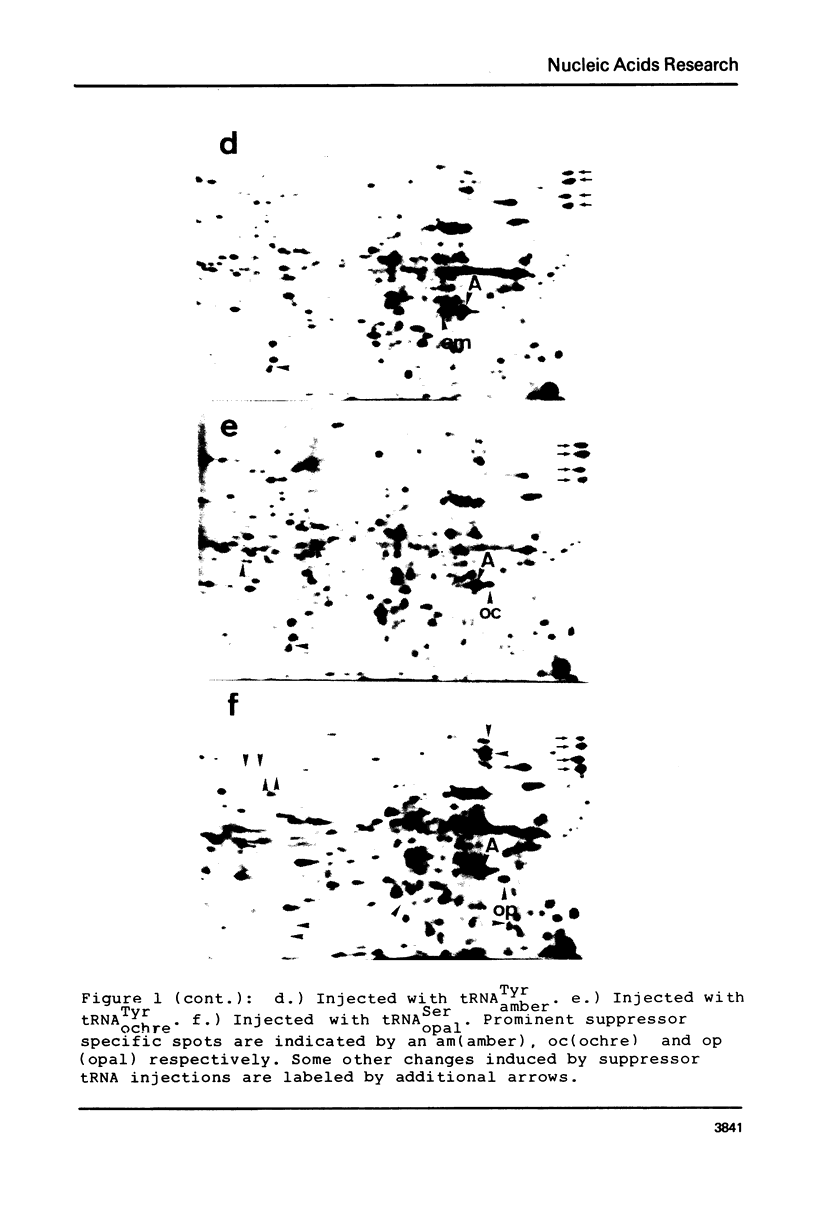

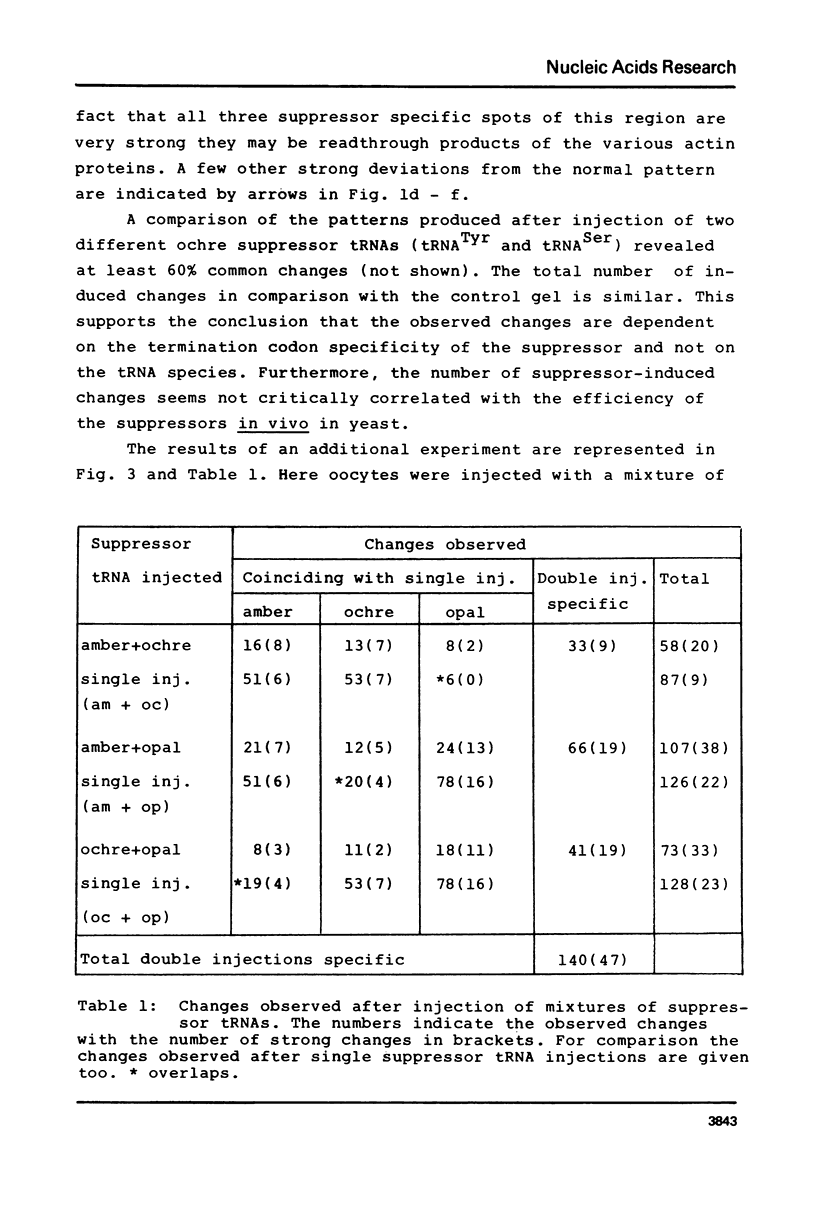







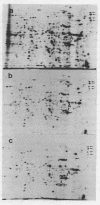
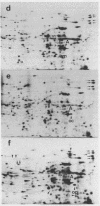
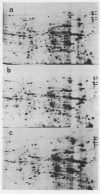
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Elseviers D., Gorini L. Low activity of -galactosidase in frameshift mutants of Escherichia coli. Proc Natl Acad Sci U S A. 1972 May;69(5):1192–1195. doi: 10.1073/pnas.69.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J. F., Gesteland R. F., Reid B. R., Anderson C. W. Normal tRNAs promote ribosomal frameshifting. Cell. 1979 Dec;18(4):1119–1131. doi: 10.1016/0092-8674(79)90225-3. [DOI] [PubMed] [Google Scholar]
- Barrell B. G., Anderson S., Bankier A. T., de Bruijn M. H., Chen E., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A. Different pattern of codon recognition by mammalian mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3164–3166. doi: 10.1073/pnas.77.6.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M., Kubli E., Kohli J., de Henau S., Grosjean H. Nonsense suppression in eukaryotes: the use of the Xenopus oocyte as an in vivo assay system. Nucleic Acids Res. 1980 Nov 25;8(22):5169–5178. doi: 10.1093/nar/8.22.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Bossi L., Ruth J. R. The influence of codon context on genetic code translation. Nature. 1980 Jul 10;286(5769):123–127. doi: 10.1038/286123a0. [DOI] [PubMed] [Google Scholar]
- Brenner S., Stretton A. O., Kaplan S. Genetic code: the 'nonsense' triplets for chain termination and their suppression. Nature. 1965 Jun 5;206(988):994–998. doi: 10.1038/206994a0. [DOI] [PubMed] [Google Scholar]
- Clarkson S. G., Birnstiel M. L., Serra V. Reiterated transfer RNA genes of Xenopus laevis. J Mol Biol. 1973 Sep 15;79(2):391–410. doi: 10.1016/0022-2836(73)90013-2. [DOI] [PubMed] [Google Scholar]
- Cremer K. J., Bodemer M., Summers W. P., Summers W. C., Gesteland R. F. In vitro suppression of UAG and UGA mutants in the thymidine kinase gene of herpes simplex virus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):430–434. doi: 10.1073/pnas.76.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ronde A., Van Loon A. P., Grivell L. A., Kohli J. In vitro suppression of UGA codons in a mitochondrial mRNA. Nature. 1980 Sep 25;287(5780):361–363. doi: 10.1038/287361a0. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H., Dekel L., Israeli-Reches M., Belfort M. The requirement of nonsense suppression for the development of several phages. Mol Gen Genet. 1979 Feb 26;170(2):155–159. doi: 10.1007/BF00337791. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H. UGA suppression by normal tRNA Trp in Escherichia coli: codon context effects. Nucleic Acids Res. 1981 Feb 25;9(4):983–991. doi: 10.1093/nar/9.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D. Five TGA "stop" codons occur within the translated sequence of the yeast mitochondrial gene for cytochrome c oxidase subunit II. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6534–6538. doi: 10.1073/pnas.76.12.6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D., Weiss-Brummer B. Leaky +1 and -1 frameshift mutations at the same site in a yeast mitochondrial gene. Nature. 1980 Nov 6;288(5786):60–63. doi: 10.1038/288060a0. [DOI] [PubMed] [Google Scholar]
- Fradin A., Gruhl H., Feldmann H. Mapping of yeast tRNAs by two-dimensional electrophoresis on polyacrylamide gels. FEBS Lett. 1975 Feb 1;50(2):185–189. doi: 10.1016/0014-5793(75)80485-6. [DOI] [PubMed] [Google Scholar]
- Geller A. I., Rich A. A UGA termination suppression tRNATrp active in rabbit reticulocytes. Nature. 1980 Jan 3;283(5742):41–46. doi: 10.1038/283041a0. [DOI] [PubMed] [Google Scholar]
- Gurdon J. B. Changes in somatic cell nuclei inserted into growing and maturing amphibian oocytes. J Embryol Exp Morphol. 1968 Nov;20(3):401–414. [PubMed] [Google Scholar]
- Gurdon J. B., De Robertis E. M., Partington G. Injected nuclei in frog oocytes provide a living cell system for the study of transcriptional control. Nature. 1976 Mar 11;260(5547):116–120. doi: 10.1038/260116a0. [DOI] [PubMed] [Google Scholar]
- Hawthorne D. C., Leupold U. Suppressors in yeast. Curr Top Microbiol Immunol. 1974;64(0):1–47. doi: 10.1007/978-3-642-65848-8_1. [DOI] [PubMed] [Google Scholar]
- Iserentant D., Van Montagu M., Fiers W. Studies on the bacteriophage MS2. XLI. Nature of the azure mutation. J Mol Biol. 1980 May 15;139(2):243–263. doi: 10.1016/0022-2836(80)90307-1. [DOI] [PubMed] [Google Scholar]
- Kaplan S. In vivo translation of amber and ochre codons in Escherichia coli. Mol Gen Genet. 1973 Feb 2;120(3):191–200. doi: 10.1007/BF00267151. [DOI] [PubMed] [Google Scholar]
- Kohli J., Kwong T., Altruda F., Söll D., Wahl G. Characterization of a UGA-suppressing serine tRNA from Schizosaccharomyces pombe with the help of a new in vitro assay system for eukaryotic suppressor tRNAs. J Biol Chem. 1979 Mar 10;254(5):1546–1551. [PubMed] [Google Scholar]
- Lu P., Rich A. The nature of the polypeptide chain termination signal. J Mol Biol. 1971 Jun 14;58(2):513–531. doi: 10.1016/0022-2836(71)90368-8. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R. Leaky UAG termination codon in tobacco mosaic virus RNA. Nature. 1978 Mar 30;272(5652):469–471. doi: 10.1038/272469a0. [DOI] [PubMed] [Google Scholar]
- Philipson L., Andersson P., Olshevsky U., Weinberg R., Baltimore D., Gesteland R. Translation of MuLV and MSV RNAs in nuclease-treated reticulocyte extracts: enhancement of the gag-pol polypeptide with yeast suppressor tRNA. Cell. 1978 Jan;13(1):189–199. doi: 10.1016/0092-8674(78)90149-6. [DOI] [PubMed] [Google Scholar]
- Piper P. W., Wasserstein M., Engbaek F., Kaltoft K., Celis J. E., Zeuthen J., Liebman S., Sherman F. Nonsense suppressors of Saccharomyces cerevisiae can be generated by mutation of the tyrosine tRNA anticodon. Nature. 1976 Aug 26;262(5571):757–761. doi: 10.1038/262757a0. [DOI] [PubMed] [Google Scholar]
- Rafalski A., Kohli J., Agris P., Söll D. The nucleotide sequence of a UGA suppressor serine tRNA from Schizosaccharomyces pombe. Nucleic Acids Res. 1979 Jun 25;6(8):2683–2695. doi: 10.1093/nar/6.8.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W., Fluck M., Epstein R. The influence of the reading context upon the suppression of nonsense codons. 3. Cold Spring Harb Symp Quant Biol. 1969;34:513–520. doi: 10.1101/sqb.1969.034.01.058. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Weber K. A single UGA codon functions as a natural termination signal in the coliphage q beta coat protein cistron. J Mol Biol. 1973 Nov 15;80(4):837–855. doi: 10.1016/0022-2836(73)90213-1. [DOI] [PubMed] [Google Scholar]
- Williams J. G., Kay R. M., Patient R. K. The nucleotide sequence of the major beta-globin mRNA from Xenopus laevis. Nucleic Acids Res. 1980 Sep 25;8(18):4247–4258. doi: 10.1093/nar/8.18.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahata H., Ocada Y., Tsugita A. Adjacent effect on suppression efficiency. II. Study on ochre and amber mutants of T4 phage lysozyme. Mol Gen Genet. 1970;106(3):208–212. doi: 10.1007/BF00340380. [DOI] [PubMed] [Google Scholar]
- Yates J. L., Gette W. R., Furth M. E., Nomura M. Effects of ribosomal mutations on the read-through of a chain termination signal: studies on the synthesis of bacteriophage lambda O gene protein in vitro. Proc Natl Acad Sci U S A. 1977 Feb;74(2):689–693. doi: 10.1073/pnas.74.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]