Abstract
The microphthalmia-associated transcription factor (Mitf) regulates gene expression required for osteoclast differentiation. Genes regulated by Mitf have been previously identified. However, proteins that interact and regulate Mitf’s activity in osteoclasts are not well known. Here, we report that POH1, a subunit of the 19S proteasome lid is a regulator of Mitf. We show that POH1 and Mitf interact in osteoclasts and that this interaction is dependent on RANKL signaling. Overexpression of POH1 increased Mitf’s activation of 5XGal4-TK and Acp5 promoters. The amino terminus of POH1 mediates the binding to Mitf and is sufficient to increase Mitf’s transcriptional activity. Finally, we show that mutations in the JAMM motif of POH1 reduced Mitf activation of promoters. In summary, our results identify a novel mechanism of Mitf regulation in osteoclasts by POH1.
Keywords: POH1, Mitf, ubiquitination, osteoclast
Osteoclasts, derived from the monocyte lineage mediate the removal of old bone and facilitate maintenance of systemic mineral homeostasis. Osteoclast differentiation requires stringent control of gene activation and suppression in response to physiological cues. The fidelity of skeletal gene expression requires an integrating spectrum of regulatory signals that govern osteoclast differentiation. To regulate expression of osteoclast genes, it is necessary to characterize the promoter regulatory elements as well as protein/DNA and protein/protein interactions to determine the extent to which genes are transcribed. Mouse models have indicated a critical role for the microphthalmia-associated transcription factor, Mitf, in regulating genes necessary for osteoclast differentiation [Luchin et al., 2001; Sharma et al., 2007]. Mitf is a basic helix-loop-helix leucine zipper transcription factor that is expressed in many cell types including melanocytes, mast cells, and osteoclasts [Hemesath et al., 1994]. Furthermore it has been demonstrated that Mitf acts synergistically with PU.1 and NFATc1 to activate Acp5, cathepsin K (Ctsk) and OSCAR promoters during osteoclast differentiation [Luchin et al., 2001; Matsumoto et al., 2004; Sharma et al., 2007].
Mitf has also been shown to be a target of both the M-CSF [Weilbaecher et al., 2001] and RANKL [Mansky et al., 2002] signaling pathways in osteoclasts. M-CSF signaling in osteoclasts induces Mitf phosphorylation on serine 73 by Erk2 with recruitment of the p300/CBP co-activator [Weilbaecher et al., 2001]. RANKL signaling leads to Mitf phosphorylation by p38 MAP kinase at amino acid residue 307, resulting in an increase in Mitf activity [Mansky et al., 2002]. Results from Sharma et al. [2007] indicate that the Mitf complex integrates signals necessary for the appropriate temporal regulation of osteoclast genes such as Ctsk and Acp5 during differentiation. M-CSF signaling alone can regulate nuclear localization and recruitment of Mitf to the target promoter [Bronisz et al., 2006]. However, the Mitf complex with just M-CSF signaling does not activate gene expression but it is the combination of M-CSF and RANKL, which increases expression of osteoclast differentiation genes [Sharma et al., 2007].
Relatively little is known about proteins that interact with and potentially regulate the ability of Mitf to activate osteoclast promoters during M-CSF and RANKL signaling. To identify proteins that interact with and regulate Mitf, we used a yeast two hybrid system designed to identify proteins that interact with transactivation domains of transcription factors such as Mitf [Hirst et al., 2001]. We identified POH1 (pad one homologue) and showed that POH1 can interact with the amino terminus of Mitf. POH1 is the human functional homologue of the fission yeast Pad1 and also called Rpn11 [Verma et al., 2002]. POH1 shares 68% identity with Pad1 and is more distantly related to the c-Jun binding protein JAB1 [Wei et al., 1998]. POH1 mRNA is expressed in a variety of tissues including osteoclasts [Verma et al., 2002]. POH1 functions as a deubiquitinating enzyme (DUB, [Verma et al., 2002]) and is part of the regulatory particle of the proteasome that binds polyubiquitinated proteins, unfolds them and feeds them into the lumen of the proteasome where they are degraded by hydrolysis [Yao and Cohen, 2002]. DUBs have also been shown to have an ubiquitin-editing activity, which can rescue particular substrates from being degraded by removing or trimming the degradation signal. A number of studies have implicated POH1 as a possible regulator of the AP-1 transcription complex [Nabhan et al., 2002; Nabhan and Ribeiro, 2006; Stitzel et al., 2001]. The deubiquitination activity of POH1 has been mapped to the JAB1/MPN/Mov34 metalloenzyme (JAMM) sequence motif also referred to as the MPN+ motif [Maytal-Kivity et al., 2002].
Ubiquitin-mediated degradation plays a role in many cellular processes including but not limited to cell cycle and division, differentiation and development, modulation of cell surface receptors and transcriptional regulation. With so many substrates, it is not a surprise to find the ubiquitin pathway underlying the pathogenesis of many diseases including cystic fibrosis, Angelman’s syndrome, Liddle syndrome and neurodegenerative conditions such as Alzheimer’s [Ciechanover, 2003; Schwartz and Ciechanover, 1999]. In multiple myeloma, proteasome inhibitors such as Velcade (PS-431) have been shown to inhibit growth, induce apoptosis and overcome drug resistance of the tumor cells [Hideshima et al., 2001] and are currently being used as a therapy during relapse of the disease. In addition, ubiquitin and the 26S proteasome complex have been linked to Paget’s disease of the bone by modulating the activity of the RANK signaling pathway [Layfield and Shaw, 2007].
In this paper, we demonstrate that POH1, a component of the 26S proteasome, and Mitf can interact in osteoclasts and that this interaction is dependent on RANKL signaling. Mitf becomes ubiquitinated following M-CSF stimulation of RAW 264.7 c4 cells, and POH1 can prevent Mitf ubiquitination. We also show that POH1 interaction with Mitf increases Mitf’s ability to activate transcription. Further, we present data that shows that the amino terminus of both Mitf and POH1 are sufficient for their interaction and that the POH1 amino terminus mediates increased Mitf activity. Finally, we demonstrate that the JAMM motif is necessary for POH1’s increase in Mitf’s activity in RAW 264.7 cells. Characterizing the interactions between Mitf and POH1 will allow us to better understand the mechanisms that regulate Mitf activity during osteoclast differentiation.
Materials and Methods
Yeast two-hybrid
Yeast two-hybrid assay was done as previously described [Hirst et al., 2001]. Briefly, yeast strain MAV108 containing plasmid pY1-MITF, which encodes the Gal4 DNA binding domain fused to amino acids 109-185 of Mitf, was transformed with the pBD Jurkat cDNA library and allowed to form colonies on YM plates lacking leucine and tryptophan. Transformants were scraped, resuspended in 50% glycerol, snap frozen in liquid nitrogen and stored at −70°C. Aliquots were thawed and plated on 5-FOA plates (lacking leucine and tryptophan). Colonies were picked and expanded in YM lacking leucine and tryptophan. Cells were harvested by centrifugation, resuspended in 100 µL YPER (Pierce) and incubated at room temperature for 10 minutes. DNA was prepared from the lysates by sequential addition of solution II and III as with standard alkaline lysis miniprep protocol. cDNA inserts were amplified by PCR using vector specific primers OKL1 (5’ TTGCCTGTGGTGTCCTCAAAC) and OKL2 (5’ TGACCAACCTCTGGCGAAG) and purified by QIA quick columns and sequenced by OKL1.
Cell Culture, Luciferase Assays and Transfections
293T and NIH 3T3 were maintained in Dulbecco’s modified Eagle medium supplemented with 10% bovine calf serum, 2% l-glutamine and 0.5% penicillin/streptomycin (Invitrogen). 293T and NIH 3T3 cells were transiently transfected by Lipofectamine Plus Reagent (Invitrogen) according to the instructions of the manufacturer. The luciferase activities were measured using 5X Cell Lysis Buffer (Promega) according to the instruction of the manufacturer. RAW 264.7 c4 cells were transfected with Fugene 6 (Roche) according to the manufacturer’s instructions. For differentiation assays, RAW 264.7 c4 cells were spilt and treated with RANKL for 24 hours before transfection with Fugene 6. After transfection, RAW 264.7 c4 cells were treated with RANKL and M-CSF for 7 days to induce differentiation. The presence of multinuclear cells was analyzed microscopically. Experiments were repeated twice in triplicate. Osteoclasts were isolated from bone marrow of mice as previously described [Sharma et al., 2007]. Briefly bone marrow was flushed from femurs. Resulting cells were cultured for 3 days in the presence of 50 ng/mL of M-CSF on non-tissue culture dishes. The adherent cell population, containing the osteoclasts, was cultured for the indicated time and amounts of M-CSF and RANKL.
Antibodies and Chemicals
POH1 antibody was purchased from Zymed. The FLAG (M2) antibody coupled to the protein G beads was purchased from Sigma. The anti c-myc monoclonal antibody (9E10) was purchased from Cell Signaling Technologies. Polyclonal Mitf antibody was generated by 21st century Biochemicals (Marlboro, Mass) using a peptide containing mouse Mitf amino acids 85-96. M-CSF and RANKL were purchased from R & D Systems (Minneapolis, MN) and used at 10 ng/mL (M-CSF) for bone marrow derived osteoclasts and 1 ng/mL for RAW 264.7c4 cells or 60 ng/mL (RANKL). MG132 was purchased from Sigma and used at 25 µM.
Immunoprecipitation and Western Blotting
At 24 hours after transfection, the cells were harvested in NP-40 lysis buffer (20 mM Tris, pH 8.0, 137 mM NaCl, 10% glycerol, 1% NP-40 and protease and phosphatase inhibitors). Extracts were incubated with target antibody and Protein A/G beads (Pierce) overnight at 4°C. Next day immunoprecipitates were precipitated and washed three times in lysis buffer. The bound proteins were resuspended in sample buffer and resolved by SDS-PAGE. The resolved proteins were transferred to nitrocellulose membrane (BioRad), blocked and blotted in primary antibody overnight at 4°C. The next day the blot was incubated with horseradish peroxidase anti-rabbit or anti-mouse (GE Healthcare) for one hour at room temperature. Antibody binding was detected using ECL system (GE Healthcare). For the in situ ubiquitination assay, RAW 264.7 c4 cells were stimulated with M-CSF or M-CSF and RANKL for 48 hours. Cells were lysed and immunoprecipitated with Mitf antibody. Immunoprecipitates were resolved by SDS-PAGE and blotted with an anti-ubiquitin antibody (P4D1, Cell Signaling).
Plasmids and Mutagenesis
FLAG Mitf was previously described [Bronisz et al., 2006]. POH1 was kindly provided to us by Dr. Chris Norbury and previously described [Spataro et al., 1997]. The POH1 N-terminus (amino acids 1-180) and C-terminus (amino acids 181-311) were subcloned into CMV3a 3X-myc (Stratagene). POH1 single or double point mutations were generated by QuikChange method (Stratagene). HA-Ubiquitin plasmid was provided by Dr. Kylie Walters. All constructs were verified by DNA sequencing.
RNA Isolation and Real Time PCR
RNA was extracted using TRIzol (Invitrogen). 2 µg of purified total RNA was reverse transcribed by iScript cDNA Synthesis kit (Bio-Rad). Quantitative real-time RT-PCR was performed using MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad) using 1 µL of the cDNA with 2X iQ SYBR Green supermix (Bio-Rad). Osteoclast genes were normalized to L4 RNA.
Results
Yeast two hybrid screen
To identify proteins that interact and regulate Mitf’s activation of osteoclast promoters, we used a novel yeast two hybrid system developed to identify proteins that interact with activation domains of transcription factors [Hirst et al., 2001]. In this system, the Gal4 DNA binding domain is fused in frame to the activation domain of Mitf (amino acids 109-185). This fusion protein activates the GAL1-URA3 gene in yeast. In the presence of the drug 5-FOA, the GAL1-URA3 gene product converts 5-FOA into a toxic substance, which kills the yeast. The cDNA library is fused in frame with TUP1, a yeast general repressor protein. If the activation domain of the activator binds to a protein expressed by the library, then TUP1 prevents the activator (Mitf) from activating expression of the GAL1-URA3 gene. This process in turn stops the conversion of 5-FOA into a toxic product, and allows the yeast to survive and grow [Hirst et al., 2001].
From our two-hybrid screen, we obtained 70 independent cDNA clones. To determine the identity of the cDNAs, we isolated DNA from the yeast and amplified by PCR. The resulting PCR product was gel extracted, sequenced and screened against the NCBI database to identify the sequence. Six out of the seventy clones were identified as POH1, and three out of seventy clones were identified as MARK3 or C-TAK1. Recently, C-TAK1 was shown to interact with and regulate the subcellular localization of Mitf [Bronisz et al., 2006].
POH1 is a Mitf binding partner in osteoclasts
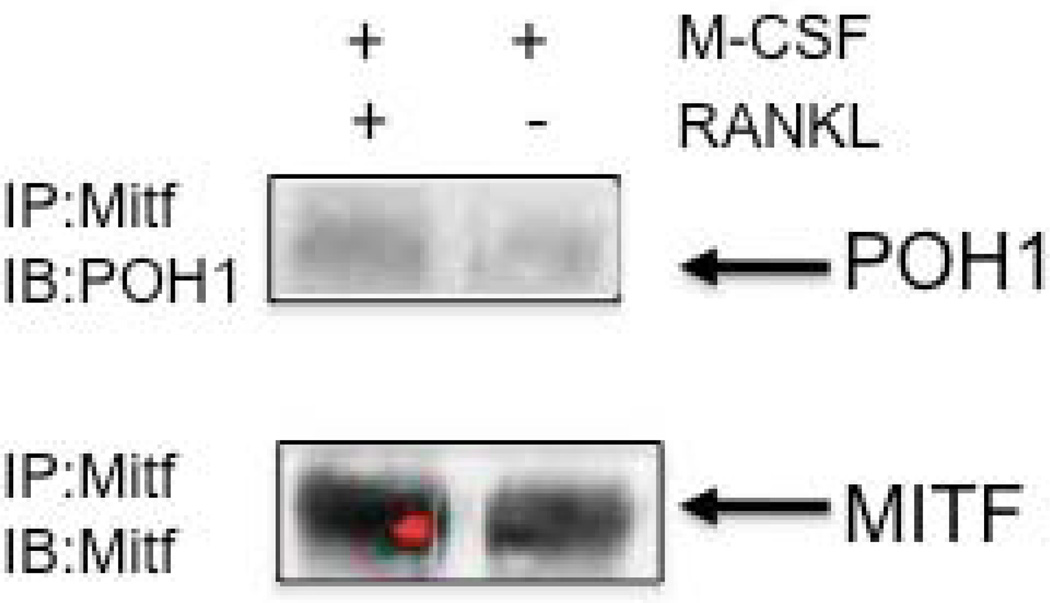
POH1 was identified as interacting with the amino terminus (amino acids 109-185) of Mitf, which contains the activation domain. POH1 is a component of the 19S lid complex of the proteasome, which may play a role in the editing of polyubiquitinated substrates to control degradation [Verma et al., 2002; Voges et al., 1999; Yao and Cohen, 2002]. POH1 was recently described as a cellular DUB. DUBs can provide ubiquitin-editing activity, which can rescue particular substrates from being degraded by removing or trimming the ubiquitinated amino acid residues [Guterman and Glickman, 2004; Soboleva and Baker, 2004]. To verify the interaction between Mitf and POH1, osteoclasts were stimulated with M-CSF or M-CSF and RANKL for 5 days. Osteoclast extracts were immunoprecipitated with an antibody that recognizes Mitf and subsequently analyzed by Western blotting for the presence of POH1. We detected an interaction between POH1 and Mitf when osteoclasts were stimulated with M-CSF and RANKL (see Figure 1). This interaction between Mitf and POH1 in osteoclasts suggests that POH1 could prevent Mitf ubiquitination during RANKL signaling in osteoclasts.
Figure 1. Mitf interacts with POH1 in osteoclasts.
Representative Western blot of Mitf immunoprecipitates from bone marrow derived osteoclasts stimulated with M-CSF (10 ng/mL) or M-CSF and RANKL (60 ng/mL) for 5 days and immunoblotted against POH1 (Zymed) and Mitf.
Mitf is ubiquitinated in osteoclasts
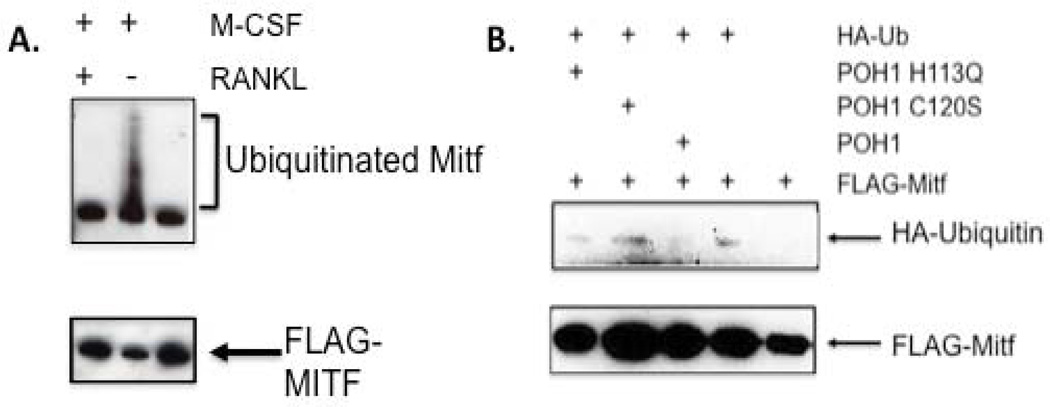
In melanocytes, Mitf is ubiquitinated in response to c-Kit stimulation leading to proteolysis [Wu et al., 2000]. To determine if Mitf is ubiquitinated in osteoclasts, Mitf was immunoprecipitated from RAW 264.7 c4 cells, a stable subclone of commercially available RAW 264.7 cells that require both M-CSF and RANKL for efficient differentiation in vitro [Bronisz et al., 2006]. Cells were left unstimulated, stimulated with either M-CSF or M-CSF + RANKL. The immunoprecipitations were then analyzed by Western blot for the presence of polyubiquitinated residues. We detected ubiquitinated Mitf when the cells were stimulated with M-CSF alone, but not after stimulation with M-CSF and RANKL (see Figure 2A), which indicates that RANKL signaling decreases Mitf ubiquitination.
Figure 2. POH1 reduces ubiquitination of Mitf.
(A) Representative Western blot of Mitf immunoprecipitates from RAW 264.7 c4 cells stimulated with M-CSF (10 ng/mL) or M-CSF (10 ng/mL) and RANKL (60 ng/mL) for 2 days and immunoblotted against ubiquitin (Cell Signaling). (B) Representative Western blot of 293T cells transiently transfected with FLAG-Mitf, HA-ubiquitin and wild type or mutant POH1. Lysates were collected 24 hours following transfection and analyzed by Western blotting with a HA-tag antibody. The lower panel was immunoblotted with a FLAG antibody.
POH1 reduces ubiquitination of Mitf
POH1 has been described as a DUB and it has been shown to reduce ubiquitination of the transcription factor, c-jun [Nabhan et al., 2002; Nabhan and Ribeiro, 2006]. We wanted to determine if POH1 could reduce ubiquitination of Mitf in an intact cellular environment by transiently transfecting 293T cells with FLAG-Mitf, wild type POH1 or C120S, H113Q mutant POH1 and HA-ubiquitin. Cell lysates were immunoprecipitated with an antibody that recognizes the FLAG-tag and analyzed by immunoblotting with a HA-tag antibody to detect ubiquitinated proteins or anti-FLAG antibody to monitor total amount of FLAG-Mitf recovered (see Figure 2B). We observed that wild type POH1 decreased ubiquitination of Mitf compared with the mock transfected cells (compare Mitf+HA-Ub and Mitf+HA-Ub+POH1). In contrast, cells transfected with mutant POH1 (C120S and H113Q) exhibited the same amount of ubiquitination as the control. The C120S and H113Q mutants are located in the JAMM motif, which has been shown to be responsible for the DUB activity of POH1 [Maytal-Kivity et al., 2002]. These data suggest that POH1 reduces ubiquitination of Mitf, which is expected to enhance Mitf protein stability.
Overexpression of POH1 and inhibition of proteolysis enhances Mitf activity
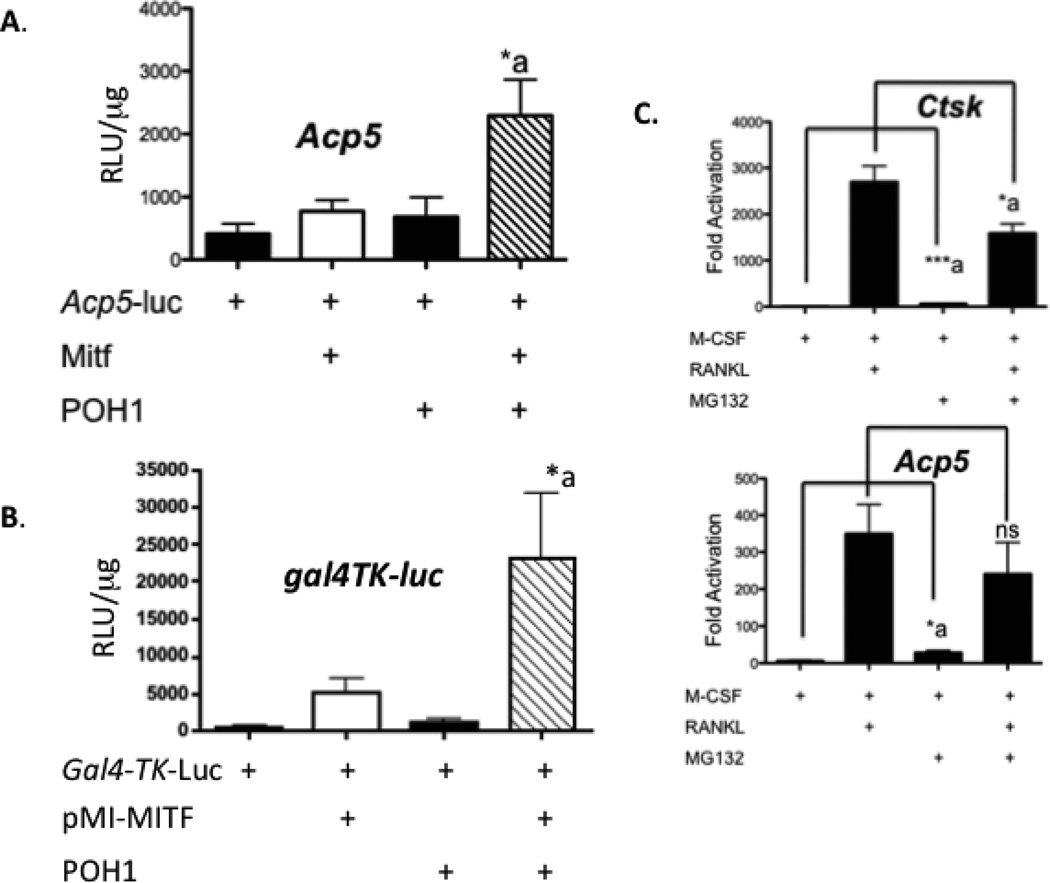
To determine if POH1 regulated Mitf’s activation of the Acp5 promoter, we co-transfected NIH 3T3 cells with an Acp5-luciferase reporter, full length Mitf and/or POH1. We detected an increase in activation of the Acp5-luciferase reporter of approximately 5 fold compared in cultures transfected with Mitf alone (Figure 3A, compare Mitf to Mitf+POH1). In our yeast two hybrid screen, we used the amino terminus (amino acids 109-185) of Mitf as the bait. To confirm that the amino terminus is sufficient for functional interactions with POH1, we cloned Mitf amino acid residues 1-185 into an expression plasmid fusing the Gal4 DNA binding domain to the amino terminus of Mitf (pMI-Mitf). Transfection of NIH 3T3 cells with pMI-Mitf, POH1 and a luciferase reporter containing 5X Gal4 DNA binding sites stimulated the reporter 22 fold over when pMI-Mitf was transfected alone (Figure 3B, compare pMI-Mitf to pMI-Mitf+POH1).
Figure 3. POH1 increases Mitf’s ability to activate transcription.
(A) NIH 3T3 cells were transiently transfected with TRAP luciferase reporter construct and/or Mitf and POH1. *p≤0.05 vs. Acp5-luc+Mitf. (B) NIH 3T3 cells were transiently transfected with 5X Gal4-TK-luciferase reporter construct and/or pMI-Mitf and full length POH1. pMI-MITF encodes amino terminus (amino acids 1-185) of Mitf fused in frame to the Gal4 DNA binding domain. Luciferase reporter activity is presented as relative luciferase units (RLU) and the results of three experiments each performed in duplicate are presented. *p≤0.05 vs.gal4-TK-luc+Mitf. (C) Real time RT-PCR analysis of osteoclasts treated with or without 25 µM MG132 6 hours before harvest for expression of Acp5 (encoding TRACP) or cathepsin K RNA. ***p≤0.0005 vs. M-CSF treatment and *p≤0.05 vs. RANKL treatment for Ctsk and *p≤0.05 vs. M-CSF treatment and NS, not significant vs. RANKL treatment for Acp5.
If Mitf activity was restricted by proteolytic degradation, we reasoned that inhibition of proteosomal activity would enhance activation of Mitf target genes including Acp5 (encodes TRACP protein) and Ctsk (cathepsin K). To test this, we stimulated osteoclasts with M-CSF or M-CSF and RANKL for 3 days. Cells were treated with MG132, an inhibitor of the proteosome complex and ubiquitination, 6 hours before harvesting the cells for RNA. Following isolation of RNA from the osteoclasts, mRNA of Acp5 (encodes for TRACP protein) and Ctsk (cathepsin K) was determined by real time RT-PCR. Mitf has been shown to regulate both the Acp5 and Ctsk promoters [Luchin et al., 2000; Motyckova et al., 2001]. We detected a significant increase in RNA from the Acp5 and the Ctsk promoter (5 fold, p <0.01 and 9 fold increase, p<0.0005) when we treated the osteoclasts with 25 µM MG132 and M-CSF (Figure 3C, compare M-CSF and M-CSF + MG132). These results suggest that inhibiting proteasome mediated degradation increased gene expression from osteoclast promoters targeted by Mitf when osteoclasts are stimulated with M-CSF. As expected when osteoclasts were treated with RANKL, we detected a significant increase in Ctsk (300 fold, p<0.0005) and Acp5 genes (50 fold, p<0.005). We detected a decrease in Ctsk (2 fold decrease, p<0.05) and Acp5 (1.5 fold decrease, ns) gene expression in cultures treated with RANKL and MG132 (see Figure 3C, compare RANKL and RANKL+MG132). Taken together, these observations indicate that POH1 promotes Mitf activity by protecting it from ubiquitin-mediated proteolysis.
Amino terminal domain of POH1 is sufficient to regulate Mitf
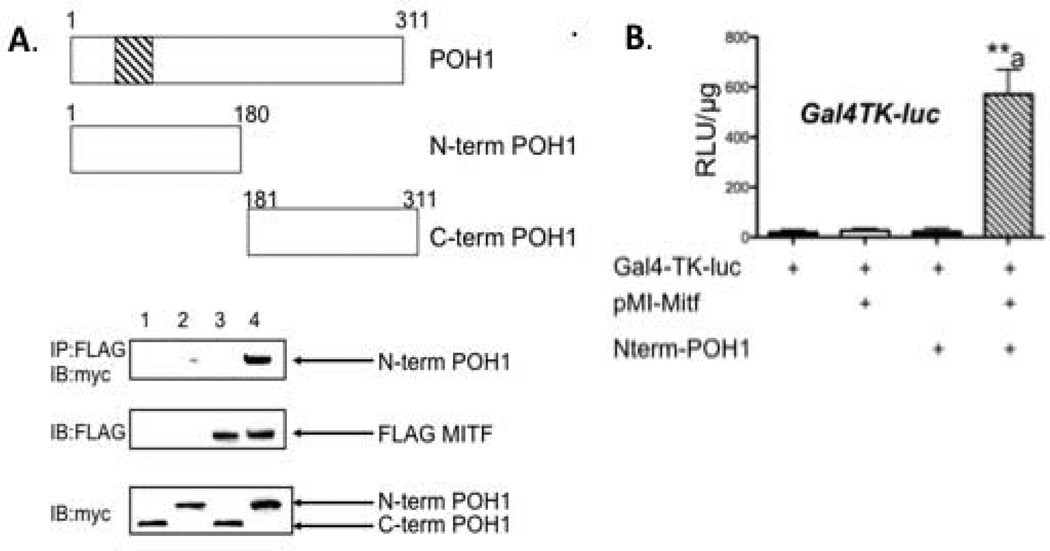
To map the domain of POH1 that interacts with Mitf, we over expressed myc-POH1 in 293T cells. We were unable to detect expression of full length myc-tagged POH1 in 293T cells so we subcloned the amino terminus into a myc-tagged expression vector (amino acid residues 1-180) and carboxy terminus (amino acid residues 181-311) of POH1. Cell lysates were immunoprecipitated with an antibody that recognizes the FLAG-tagged Mitf and analyzed by Western blot for the presence of myc-tagged POH1. We could only detect Mitf in the presence of the amino terminus region of POH1 (see Figure 4A, top panel). We also cotransfected NIH 3T3 cells with 5XGal4 DNA binding luciferase reporter, pMI-Mitf and myc-tagged amino terminus of POH1. We detected a 22 fold increase in Mitf’s activation of the Gal4 luciferase promoter when the cells were cotransfected with the amino terminus region of POH1 and Mitf compared to just Mitf alone (Figure 4B, compare pMI-Mitf and pMI-Mitf+N-term POH1).
Figure 4. Amino terminus of POH1 is sufficient to interact with Mitf.
(A) Myc-tagged POH1 constructs and FLAG-tagged Mitf were expressed in 293T cells and complexes from whole cell lysates were immunoprecipitated with FLAG antibody and analyzed by Western blot with the indicated antibodies. (B) NIH 3T3 cells were transiently transfected with 5X Gal4-TK-luciferase reporter construct and/or pMI-Mitf and amino terminus of POH1 (amino acids 1-180). Luciferase reporter activity is presented as relative luciferase units (RLU) and the results of three experiments each performed in duplicate are presented. **p≤0.005 vs. Gal4-TK-luc+pMI-Mitf.
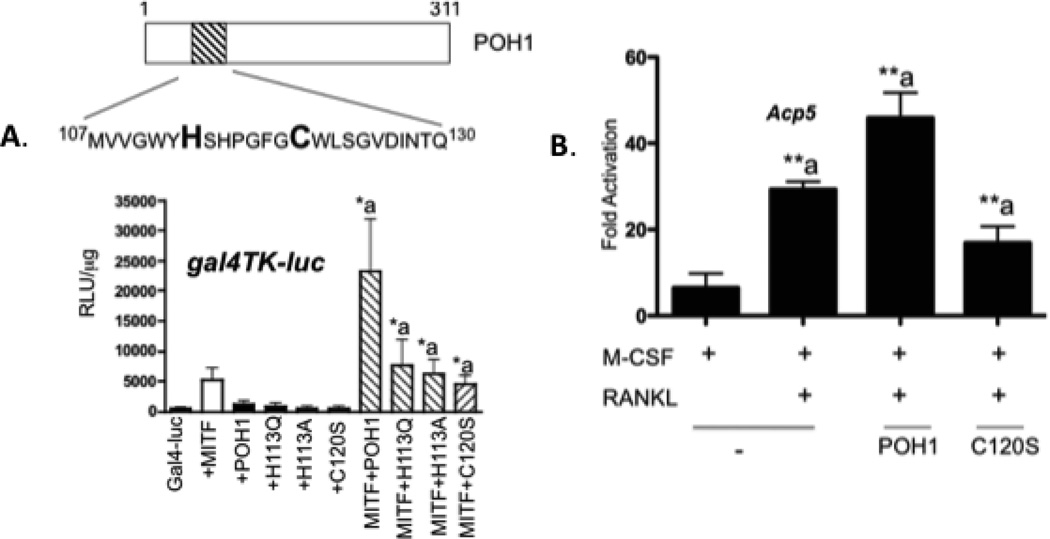
JAMM motif of POH1 is required to regulate Mitf activity
One of the most important functional motifs in POH1 is the MPN+ (Mpr1 pad1 N-terminal plus) or JAMM (JAB1/MPN/Mov34) motif [Maytal-Kivity et al., 2002]. This motif is primarily responsible for the DUB activity of POH1 [Maytal-Kivity et al., 2002]. The JAMM consensus sequence is EXnHS/THX7SXXD [Maytal-Kivity et al., 2002] and located in the amino terminus of POH1 between amino acid residues 107-130 (see Figure 5A). We co-transfected NIH 3T3 cells with wild type or mutant POH1 and pMI-Mitf to determine if the POH1 mutated in the JAMM motif is still able to increase Mitf’s activation of the 5XGal4 DNA binding luciferase reporter. As shown in figure 5A, wild type POH1 was able to increase Mitf’s activation of the promoter approximately 5 fold; however, none of the POH1 mutants were able to increase Mitf’s activation of the Gal4 DNA luciferase reporter compared to when Mitf was transfected alone (compare Mitf+POH1 to Mitf+H113Q, Mitf+H113A, Mitf+C120S).
Figure 5. MPN+ domain of POH1 is required for regulation of Mitf’s activity.
(A) NIH 3T3 cells were transiently transfected with 5X Gal4-TK-luciferase reporter construct and/or pMI-Mitf POH1 point mutants in the MPN+/JAMM motif. Luciferase reporter activity is presented as relative luciferase units (RLU) and the results of three experiments each performed in duplicate are presented. *p≤ 0.05 vs. Gal4-TK-luc+pMI-Mitf. (B) Real time RT-PCR from RAW 264.7 c4 cells that were either mock transfected, transiently transfected with full length POH1 or full length POH1 containing the point mutant C120S. Cells were stimulated with M-CSF (10 ng/mL) or M-CSF and RANKL (60 ng/mL) for 5 days. **p≤0.005 vs. M-CSF treatment.
To further characterize the activity of POH1, RAW 264.7 c4 cells were mock transfected or transfected with either wild type POH1 or C120S POH1. Cells were stimulated with M-CSF or M-CSF and RANKL for 3 days. Total RNA was isolated and real time PCR was performed to measure expression of the Acp5 mRNA. In mock-transfected RAW 264.7 c4 cells stimulated with M-CSF and RANKL, we detected a 3 fold increase in Acp5 expression, further increased with the transfection of wild type POH1. Transfection of the C120S POH1 mutant increased Acp5 expression only 2 fold compared to M-CSF treated cells (Figure 5B).
POH1 enhances differentiation of RAW 264.7 cells
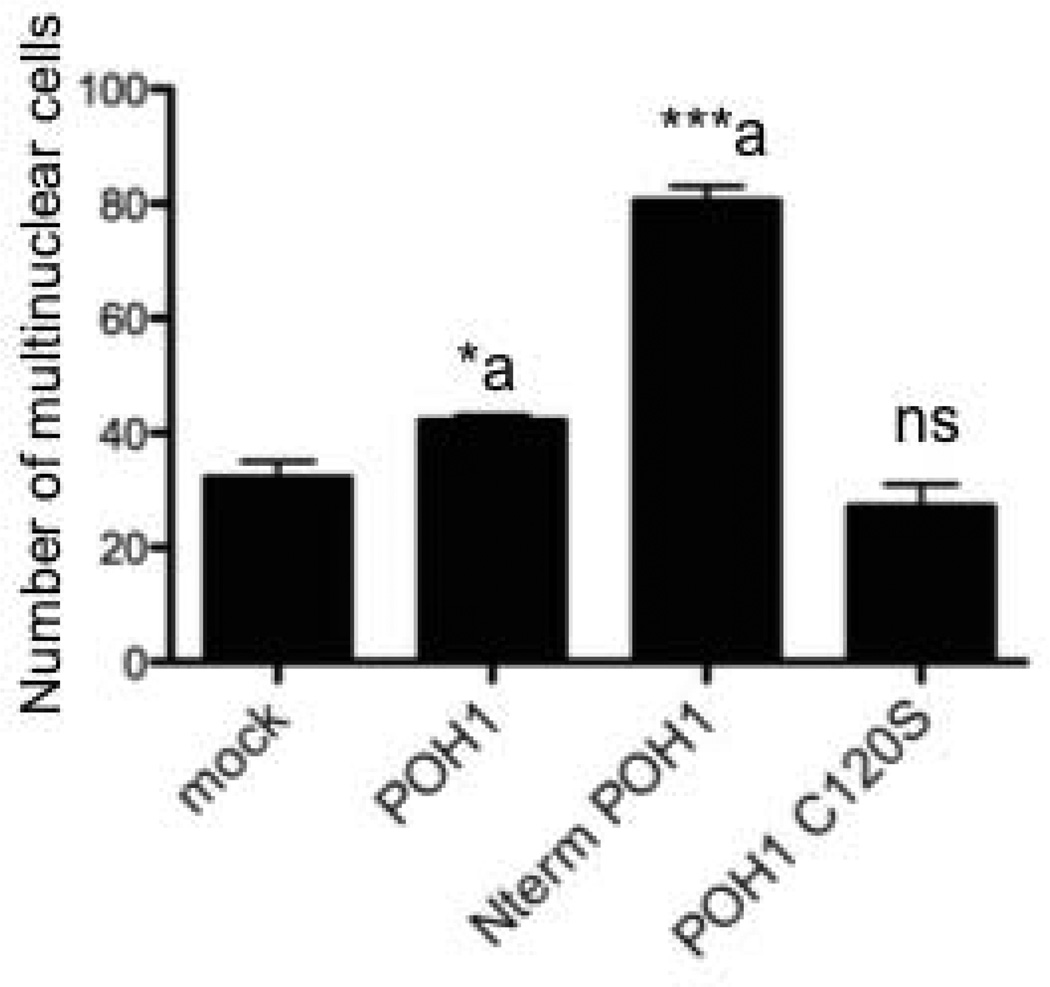
Since we had demonstrated that POH1 could enhance Mitf’s activation of osteoclast genes, we asked whether POH1 could enhance differentiation of RAW 264.7 cells. The RAW 264.7 preosteoclast cell line was transfected with full length POH1, C120S mutant POH1 or the amino terminal portion of POH1 (see Figure 4) and grown in presence of M-CSF and RANKL for 7days to induce formation of multinuclear cells. As shown in Figure 6, full length (approximately a 1.5 fold increase, p<0.03) as well as the amino terminal POH1 construct (2.5 fold increase, p<0.002) significantly induced multinuclear cells whereas the C120S POH1 mutant failed to enhance multinuclear cell formation over mock transfected RAW 264.7 c4 cells.
Figure 6. POH1 enhances RAW 264.7c4 differentiation.
RAW 264.7c4 cells were either mock transfected or transfected with wild type full length POH1, amino terminal portion of POH (amino acids 1-180) or full length C120S POH1. Cells were cultured in M-CSF and RANKL for 7 days. The number of multinuclear cells (greater than 2 nuclei/cell) was counted and the results of two experiments performed in triplicate are presented. *p≤0.05 and ***p≤0.0005 vs. mock transfected
Discussion
Osteoclast gene expression switches from “off” to “on” or vice versa during the process of differentiation. Stimulation by receptors on the cell surface link transcription factors and the change in gene expression to those signals received on the cell surface. Mitf belongs to the basic helix-loop-helix leucine zipper family of transcription factors, which affects differentiation and survival of osteoclasts [Luchin et al., 2000; McGill et al., 2002; Motyckova et al., 2001; So et al., 2003]. Recent studies from other labs have shown that Mitf interacts with other transcription factors such as PU.1 to activate osteoclast gene expression [Luchin et al., 2001; Sharma et al., 2007]. Genes that Mitf targets in osteoclasts have been identified and more continue to be discovered; however, very few proteins that interact and potentially regulate Mitf activation of target promoters have been identified.
POH1 is a well characterized deubiquitinase of the 19S lid particle and is required for a functional proteasome [Glickman et al., 1999]. POH1 is a DUB that removes the polyubiquitin chains from incoming substrates. POH1 is believed to function in a dual capacity: the removal of the ubiquitin tag can facilitate substrate translocation into the 20S chamber and thus facilitate proteolysis [Verma et al., 2002; Yao and Cohen, 2002] or it can cause the protein to be released from the proteasome allowing it to escape degradation. It is hypothesized that POH1 works as a proteasomal “proofreading” protein that determines the fate of incoming substrates as to whether they will be degraded or rescued [Glickman et al., 1998]. The data presented in this study suggests that POH1 plays a role in Mitf’s ability to activate osteoclast gene expression and differentiation. Similar to POH1’s interaction with c-jun, we hypothesize that POH1 deubiquitinates Mitf in osteoclasts allowing for Mitf to be more stably expressed on osteoclast promoters. This hypothesis is supported by our results in 293T cells where POH1 prevents Mitf from being ubiquitinated, and POH1’s activity was dependent on an intact JAMM motif, the region responsible for the DUB catalytic activity [Maytal-Kivity et al., 2002]. POH1 enhances Mitf’s activation of both the Acp5 and 5X Gal4-TK luciferase reporters, and this activity was again dependent on the ability of POH1 to function as a DUB. The amino terminus of POH1 (amino acids 1-180), the region containing the JAMM motif, was sufficient to interact with Mitf, was sufficient to enhance Mitf’s activation of a 5X Gal4 promoter and was sufficient to enhance differentiation of RAW 264.7 cells.
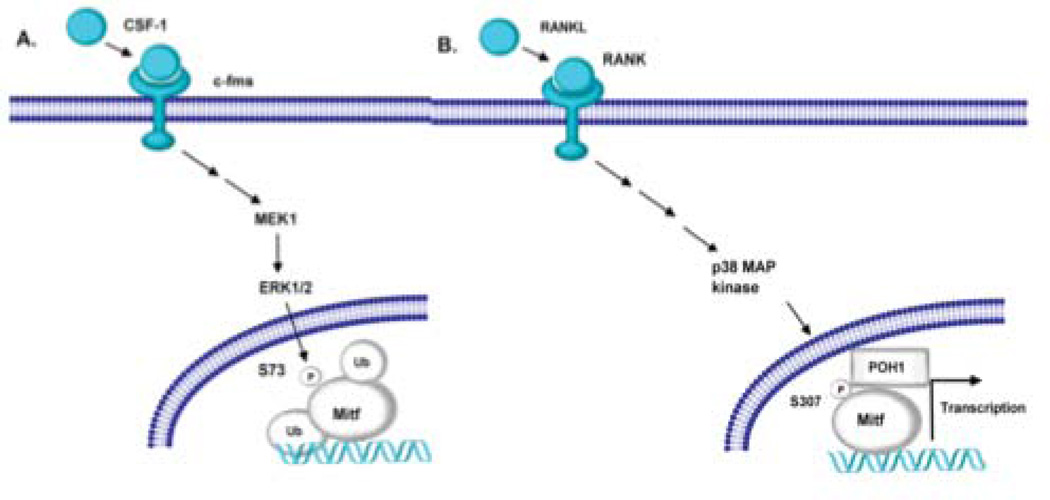
We only detect polyubiquitinated Mitf when RAW 264.7 c4 cells were stimulated with M-CSF and not when the cells were treated with M-CSF and RANKL. It has been shown by others that serine 73 phosphorylation is essential for ubiquitination of Mitf [Xu et al., 2000]. The authors further showed that lysine at residue 201is a potential ubiquitination site [Xu et al., 2000]. M-CSF signaling in osteoclasts has been linked to phosphorylation of Mitf amino acid residue 73 [Weilbaecher et al., 2001]. Sharma et al. presented data that the Mitf complex is recruited to target genes with M-CSF treatment [Sharma et al., 2007]. However, it is the combination of M-CSF and RANKL signaling that leads to robust osteoclast gene expression. In our study, treatment of bone marrow derived osteoclasts with the proteasome inhibitor MG132 increased gene expression from the Acp5 and Ctsk promoter when the cells were stimulated with M-CSF. Therefore, results from other laboratories as well as those presented here suggest that Mitf is ubiquitinated and potentially degraded during M-CSF signaling and when osteoclasts are stimulated with RANKL, Mitf interacts with POH1, which prevents Mitf’s ubiquitination and allows for osteoclast gene expression (see Figure 7).
Figure 7. Model of Mitf and POH1 interaction.
(A) M-CSF stimulation of osteoclast activates ERK1/2, which phosphorylates Mitf on serine residue 73 [Weilbaecher et al., 2001]. The serine at residue 73 has been shown to be necessary for Mitf to be ubiquinated [Xu et al., 2000]. (B) RANKL stimulation activates the p38MAPK pathway resulting in Mitf phosphorylated on serine residue 307 [Mansky et al., 2002]. We hypothesize that POH1 interacts with Mitf when it is phosphorylated on serine residue 307. Mitf’s interaction with POH1 prevents Mitf’s ubiquitination allowing for persistent activation of genes necessary for osteoclast differentiation.
We detect an interaction between POH1 and Mitf only when osteoclasts were stimulated with both M-CSF and RANKL suggesting that phosphorylation status of Mitf may play a role in the ability of Mitf and POH1 to interact. Our data suggests that we will detect POH1 at the promoter with Mitf only with RANKL signaling. We have been able to detect POH1 in the nucleus of osteoclasts by Western blot (data not shown). RANKL signaling leads to phosphorylation by p38 MAP kinase at amino acid residue 307, resulting in an increase in Mitf activity [Mansky et al., 2002]. We predict that Mitf mutated at amino acid residue 307 would not be able to interact with POH1 if p38 phosphorylation of Mitf is necessary for Mitf and POH1’s interaction as predicted by our data in Figure 1. The lysine at residue 201 has been predicted to be an ubiquitination site in Mitf. It would be interesting to determine if Mitf mutated at lysine residue 201, which cannot be ubiquitinated in vitro, can still be enhanced by POH1. In conclusion this study suggests that Mitf can interact with POH1 and this interaction enhances Mitf’s ability to activate gene expression.
Acknowledgments
We acknowledge A. Ian Cassady for the gift of the RAW 264.7 c4 cells clones. We would like to thank Dr. Sadowski for the use of the yeast two hybrid system. This work was supported by National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant 5R21-AR053946 (to K.C.M).
References
- Bronisz A, Sharma SM, Hu R, Godlewski J, Tzivion G, Mansky KC, Ostrowski MC. Microphthalmia-associated transcription factor interactions with 14-3-3 modulate differentiation of committed myeloid precursors. Mol Biol Cell. 2006;17:3897–3906. doi: 10.1091/mbc.E06-05-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin proteolytic system and pathogenesis of human diseases: a novel platform for mechanism-based drug targeting. Biochem Soc Trans. 2003;31:474–481. doi: 10.1042/bst0310474. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Fu H, Larsen CN, Coux O, Wefes I, Pfeifer G, Cjeka Z, Vierstra R, Baumeister W, Fried V, Finley D. Functional analysis of the proteasome regulatory particle. Mol Biol Rep. 1999;26:21–28. doi: 10.1023/a:1006928316738. [DOI] [PubMed] [Google Scholar]
- Guterman A, Glickman MH. Deubiquitinating enzymes are IN/(trinsic to proteasome function) Curr Protein Pept Sci. 2004;5:201–211. doi: 10.2174/1389203043379756. [DOI] [PubMed] [Google Scholar]
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliott PJ, Adams J, Anderson KC. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001;61:3071–3076. [PubMed] [Google Scholar]
- Hirst M, Ho C, Sabourin L, Rudnicki M, Penn L, Sadowski I. A two-hybrid system for transactivator bait proteins. Proc Natl Acad Sci U S A. 2001;98:8726–8731. doi: 10.1073/pnas.141413598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layfield R, Shaw B. Ubiquitin-mediated signalling and Paget's disease of bone. BMC Biochem. 2007;8 Suppl 1:S5. doi: 10.1186/1471-2091-8-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchin A, Purdom G, Murphy K, Clark MY, Angel N, Cassady AI, Hume DA, Ostrowski MC. The microphthalmia transcription factor regulates expression of the tartrate-resistant acid phosphatase gene during terminal differentiation of osteoclasts. J Bone Miner Res. 2000;15:451–460. doi: 10.1359/jbmr.2000.15.3.451. [DOI] [PubMed] [Google Scholar]
- Luchin A, Suchting S, Merson T, Rosol TJ, Hume DA, Cassady AI, Ostrowski MC. Genetic and physical interactions between Microphthalmia transcription factor and PU.1 are necessary for osteoclast gene expression and differentiation. J Biol Chem. 2001;276:36703–36710. doi: 10.1074/jbc.M106418200. [DOI] [PubMed] [Google Scholar]
- Mansky KC, Sankar U, Han J, Ostrowski MC. Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-kappa B ligand signaling. J Biol Chem. 2002;277:11077–11083. doi: 10.1074/jbc.M111696200. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Kogawa M, Wada S, Takayanagi H, Tsujimoto M, Katayama S, Hisatake K, Nogi Y. Essential Role of p38 Mitogen-activated Protein Kinase in Cathepsin K Gene Expression during Osteoclastogenesis through Association of NFATc1 and PU.1. Journal of Biological Chemistry. 2004;279:45969–45979. doi: 10.1074/jbc.M408795200. [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, Lin YL, Ramaswamy S, Avery W, Ding HF, Jordan SA, Jackson IJ, Korsmeyer SJ, Golub TR, Fisher DE. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- Motyckova G, Weilbaecher KN, Horstmann M, Rieman DJ, Fisher DZ, Fisher DE. Linking osteopetrosis and pycnodysostosis: regulation of cathepsin K expression by the microphthalmia transcription factor family. Proc Natl Acad Sci U S A. 2001;98:5798–5803. doi: 10.1073/pnas.091479298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Hamdan FF, Ribeiro P. A Schistosoma mansoni Pad1 homologue stabilizes c-Jun. Mol Biochem Parasitol. 2002;121:163–172. doi: 10.1016/s0166-6851(01)00450-9. [DOI] [PubMed] [Google Scholar]
- Nabhan JF, Ribeiro P. The 19 S proteasomal subunit POH1 contributes to the regulation of c-Jun ubiquitination, stability, and subcellular localization. J Biol Chem. 2006;281:16099–16107. doi: 10.1074/jbc.M512086200. [DOI] [PubMed] [Google Scholar]
- Schwartz AL, Ciechanover A. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu Rev Med. 1999;50:57–74. doi: 10.1146/annurev.med.50.1.57. [DOI] [PubMed] [Google Scholar]
- Sharma SM, Bronisz A, Hu R, Patel K, Mansky KC, Said S, Ostrowski MC. Mitf and PU.1 Recruit p38 MAPK and NFATc1 to Target Genes during Osteoclast Differentiation. Journal of Biological Chemistry. 2007a;282:15921–15929. doi: 10.1074/jbc.M609723200. [DOI] [PubMed] [Google Scholar]
- So H, Rho J, Jeong D, Park R, Fisher DE, Ostrowski MC, Choi Y, Kim N. Microphthalmia transcription factor and PU.1 synergistically induce the leukocyte receptor osteoclast-associated receptor gene expression. J Biol Chem. 2003;278:24209–24216. doi: 10.1074/jbc.M302940200. [DOI] [PubMed] [Google Scholar]
- Soboleva TA, Baker RT. Deubiquitinating enzymes: their functions and substrate specificity. Curr Protein Pept Sci. 2004;5:191–200. doi: 10.2174/1389203043379765. [DOI] [PubMed] [Google Scholar]
- Spataro V, Toda T, Craig R, Seeger M, Dubiel W, Harris AL, Norbury C. Resistance to diverse drugs and ultraviolet light conferred by overexpression of a novel human 26 S proteasome subunit. J Biol Chem. 1997;272:30470–30475. doi: 10.1074/jbc.272.48.30470. [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Durso R, Reese JC. The proteasome regulates the UV-induced activation of the AP-1-like transcription factor Gcn4. Genes Dev. 2001;15:128–133. doi: 10.1101/gad.863801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Wei N, Tsuge T, Serino G, Dohmae N, Takio K, Matsui M, Deng XW. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr Biol. 1998;8:919–922. doi: 10.1016/s0960-9822(07)00372-7. [DOI] [PubMed] [Google Scholar]
- Weilbaecher KN, Motyckova G, Huber WE, Takemoto CM, Hemesath TJ, Xu Y, Hershey CL, Dowland NR, Wells AG, Fisher DE. Linkage of M-CSF signaling to Mitf, TFE3, and the osteoclast defect in Mitf(mi/mi) mice. Mol Cell. 2001;8:749–758. doi: 10.1016/s1097-2765(01)00360-4. [DOI] [PubMed] [Google Scholar]
- Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, Fisher DZ, Fisher DE. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 2000;14:301–312. [PMC free article] [PubMed] [Google Scholar]
- Xu W, Gong L, Haddad MM, Bischof O, Campisi J, Yeh ET, Medrano EE. Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzyme hUBC9. Exp Cell Res. 2000;255:135–143. doi: 10.1006/excr.2000.4803. [DOI] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]