Abstract
Many pollinator populations are declining, with large economic and ecological implications. Parasites are known to be an important factor in the some of the population declines of honey bees and bumblebees, but little is known about the parasites afflicting most other pollinators, or the extent of interspecific transmission or vectoring of parasites. Here we carry out a preliminary screening of pollinators (honey bees, five species of bumblebee, three species of wasp, four species of hoverfly and three genera of other bees) in the UK for parasites. We used molecular methods to screen for six honey bee viruses, Ascosphaera fungi, Microsporidia, and Wolbachia intracellular bacteria. We aimed simply to detect the presence of the parasites, encompassing vectoring as well as actual infections. Many pollinators of all types were positive for Ascosphaera fungi, while Microsporidia were rarer, being most frequently found in bumblebees. We also detected that most pollinators were positive for Wolbachia, most probably indicating infection with this intracellular symbiont, and raising the possibility that it may be an important factor in influencing host sex ratios or fitness in a diversity of pollinators. Importantly, we found that about a third of bumblebees (Bombus pascuorum and Bombus terrestris) and a third of wasps (Vespula vulgaris), as well as all honey bees, were positive for deformed wing virus, but that this virus was not present in other pollinators. Deformed wing virus therefore does not appear to be a general parasite of pollinators, but does interact significantly with at least three species of bumblebee and wasp. Further work is needed to establish the identity of some of the parasites, their spatiotemporal variation, and whether they are infecting the various pollinator species or being vectored. However, these results provide a first insight into the diversity, and potential exchange, of parasites in pollinator communities.
Introduction
Pollinators are of great ecological and economic importance. They pollinate a wide variety of crops with an estimated global value of $153 billion pa [1], [2], and are also essential for the reproduction of at least two thirds of flowering plant species, including many which are now endangered [3], [4], [5], [6]. Although much attention has focused on managed populations of honey bees, pollinators include a diversity of insects, with the main groups in temperate areas being bumblebees, social wasps, hoverflies and solitary bees. However, many pollinator populations appear to be decreasing, although declines are by no means universal [7], [8]. The species richness of bees in the UK and Holland has decreased substantially [9], [10], and around a third of honey bee colonies in the US have been lost each year since 2006, in part due to a syndrome termed ‘Colony Collapse Disorder’ [11]. Of the UK's 25 bumblebee species, 3 are now extinct, 7 are threatened and 15 have undergone major range contractions in recent years [10], while North American bumblebees are also declining in abundance and species richness [12], [13]. There are a multitude of factors that are responsible for this, including land use change, pesticide exposure, reductions in population genetic diversity and climate change [14], [15], [16].
One factor that may be particularly important in pollinator declines is parasites. Parasites are a key selection pressure for most organisms, including insects. Some insect parasites are highly virulent and cause obvious disease symptoms, such as the obligate killer fungal parasites Metarhizium and Ascosphaera [17], [18], [19], [20]. Other parasites are far less obvious, such as Wolbachia intracellular bacteria, which are widespread in insects and can have a major impact on host fitness by manipulating host sex ratios or negatively affecting host survival [21], [22]. The parasites of honey bees are relatively well known and have been implicated in the recent colony losses seen in the US and elsewhere [14], [23], [24], [25], [26]. They include apparently long-established host-parasite relationships such as the Ascosphaera apis fungal parasite which causes chalkbrood disease, the microsporidian Nosema apis which causes dysentery, and many viruses [27]. However, they are also characterised by a number of emerging parasites, the appearance of which can have large impacts on honey bee populations, such as the microsporidian Nosema ceranae in Spain and Portugal [25], [28], [29], [30]. Bumblebees too may be infected by N. ceranae [31], as well as suffering from their own microsporidian, Nosema bombi, which can have major effects on their fitness [32], [33], [34].
However, our knowledge of the parasites that afflict other pollinators is far more limited. The economically important alfalfa leaf-cutter bee (Megachile rotundata) is well known to suffer from Ascosphaera fungi [35], [36], and the solitary bee Andrena scotica has been found to have high prevalence infections by the Antonospora scoticae microsporidian [37], [38], but nothing is known of the parasites of the vast majority of pollinators. In addition, the shared use of flowers by communities of pollinator species represents a significant opportunity from the perspective of a parasite for interspecific transmission or at least vectoring (i.e. transport of a parasite without infection). The discoveries of Nosema ceranae in Argentinean bumblebees [31], deformed wing virus (DWV) in German Bombus pascuorum and commercial Bombus terrestris [39], and of acute bee paralysis virus (ABPV) in UK bumblebees [40], suggest that even intergeneric pathogen spillover can occur. Most recently, molecular screening of bees and wasps collected near apiaries in the USA found that many were positive for various honey bee viruses [41], although the limited sampling effort did not provide any information on prevalence. Far more information is therefore needed on the frequency of parasites in pollinators, including those known to cause disease in honey bees.
Here we carry out a preliminary examination of the occurrence of parasites in a variety of pollinators from across the UK. We collected samples of social wasps, bumblebees, hoverflies, honey bees and other bees (all of which pollinate flowers [9], [42], [43], [44]) and used molecular methods to screen them for the presence of the six most common honey bee viruses, Ascosphaera fungi, Microsporidia, and the Wolbachia intracellular bacterium.
Results
Out of 325 pollinator samples collected, DNA and RNA was successfully extracted from 272 individuals and amplified in both conventional and real-time PCRs, based on the internal control gene (Table S1). Pollinators were found to be frequently positive for Wolbachia and Ascosphaera, and less frequently for Microsporidia and DWV parasites (Figure 1). One Vespula vulgaris wasp was positive for black queen cell virus and another for sacbrood virus. ABPV, chronic bee paralysis virus and Israeli acute paralysis virus were not detected in any samples. The CoxA sequences obtained from five hoverflies and 7 bees were all confirmed to be Wolbachia. Eight bumblebees, three wasps and one Lasioglossum bee were sequenced for the V1f/530r microsporidian gene, with the bumblebee sequences all matching most closely (96–100%) that of Nosema bombi. The microsporidians from wasps most closely matched Nosema bombi (95–97%), while that from the Lasioglossum bee had no strong match. The Ascosphaera sequences of four bees were examined and two (one Andrena and one Halictus) closely matched that of Ascosphaera apis (98%), while the other two (one Lasioglossum and one Halictus) had no strong match.
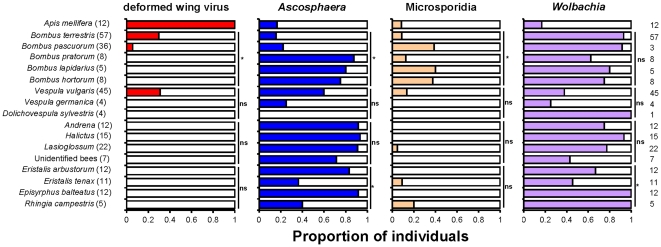
Figure 1. Incidence of parasites in pollinators.
Overall proportions of honey bees (Apis mellifera), five species of bumblebees (Bombus spp.), three species of wasps (Vespula vulgaris, V. germanica, Dolichovespula sylvestris), three genera of other bees (Andrena, Halcitus, Lasioglossum; there were also some that could not be identified) and four species of hoverflies (Eristalis arbustorum, E. tenax, Episyrphus balteatus, Rhingia campestris) which were positive (coloured) or negative (white) for the deformed wing virus (DWV), Ascosphaera fungus, Microsporidia and Wolbachia parasites. Sample sizes for each host are given in parentheses. Significant differences in incidence between species within each of the five host types (honey bees, bumblebees, wasps, other bees, hoverflies) are indicated to the right of the relevant bars (* = P<0.05; ns = P>0.05).
The pollinator types differed significantly in the numbers of individuals that were positive for DWV (χ2 = 82, df = 4, P<0.001), Ascosphaera (χ2 = 69.8, df = 4, P<0.001), Microsporidia (χ2 = 19.4, df = 4, P = 0.001) and Wolbachia (χ2 = 60.3, df = 4, P<0.001). All honey bees were positive for DWV, as were about a third of Vespula vulgaris wasps, while bumblebee species interestingly varied significantly, with a third of Bombus terrestris bumblebees and also a small number of B. pascuorum being positive, but all other species being entirely negative (Figure 1). Ascosphaera was common in Andrena, Halictus and Lasioglossum bees, of moderate frequency in wasps and honey bees, while its incidence varied between both bumblebee species and hoverfly species (Figure 1). Microsporidia were in general relatively rare, being found at the greatest frequency in bumblebees where a fifth of samples overall were positive and incidence differed between species (Figure 1). Wolbachia was very common in bumblebees, Andrena, Halictus, and Lasioglossum bees, but infected only 17% of honey bees, and differed in incidence between the four hoverfly species (Figure 1).
Discussion
Our use of molecular methods revealed that many pollinators were positive for the parasites that we screened for. Both Ascosphaera and Wolbachia were very prevalent, being found in most pollinators of all types. Microsporidia were much rarer, mainly being found in bumblebees. We found no evidence of acute bee paralysis virus, chronic bee paralysis virus or Israeli acute paralysis virus in any of the pollinators, and only found a single V. vulgaris wasp positive for sacbrood virus and another for black queen cell virus. However, we did find that a third of Bombus terrestris and a third of Vespula vulgaris wasps were positive for deformed wing virus, as well as all of the honey bees in our sample. Inefficiencies in the Chelex extraction method used [45], [46], mean that true levels of the viruses may be greater still.
All of the pollinator taxa we examined included individuals that were positive for Wolbachia. As Wolbachia are intracellular bacteria, the positive amplification of it means these individuals were most probably infected by the bacteria rather than vectoring them. Wolbachia is a widespread parasite of insects [22], including many ants [47], [48], [49], but has been surprisingly little investigated in social bees and wasps. It has previously been found in the Apis mellifera scutellata and A. m. capensis African subspecies of the Western honey bee [50], [51], [52], and in Osmia (Megachilidae), Rediviva (Melittidae), Agapostemon (Halictidae), Colletes (Colletidae) and Diadasia (Apidae) solitary bees [53], [54]. Our results therefore confirm that Wolbachia infects honey bees, and also expand its host range to include bumblebees, Andrena (Andrenidae), Halictus (Halictidae) and Lasioglossum (Halictidae) bees, as well as wasps and hoverflies. In other insects, Wolbachia can have profound effects on host sex ratios, as well as negatively or positively affecting other aspects of host fitness [22], [55], [56], [57], [58], [59]. It will therefore be important to discover what effects Wolbachia has on its pollinator hosts.
The Ascosphaera fungi, which cause chalkbrood disease, were found very commonly in many of the pollinator taxa that we sampled. Ascosphaera has previously only been recorded as a parasite of honey bees, Megachile, Osmia and Coelioxys megachilid solitary bees [18], [60], [61], [62], [63], and as an apparently non-pathogenic symbiont of Nomia (Halictidae) solitary bees [64]. In all cases it specifically infects the larval life-stage of the host insect and has not previously been recorded from the adult life-stage that made up our samples. It is unlikely that Ascosphaera would be able to infect hoverflies given their larval biology differs so much from that of bees. It is also unlikely that Ascosphaera infects bumblebees given that it has never been recorded parasitizing them in spite of the intensive research into their host-parasite interactions [33], [65]. It therefore seems most probable that the high incidence of Ascosphaera in our samples of adult pollinators reflects vectoring of the fungal spores, either on the body surface of the insects or, more probably given our sampling protocol, in their guts. The results suggest that Ascosphaera is very common in the environment of pollinators and that vectoring by non-host pollinators may have an important role to play in its movement around the environment.
Microsporidian parasites were relatively rare in our samples. They were most frequently found in bumblebees, with only a small number of honey bees, wasps, Lasioglossum bees and hoverflies also being positive for Microsporidia, and the remaining pollinators all being negative. In the case of bumblebees, the Microsporidia were Nosema bombi, but the identity of the microsporidians in the other pollinators is unknown. Microsporidia are common parasites in both honey bees and bumblebees, and the incidence in our samples was lower than has often been found previously [27], [33]. However, there can be substantial seasonal or population variation in the incidence of microsporidian parasites, which may explain the low levels we found [33], [66], [67].
Possibly the most important result was the discovery that DWV was present in a third of Bombus terrestris bumblebees and Vespula vulgaris wasps, as well as a few B. pascuorum. Symptomatic DWV infections in honey bees result in bees developing with deformed wings and thus being unable to fly, although most infections do not result in such obvious symptoms [27]. DWV has previously been found infecting, and causing symptoms in, a colony of B. pascuorum in Germany, as well as possibly in 10% of commercially-reared B. terrestris [39]. A similar molecular screening to ours detected it in every bee and wasp species examined [41], although the limited sampling in that study gave little information on prevalence. Our results indicate that DWV is in fact quite widespread in bumblebees, at least in B. terrestris, and is also common in V. vulgaris wasps. As our samples were of foraging bees and wasps collected at flowers or in pan traps, the DWV clearly had not caused the deformed wing symptoms. It therefore represented either asymptomatic infections or was being vectored. Further work will be needed to establish whether DWV is a natural parasite of these species, or has spilled over from honey bees, or was simply being vectored by them. We sampled only very few individuals for the other bumblebee and wasp species, and so the lack of DWV in these species may be due to the low sample size, and the same may be true of the other pollinators. The bumblebees and wasps were collected at different sites from most of the other bees and hoverflies, so spatial variation could also explain the differences, although it is notable that the single bumblebee sampled in the south of England was positive for DWV. Nevertheless, the complete lack of DWV in any of the other bees or hoverflies that we sampled suggests that it is not a general parasite of pollinating insects and that bumblebees and Vespula wasps are more likely to be infected or contaminated with DWV than other pollinators. Unlike other pollinators, both bumblebees and Vespula wasps rob honey from honey bee colonies. Honey has been shown to contain infective particles of DWV [41], so this may therefore be the main route by which bumblebees and Vespula wasps become infected or contaminated with DWV.
These preliminary data demonstrate that a wide variety of pollinators carry Wolbachia, Ascosphaera, microsporidian and DWV parasites. Regardless of whether the results represent vectoring or infection, it appears there may be significant interaction between host species in the movement of parasites. All of these parasites have the potential to substantially reduce host fitness and the results thus emphasise the importance of determining the diversity and impact of parasites in order to inform the conservation of pollinator populations. The shared use of flowers by multiple pollinator species, as well as robbing of food stores in some, has the potential to make the transmission or vectoring of parasites between taxa relatively frequent. Incorporating multi-species pollinator interactions will therefore be essential to accurately model and predict the population-level dynamics of pollinator parasites.
Materials and Methods
Sample collection
A total of 325 pollinators were collected by hand from flowers and by pan-trapping from 83 locations within 18 urban and arable sites (up to 5 km2) across the UK, during June/July 2007 and 2008 (Figure 2, Table S1). Hand-collected samples were stored immediately in 96% ethanol. Pan traps were checked every 48 h and samples then transferred to 96% ethanol. All samples were stored at −20°C. The samples collected included representatives of five species of bumblebees (Bombus terrestris, B. pascuorum, B. lapidarius, B. pratorum, B. hortorum), honey bees (Apis mellifera), three genera of solitary bees (Andrena, Halictus, Lasioglossum), three species of social wasps (Vespula vulgaris, V. germanica, Dolichovespula sylvestris) and four species of hoverflies (Eristalis arbustorum, E. tenax, Episyrphus balteatus, Rhingia campestris). The species of white-tailed bumblebee cannot be distinguished morphologically, so we sequenced the host CO1 gene for a subset of these individuals from each site. We compared their sequences to sequences in Genbank using BLASTN and found that all were B. terrestris.
Figure 2. Locations of the sites at which pollinator samples were collected.
Samples were collected within a 5 km area at each site. See Table 1 for the precise location of each of the numbered sites and the numbers of pollinators collected at each.
Molecular analysis
The midgut, ovaries and fat bodies were dissected out from the pollinators, as these seemed the tissues most likely to contain the parasites of interest. Wolbachia is generally considered a vertically transmitting symbiont so is most likely to be found in ovaries, although it can also occur at high intensities in the fat body and other tissues [22]. Microsporidia infect via the faecal-oral route, while the viruses and the Ascosphaera fungi also infect via ingestion, making them appear most likely to be found in the gut [27]. A small sample of the three tissues for each individual was combined, rehydrated in ddH20 and homogenised using a sterile pestle. DNA and RNA was extracted by boiling the sample in 5% Chelex solution for 15 minutes. Samples were then centrifuged at 3500 rpm (1060 g) for 10 minutes and the supernatant stored at −20°C. PCR amplification of the DNA was carried out using ABI 3700 thermal cyclers in 10 µl volumes containing 1 µl DNA, 0.2 µl of each forward and reverse primer, 2 µl PCR buffer and 0.05 µl of 5 U/µl Taq (Promega). Reactions contained primer specific quantities of 25 mM MgCl2 and 10 mM dNTPs and made up to 10 µl with ddH20. Samples were amplified for: 1) The CO1 host control gene with LCO-Hym/HCOout primers [68], [69] using 1.5 µl MgCl2 and 1 µl dNTPs, with an initial denaturation of 2 min at 94°C followed by 35 cycles of 30 s at 94°C, 45 s at 50°C and 2 min at 72°C, and a final extension step of 72°C for 7 min. 2) Microsporidia with the V1f/530r primers [70] using 1.5 µl MgCl2 and 0.5 µl dNTPs, with an initial denaturation of 1 min at 95°C followed by 35 cycles of 1 min at 95°C, 1 min at 60°C and 1 min at 72°C, and a final extension step of 72°C for 7 min. 3) Ascosphaera fungi with the AscoAll1/AscoAll2 primers [71] using 1 µl MgCl2 and 1.5 µl dNTPs, with an initial denaturation of 10 min at 94°C followed by 30 cycles of 45 s at 94°C, 45 s at 62°C and 1 minute at 72°C, and a final extension step of 72°C for 5 min. 4) Wolbachia intracellular bacteria with CoxA f/r primers [72] using 1 µl MgCl2 and 1 µl dNTPs, with an initial denaturation of 2 min at 94°C followed by 30 cycles of 30 s at 94°C, 45 s at 55°C and 2 min at 72°C, and ending with a final extension step of 72°C for 7 min. PCR products were visualised under UV using 1% agarose gels stained with ethidium bromide and compared to a 100 bp size ladder. Positive and negative controls were included in every PCR. The DNA from a subset of positive samples were subsequently amplified in 50 µl PCR reactions and purified using the Qiaquick PCR purification kit (Qiagen). Products were sequenced using the ABI Dye Terminator Labelled Sequencing sytem with an ABI3030xl capillary sequencer, and the resulting sequences compared with existing sequences in Genbank using a BLASTN search.
The Chelex extractions of all samples were in addition screened for the presence of six honey bee viruses using Taqman real-time PCR assays (Table 1). Probes for the detection of black queen cell virus (BQCV), chronic bee paralysis virus (CBPV), deformed wing virus (DWV) and Israeli acute paralysis virus (IAPV) were duel labelled with the fluorescent reporter dye FAM (6-carboxyfluorescein) at the 5′ end and with the fluorescent quencher dye TAMRA at the 3′ end. Probes for the detection of acute bee paralysis virus (ABPV) and sacbrood virus (SBV) substituted TAMRA for a minor groove binder (MGB) at the 3′ end. Each sample was screened in duplicate 25 µl reactions comprising 10× Buffer A (50 mMKCl, 10 mM Tris–HCl, pH 8.3, carboxy-X-rhodamine [ROX] passive reference dye), 0.025 U AmpliTaq Gold, 0.2 mM (each) deoxynucleoside triphosphate, 5.5 mM MgCl2, 0.016 U MMLV, 300 nM of each primer and 100 nM of duel-labelled probe. Reactions were run on 384-well plates and cycled using generic system conditions (48°C for 30 min, 95°C for 10 min and 40 cycles of 60°C for 1 min plus 95°C for 15 s) within the 7900 Sequence Detection System (Applied Biosystems, Branchburg, New Jersey, USA) using real-time data collection. Positive and negative controls were included in all real-time PCR assays. It should be noted that while the Chelex method has been used successfully for extraction of RNA viruses [73], it is of limited effectiveness [45], [46], and so our protocol may underestimate the true levels of viruses present.
Table 1. Real-time Taqman PCR primers and probes used to detect acute bee paralysis virus (ABPV), black queen cell virus (BQCV), chronic bee paralysis virus (CBPV), deformed wing virus (DWV), Israeli acute paralysis virus (IAPV) and sacbrood virus (SBV).
| Virus | Forward primer | Reverse primer | Probe |
| ABPV | ABPV 5436F:TAA CCA ATG AAG TRT CCA TAG GAA CTA | ABPV 5481R:TCT CCT GCR ATA ACC TTG GGT | ABPV 5515TMGB:TGT TTA TTC CCA AGA TTG |
| BQCV 1 , 3 | BQCV 9195F:GGT GCG GGA GAT GAT ATG GA | BQCV 8265R:GCC GTC TGA GAT GCA TGA ATA C | BQCV 8217T:TTT CCA TCT TTA TCG GTA CGC CGC C |
| CBPV 2 , 3 | CBPVF:CGC AAG TAC GCC TTG ATA AAG AAC | CBPVR:ACT ACT AGA AAC TCG TCG CTT CG | CBPVT:TCA AGA ACG AGA CCA CCG CCA AGT TC |
| DWV 1 , 3 | DWV 9587F:CCT GGA CAA GGT CTC GGT AGA A | DWV 9711R:ATT CAG GAC CCC ACC CAA AT | DWV 9627T:CAT GCT CGA GGA TTG GGT CGT CGT |
| IAPV 2 , 3 | IAPV B4S0427_R130M:RCR TCA GTC GTC TTC CAG GT | IAPV B4S0427_L17M:CGA ACT TGG TGA CTT GAR GG | IAPVT:TTG CGG CAA TCC AGC CGT GAA AC |
| SBV 2 , 3 | SBV 311F:AAG TTG GAG GCG CGY AAT TG | SBV 380R:CAA ATG TCT TCT TAC DAG AGG YAA GGA TTG | SBV 331TMGB:CGG AGT GGA AAG AT |
Statistical analysis
We analysed our data using Generalized Linear Models for binomially distributed data with a logit link function. For each parasite, we compared the numbers of samples that were positive and negative between the five pollinator types (honey bees, bumblebees, other bees, wasps and hoverflies). In addition, we compared the occurrence of parasites between host species within each host type, to determine whether certain species were more likely to have the parasite than others.
Ethics
No specific permits were required for the described field studies. Permission to collect samples on their private land was provided by Tom Cameron, Rebecca Neal, Alex Bateman, Kenneth McDowall, Jane Device, Iain Manfield, Will Patterson, Thomas Edwards, Paul Drake, Dick Hobson, Stan Burgess, Andy Ford, Lars Jeuken, Mark Harris, Hayley Lynch, Jenny Dunn, John Illingworth, Sam Mason, Paul Millner, Neal Haddaway, Alison Dunn, Terry McAndrew, Peter Henderson, Dave Adams, Carol Davison, Ben Chapman, Brenda Frater, Steffi Jourdan, Tim Johnson, Nicky Spencer Jones, Lesley Hooper, Anne Proud, Emma Black, Fiona Moulton, Teegan Docherty, Roberta Pagliarini, Pat Shore, Liz Paget, Richard Rodway and James Rosindell. All other samples were collected on public land that was not protected in any way and for which no specific permissions were required. The field studies did not involve endangered or protected species.
Supporting Information
Details of the samples collected and parasites found, including map point (referring to Figure 1), location and date of sampling, numbers of individuals of each host species collected at each site, and numbers of each of these species that were found by molecular screening to be positive for each of the parasites.
(DOCX)
Acknowledgments
We are grateful to Tanya Carey and Mark Goddard for assistance with collecting the samples, and Crystal Frost for technical assistance. We thank the following for permission to collect pollinator samples on their private land: Tom Cameron, Rebecca Neal, Alex Bateman, Kenneth McDowall, Jane Device, Iain Manfield, Will Patterson, Thomas Edwards, Paul Drake, Dick Hobson, Stan Burgess, Andy Ford, Lars Jeuken, Mark Harris, Hayley Lynch, Jenny Dunn, John Illingworth, Sam Mason, Paul Millner, Neal Haddaway, Alison Dunn, Terry McAndrew, Peter Henderson, Dave Adams, Carol Davison, Ben Chapman, Brenda Frater, Steffi Jourdan, Tim Johnson, Nicky Spencer Jones, Lesley Hooper, Anne Proud, Emma Black, Fiona Moulton, Teegan Docherty, Roberta Pagliarini, Pat Shore, Liz Paget, Richard Rodway and James Rosindell.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding for this study was provided by the UKPopNet (awarded to WOHH, JCB and JES), the Biotechnology and Biological Sciences Research Council (awarded to KR), the Leverhulme Foundation (awarded to WOHH) and the Waterloo Foundation (awarded to GB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Klein A-M, Vaissière BE, Cane JH, Steffan-Dewenter I, Cunningham SA, et al. Importance of pollinators in changing landscapes for world crops. Proc R Soc Lond B. 2007;274:303–313. doi: 10.1098/rspb.2006.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallai N, Salles J-M, Settele J, Vaissière BE. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol Econ. 2009;68:810–821. [Google Scholar]
- 3.Fontaine C, Dajoz I, Meriguet J, Loreau M. Functional diversity of plant-pollinator interaction webs enhances the persistence of plant communities. PLoS Biol. 2005;4:e1. doi: 10.1371/journal.pbio.0040001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kearns CA, Inouye DW. Pollinators, flowering plants, and conservation biology. Bioscience. 1997;47:297–307. [Google Scholar]
- 5.Knight TM, Steets JA, Vamosi JC, Mazer SJ, Burd M, et al. Pollen limitation of plant reproduction: Pattern and process. Ann Rev Ecol Syst. 2005;36:467–497. [Google Scholar]
- 6.Ollerton J, Winfree R, Tarrant S. How many flowering plants are pollinated by animals? Oikos. 2011;120:321–326. [Google Scholar]
- 7.Aizen MA, Harder LD. The global stock of domesticated honey bees is growing slower than agricultural demand for pollination. Curr Biol. 2009;19:915–918. doi: 10.1016/j.cub.2009.03.071. [DOI] [PubMed] [Google Scholar]
- 8.Williams PH, Osborne JL. Bumblebee vulnerability and conservation world-wide. Apidol. 2009;40:367–387. [Google Scholar]
- 9.Biesmeijer JC, Roberts SPM, Reemer M, Ohlemuller R, Edwards M, et al. Parallel declines in pollinators and insect-pollinated plants in Britain and the Netherlands. Science. 2006;313:351–354. doi: 10.1126/science.1127863. [DOI] [PubMed] [Google Scholar]
- 10.Goulson D, Lye GC, Darvill B. Decline and conservation of bumble bees. Annu Rev Entomol. 2008;53:191–208. doi: 10.1146/annurev.ento.53.103106.093454. [DOI] [PubMed] [Google Scholar]
- 11.Hackett K, Meyer R, Purcell-Miramontes M, Rose R, Holy D, et al. Colony Collapse Disorder Progress Report. USDA; 2009. [Google Scholar]
- 12.Colla S, Packer L. Evidence for decline in eastern North American bumblebees (Hymenoptera: Apidae), with special focus on Bombus affinis Cresson. Biodiv Conserv. 2008;17:1379–1391. [Google Scholar]
- 13.Cameron SA, Lozier JD, Strange JP, Koch JB, Cordes N, et al. Patterns of widespread decline in North American bumble bees. Proc Natl Acad Sci USA. 2011;108:662–667. doi: 10.1073/pnas.1014743108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratnieks FLW, Carreck NL. Clarity on honey bee collapse? Science. 2009;327:152–153. doi: 10.1126/science.1185563. [DOI] [PubMed] [Google Scholar]
- 15.Brown MJF, Paxton RJ. The conservation of bees: a global perspective. Apidol. 2009;40:410–416. [Google Scholar]
- 16.Potts SG, Biesmeijer JC, Kremen C, Neumann P, Schweiger O, et al. Global pollinator declines: trends, impacts and drivers. TREE. 2010;25:345–353. doi: 10.1016/j.tree.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Berenos C, Schmid-Hempel P, Wegner KM. Evolution of host resistance and trade-offs between virulence and transmission potential in an obligately killing parasite. J Evol Biol. 2009;22:2049–2056. doi: 10.1111/j.1420-9101.2009.01821.x. [DOI] [PubMed] [Google Scholar]
- 18.Aronstein KA, Murray KD. Chalkbrood disease in honey bees. J Invert Pathol. 2010;130:S20–S29. doi: 10.1016/j.jip.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 19.Hughes WOH, Petersen K, Ugelvig L, Pedersen D, Thomsen L, et al. Density-dependence and within-host competition in a semelparous parasite of leaf-cutting ants. BMC Evol Biol. 2004;4:45. doi: 10.1186/1471-2148-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker TN, Hughes WOH. Arboreality and the evolution of disease resistance in ants. Ecol Entomol. 2011;36:588–595. [Google Scholar]
- 21.Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia? - a statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–220. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werren JH. Biology of Wolbachia. Annu Rev Entomol. 1997;42:587–609. doi: 10.1146/annurev.ento.42.1.587. [DOI] [PubMed] [Google Scholar]
- 23.Johnson RM, Evans JD, Robinson GE, Berenbaum MR. Changes in transcript abundance relating to colony collapse disorder in honey bees (Apis mellifera). Proc Natl Acad Sci USA. 2009;106:14790–14795. doi: 10.1073/pnas.0906970106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, et al. A metagenomic survey of microbes in honey bee Colony Collapse Disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 25.Higes M, Martin-Hernandez R, Martinez-Salvador A, Garrido-Bailon E, Gonzalez-Porto AV, et al. A preliminary study of the epidemiological factors related to honey bee colony loss in Spain. Env Microbiol Rep. 2009;2:243–250. doi: 10.1111/j.1758-2229.2009.00099.x. [DOI] [PubMed] [Google Scholar]
- 26.vanEngelsdorp D, Evans JD, Saegerman C, Mullin C, Haubruge E, et al. Colony Collapse Disorder: a descriptive study. PLoS ONE. 2009;4:e6481. doi: 10.1371/journal.pone.0006481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morse RA, Flottum K. Honey bee pests, predators and diseases. Medina, Ohio: A.I. Root Company; 1997. [Google Scholar]
- 28.Paxton RJ, Klee J, S. K, Fries I. Nosema ceranae has infected Apis mellifera in Europe since at least 1998 and may be more virulent than Nosema apis. Apidol. 2008;38:558–565. [Google Scholar]
- 29.Higes M, Martín-Hernández R, C. B, Bailón EG, González-Porto AV, et al. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ Microbiol. 2008;10:2659–2669. doi: 10.1111/j.1462-2920.2008.01687.x. [DOI] [PubMed] [Google Scholar]
- 30.Higes M, Garcia-Palencia P, Martin-Hernandez R, Meana A. Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia). J Invert Pathol. 2007;94:211–217. doi: 10.1016/j.jip.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Plischuk S, Martín-Hernández R, Prieto P, Lucía M, Botías C, et al. South American native bumblebees (Hymenoptera: Apidae) infected by Nosema ceranae (Microsporidia), an emerging pathogen of honeybees (Apis mellifera). Env Microbiol Rep. 2009;1:131–135. doi: 10.1111/j.1758-2229.2009.00018.x. [DOI] [PubMed] [Google Scholar]
- 32.Otti O, Schmid-Hempel P. Nosema bombi: a pollinator parasite with detrimental fitness effects. J Invert Pathol. 2007;96:118–124. doi: 10.1016/j.jip.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Schmid-Hempel P. On the evolutionary ecology of host-parasite interactions: addressing the question with regard to bumblebees and their parasites. Naturwiss. 2001;88:147–158. doi: 10.1007/s001140100222. [DOI] [PubMed] [Google Scholar]
- 34.Meeus I, Brown MJF, De Graaf DC, Smagghe GUY. Effects of invasive parasites on bumble bee declines. Conserv Biol. 2011;25:662–671. doi: 10.1111/j.1523-1739.2011.01707.x. [DOI] [PubMed] [Google Scholar]
- 35.Anderson DL, Gibbs AJ, Gibson NL. Identification and phylogeny of spore-cyst fungi (Ascosphaera spp.) using ribosomal DNA sequences. Mycol Res. 1998;102:541–547. [Google Scholar]
- 36.Vandenberg JD. Chalkbrood susceptibility among larvae of the alfalfa leafcutting bee, Megachile rotundata, from different source populations. J Invert Pathol. 1992;60:213–214. [Google Scholar]
- 37.Fries I, Paxton RJ, Tengo J, Slemenda SB, da Silva AJ, et al. Morphological and molecular characterization of Antonospora scoticae n. gen., n. sp (Protozoa, Microsporidia) a parasite of the communal bee, Andrena scotica Perkins, 1916 (Hymenoptera, Andrenidae). Eur J Protist. 1999;35:183–193. [Google Scholar]
- 38.Paxton RJ, Fries I, Pieniazek NJ, Tengo J. High incidence of infection of an undescribed microsporidium (Microspora) in the communal bee Andrena scotica (Hymenoptera, Andrenidae). Apidol. 1997;28:129–141. [Google Scholar]
- 39.Genersch E, Yue C, Fries I, de Miranda JR. Detection of Deformed wing virus, a honey bee viral pathogen, in bumble bees (Bombus terrestris and Bombus pascuorum) with wing deformities. J Invert Pathol. 2006;91:61–63. doi: 10.1016/j.jip.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Bailey L, Gibbs AJ. Acute infection of bees with paralysis virus. J Ins Pathol. 1964;6:395–407. [Google Scholar]
- 41.Singh R, Levitt AL, Rajotte EG, Holmes EC, Ostiguy N, et al. RNA viruses in hymenopteran pollinators: evidence of inter-taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS ONE. 2010;5:e14357. doi: 10.1371/journal.pone.0014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goulson D. Bumblebees: behaviour and ecology. Oxford: Oxford Univ. Press; 2003. [Google Scholar]
- 43.Breeze TD, Bailey AP, Balcombe KG, Potts SG. Pollination services in the UK: How important are honeybees? Agriculture, Ecosystems & Environment. 2011;142:137–143. [Google Scholar]
- 44.Jacobs JH, Clark SJ, Denholm I, Goulson D, Stoate C, et al. Pollinator effectiveness and fruit set in common ivy, Hedera helix (Araliaceae). Arthropod-Plant Interact. 2010;4:19–28. [Google Scholar]
- 45.Hale AD, Green J, Brown DWG. Comparison of four RNA extraction methods for the detection of small round structured viruses in faecal specimens. J Virol Methods. 1996;57:195–201. doi: 10.1016/0166-0934(95)01966-9. [DOI] [PubMed] [Google Scholar]
- 46.Arnal C, Ferré-Aubineau V, Besse B, Mignotte B, Schwartzbrod L, et al. Comparison of seven RNA extraction methods on stool and shellfish samples prior to hepatitis A virus amplification. J Virol Methods. 1999;77:17–26. doi: 10.1016/s0166-0934(98)00083-4. [DOI] [PubMed] [Google Scholar]
- 47.Russell JA, Goldman-Huertas B, Moreau CS, Baldo L, Stahlhut JK, et al. Specialization and geographic isolation among Wolbachia symbionts from ants and lycaenid butterflies. Evolution. 2009;63:624–640. doi: 10.1111/j.1558-5646.2008.00579.x. [DOI] [PubMed] [Google Scholar]
- 48.Frost CL, Fernández-Marín H, Smith JE, Hughes WOH. Multiple gains and losses of Wolbachia symbionts across a tribe of fungus-growing ants. Mol Ecol. 2010;19:4077–4085. doi: 10.1111/j.1365-294X.2010.04764.x. [DOI] [PubMed] [Google Scholar]
- 49.Wenseleers T, Ito F, Borm SV, Huybrechts R, Volckaert F, et al. Widespread occurrence of the microorganism Wolbachia in ants. Proc R Soc Lond B. 1998;265:1447–1452. doi: 10.1098/rspb.1998.0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeyaprakash A, Hoy MA, Allsopp MH. Multiple Wolbachia strains in Apis mellifera capensis from South Africa. Apidol. 2009;40:178–183. [Google Scholar]
- 51.Hoy MA, Jeyaprakash A, Alvarez JM, Allsopp MH. Wolbachia is present in Apis mellifera capensis, A. m. scutellata, and their hybrid in Southern Africa. Apidol. 2003;34:53–60. [Google Scholar]
- 52.Jeyaprakash A, Hoy MA, Allsopp MH. Bacterial diversity in worker adults of Apis mellifera capensis and Apis mellifera scutellata (Insecta: Hymenoptera) assessed using 16S rRNA sequences. J Invert Pathol. 2003;84:96–103. doi: 10.1016/j.jip.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 53.Jeong G, Lee K, Choi J, Hwang S, Park B, et al. Incidence of Wolbachia and Cardinium endosymbionts in the Osmia community in Korea. J Microbiol. 2009;47:28–32. doi: 10.1007/s12275-009-0198-3. [DOI] [PubMed] [Google Scholar]
- 54.Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, et al. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol. 2011;20:619–628. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- 55.Hedges LM, Brownlie JC, O'Neill SL, Johnson KN. Wolbachia and virus protection in insects. Science. 2008;322:702-. doi: 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]
- 56.Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6:e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Min K-T, Benzer S. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc Natl Acad Sci USA. 1997;94:10792–10796. doi: 10.1073/pnas.94.20.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fytrou A, Schofield PG, Kraaijeveld AR, Hubbard SF. Wolbachia infection suppresses both host defence and parasitoid counter-defence. Proc R Soc Lond B. 2006;273:791–796. doi: 10.1098/rspb.2005.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Snook RR, Cleland SY, Wolfner MF, Karr TL. Offsetting effects of Wolbachia infection and heat shock on sperm production in Drosophila simulans: analyses of fecundity, fertility and accessory gland proteins. Genetics. 2000;155:167–178. doi: 10.1093/genetics/155.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McManus WR, Youssef NN. Life-cycle of the chalkbrood fungus, Ascosphaera aggregata, in the alfalfa leafcutting bee, Megachile rotundata, and its associated symptomatology. Mycologia. 1984;76:830–842. [Google Scholar]
- 61.Torchio PF. Effects of spore dosage and temperature on pathogenic expressions of chalkbrood syndrome caused by Ascosphaera torchiori within larvae of Osmia lignaria propinqua (Hymenoptera, Megachilidae). Environ Entomol. 1992;21:1086–1091. [Google Scholar]
- 62.Youssef NN, McManus WR. Ascosphaera torchioi sp nov., a pathogen of Osmia lignaria propinqua Cresson (Hymenoptera). Mycotaxon. 2001;77:7–13. [Google Scholar]
- 63.Demendoza MH, Demendoza JH, Peuerta F, Asensio E, Bustos M, et al. Ascosphaeriosis of the parasitic bee, Coelioxys rufocaudata, by Ascosphaera aggregata. J Apic Res. 1989;28:61–65. [Google Scholar]
- 64.Cross EA, Bohart GE. The biology of Imaripes apicola (Acari, Scutacaridae) and its relationships to the alkali bee, Nomia melanderi (Hymenoptera, Halictidae), and to certain fungi in the bee cell ecosystem. J Kansas Entomol Soc. 1992;65:157–173. [Google Scholar]
- 65.Schmid-Hempel P. Parasites in social insects. Princeton: Princeton Univ. Press; 1998. [Google Scholar]
- 66.Schmid-Hempel P, Loosli R. A contribution to the knowledge of Nosema infections in bumble bees, Bombus spp. Apidol. 1998;29:525–535. [Google Scholar]
- 67.Durrer S, Schmid-Hempel P. Parasites and the regional distribution of bumblebee species. Ecography. 1995;18:114–122. [Google Scholar]
- 68.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 69.Prendini L, Weygoldt P, Wheeler WC. Systematics of the Damon variegatus group of African whip spiders (Chelicerata: Amblypygi): evidence from behaviour, morphology and DNA. Org Div Evol. 2005;5:203–236. [Google Scholar]
- 70.Terry RS, EJ, Sharpe RG, Rigaud T, Littlewood DTJ, et al. Widespread vertical transmission and associated host sex-ratio distortion within the eukaryotic phylum Microspora. Proc R Soc Lond B. 2004;271:1783–1789. doi: 10.1098/rspb.2004.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.James RR, Skinner JS. PCR diagnostic methods for Ascosphaera infections in bees. J Invert Pathol. 2005;90:98–103. doi: 10.1016/j.jip.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 72.Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Env Microb. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rotenberg D, Krishna Kumar NK, Ullman DE, Montero-Astua M, Willis DK, et al. Variation in tomato spotted wilt virus titer in Frankliniella occidentalis and its association with frequency of transmission. Phytopathol. 2009;99:404–410. doi: 10.1094/PHYTO-99-4-0404. [DOI] [PubMed] [Google Scholar]
- 74.Chantawannakul P, Ward L, Boonham N, Brown M. A scientific note on the detection of honeybee viruses using real-time PCR (TaqMan) in Varroa mites collected from a Thai honeybee (Apis mellifera) apiary. J Invert Pathol. 2006;91:69–73. doi: 10.1016/j.jip.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 75.Blanchard P, Ribiere M, Celle O, Lallemand P, Schurr F, et al. Evaluation of a real-time two-step RT-PCR assay for quantitation of Chronic bee paralysis virus (CBPV) genome in experimentally-infected bee tissues and in life stages of a symptomatic colony. J Virol Methods. 2007;141:7–13. doi: 10.1016/j.jviromet.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 76.Kajobe R, Marris G, Budge G, Laurenson L, Cordoni G, et al. First molecular detection of a viral pathogen in Ugandan honey bees. J Invert Pathol. 2010;104:153–156. doi: 10.1016/j.jip.2010.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of the samples collected and parasites found, including map point (referring to Figure 1), location and date of sampling, numbers of individuals of each host species collected at each site, and numbers of each of these species that were found by molecular screening to be positive for each of the parasites.
(DOCX)