Summary
Increasing evidence suggests tumors are maintained by cancer stem cells; however, their nature remains controversial. In a HoxA9-Meis1 (H9M) model of acute myeloid leukemia (AML), we found that tumor-initiating activity existed in three, immunophenotypically distinct compartments, corresponding to disparate lineages on the normal hematopoietic hierarchy—stem/progenitor cells (Lin−kit+), and committed progenitors of the myeloid (Gr1+kit+) and lymphoid lineages (Lym+kit+). These distinct tumor-initiating cells (TIC) clonally recapitulated the immunophenotypic spectrum of the original tumor in vivo (including cells with a less-differentiated immunophenotype) and shared signaling networks, such that in vivo pharmacologic targeting of conserved TIC survival pathways (DNA methyltransferase and MEK phosphorylation) significantly increased survival. Collectively, H9M AML is organized as an atypical hierarchy that defies the strict lineage marker boundaries and unidirectional differentiation of normal hematopoiesis. Moreover, this suggests that in certain malignancies tumor-initiation activity (or “cancer-stemness”) can represent a cellular state that exists independently of distinct immunophenotypic definition.
Introduction
The cellular and molecular heterogeneity of tumors poses a challenge for therapy, and has been classically interpreted through two opposing models. The stochastic model states all tumor cells have an equal probability of regenerating the disease and must be targeted for curative therapy (Shackleton et al., 2009). The cancer stem cell (CSC) model posits that tumors are hierarchically organized, and maintained by CSC (also called tumor-initiating cells, or TIC), which uniquely self-renew, produce the heterogeneous lineages composing the bulk tumor, and must be eradicated for curative therapy (Clevers, 2011).
Original support for the CSC model came from human AML where only the rare subset of cells resembling hematopoietic stem/progenitor cells (HSPC) could initiate disease upon xenotransplantation (Bonnet and Dick, 1997). Putative CSC were also identified in solid tumors, and CSC were thought to be rare and have an undifferentiated phenotype, like tissue-specific stem cells (Ricci-Vitiani et al., 2007). Other work challenged this paradigm, showing TIC need not be rare (Kelly et al., 2007; Quintana et al., 2008), and that tumor cells with immunophenotypes of mature progenitors or lineage cells could perform as functional TIC in human and mouse AML (Deshpande et al., 2006; Eppert et al., 2011; Goardon et al., 2011; Sarry et al., 2010; Somervaille and Cleary, 2006; Taussig et al., 2010). These conflicting data underscore the challenge of defining CSC, and call into question whether the developmental structure of normal stem cell systems is maintained, or mimicked, in cancer (Dick, 2008; Jordan, 2009).
Independent of these hypotheses, incontrovertible links exist between cancer and the dysregulation of stem cell developmental pathways. Homeobox (Hox) genes are important for hematopoietic development and stem cell self-renewal (Abramovich and Humphries, 2005), and ectopic expression of HoxA9 and Meis1 (H9M) in hematopoietic progenitors leads to rapid onset AML (Kroon et al., 1998). This H9M AML model has yielded insights into the molecular (Bmi-1) and extracellular factors (Wnt/β-catenin signaling) underlying CSC maintenance (Lessard and Sauvageau, 2003; Wang et al., 2010). We demonstrate that enforced H9M expression leads to a reproducible breakdown of the normal flow of differentiation, driving the formation of multiple, phenotypically distinct TIC, and resulting in a malignancy organized as an atypical, dynamic hierarchy where tumor-initiating activity (TIA) exists as a mobile cellular “state” that is largely independent of immunophenotype.
Results
Multiple TICs in H9M AML
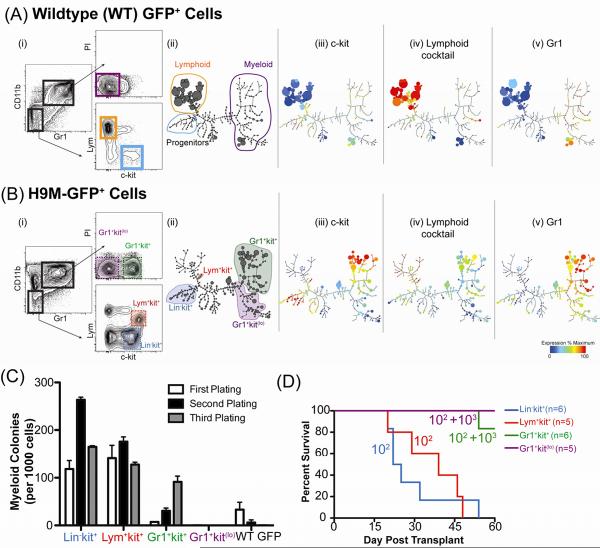
Normal HSPC express c-kit, lack mature lineage markers, and produce c-kit expressing myeloid or lymphoid progenitors that give rise to mature cells (Fig S1A). Four unique/expanded populations in H9M mice were assessed for TIA: (i) Lin−kit+, lacking mature markers and expressing c-kit, analogous to an HSPC, (ii) Lym+kit+, co-expressing lymphoid markers and c-kit, analogous to a lymphocyte precursor, (iii) Gr1+kit+, co-expressing c-kit and myeloid markers, representing an immature myeloid cell, and (iv) Gr1+kitlo, resembling a mature myeloid population (compare WT & H9M immunophenotypes, Fig. 1Ai & 1Bi and Fig S1E–J). The difference between the hematopoietic continuum of WT and H9M marrow was also visualized using SPADE, a density normalization, agglomerative clustering, and minimum-spanning tree algorithm that distills multidimensional single-cell data to interconnected “relatedness” clusters (with corresponding populations in conventional flow plots) of metacell populations displayed as a 2D tree plot (Fig 1A,Bii–v) (Qiu et al., 2011). The SPADE tree, drawn using composite data from WT-GFP, primary and secondary H9M-GFP cells, facilitated comparison of common and distinct metacells in each condition (size indicates relative frequency; compare Fig 1Aii, 1Bii). As expected, in WT-GFP, the rare, c-kit expressing progenitors lacked lineage marker expression, lymphoid metacells largely cluster together and are distinct from the myeloid region (Fig 1Aiii–v). In contrast, H9M-GFP cells have increased c-kit expression in the progenitor and myeloid regions, and a diminished lymphoid population (Fig 1Biii–v).
Figure 1. Multiple, Immunophenotypically Distinct Compartments Possess Tumor-Initiating Activity (TIA).
(A,B) Immunophenotypic comparison of bone marrow from mice transplanted with (A) GFP or (B) H9M-GFP transduced progenitors. (i) Flow cytometric analysis. “Lym” is a cocktail of B- and T-cell markers (B220, CD19, CD3ε, CD4, CD8a, and TCR-β). PI is propidium iodide. (ii) SPADE analysis of immunophenotypic progression. Size of each circle (metacell) indicates relative cell frequency. Metacell clusters were manually annotated based on (iii) c-kit, (iv) lymphoid cocktail, and (v) Gr1 expression. These parameters and CD11b used for tree drawing. (C) FACS-purified populations from H9M-GFP marrow were sorted and plated in methylcellulose with SCF, GM-CSF, IL-3, and IL-6 (S36GM). Colonies were scored after seven days, dissociated and replated. Numbers per 1000 cells plated (in triplicate ± SEM). Total WT-GFP cells shown as control. (D) Survival curves of mice lethally irradiated and transplanted with indicated numbers of cells (plus 105 helper marrow) from following compartments: Lin-kit+ (blue, 102 cells), Lym+kit+ (red, 102 cells), Gr1+kit+ (green, 102 or 103 cells), or Gr1+kitlo (purple, 102 or 103 cells). See also Figures S1–2.
The Lin−kit+, Lym+kit+, Gr1+kit+, and Gr1+kitlo compartments were assessed for TIA. In retroviral transplant models, in vitro myeloid colony-forming cells are TICs (Somervaille and Cleary, 2006). Each population was sorted to high purity and colony-forming activity (CFA) was assessed. The Lin−kit+, and unexpectedly, Lym+kit+ compartments had robust myeloid CFA (>10%; Fig 1C). In contrast, the Gr1+kit+ cells had a low initial CFA (<1%) which increased upon replating; Gr1+kitlo cells lacked any CFA. The most stringent test of TIA is transplanting disease with limiting cell numbers in vivo (Clarke et al., 2006). All recipients transplanted with 100 primary Lin−kit+ or Lym+kit+ cells succumbed rapidly to disease while more than 1000 Gr1+kit+ and 10,000 Gr1+kitlo cells were required to initiate disease in secondary recipients (Fig 1D & Fig S2A–B). Therefore, primary H9M AML is organized as a hierarchy in which multiple, phenotypically distinct TICs (Lin−kit+, Lym+kit+) are enriched for TIA and give rise to differentiated blasts (Gr1+kitlo) lacking TIA. The Gr1+kit+ compartment in primary AML has low, but seemingly selectable TIA.
Stage-dependent Differentiation Is Separate from TIA
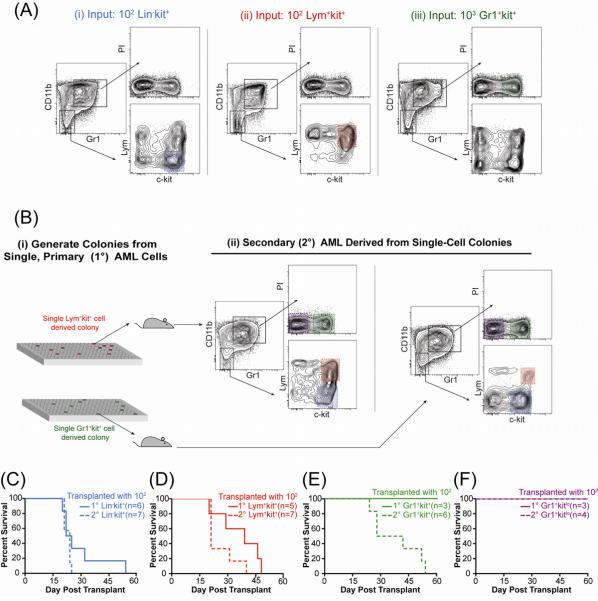
Constitutive GFP expression allowed in vivo lineage tracking of progeny. Each c-kit+ compartment recapitulated the entire immunophenotypic spectrum of primary AML in secondary recipients including cells (for the Lym+kit+ and Gr1+kit+ compartments) of a less differentiated immunophenotype (Lin−kit+) as well as cells with a phenotype corresponding to mature cells of the opposite lineage (i.e., Gr1+kit+ cells produced cells expressing lymphoid markers and vice versa; Fig 2A). To determine if this immunophenotypic recapitulation was clonal, single Lym+kit+ and Gr1+kit+ cells were expanded into colonies that were injected into secondary recipients (Fig 2Bi). Two of twenty recipients, transplanted with colonies derived from single Lym+kit+ and Gr1+kit+ cells respectively, developed AML and these leukemias again recapitulated the complete immunophenotypic spectrum (Fig 2Bii). Moreover, pre-leukemic H9M-transduced progenitors expressing myeloid markers (CD11b+Gr1+) gave rise to the immunophenotypic spectrum when transplanted to primary recipients (Fig S3), confirming that enforced H9M expression permits cells with defined lineage commitment to assume immunophenotypes of “earlier” or alternate lineages, and that traditional phenotypes associated with developmental stage specificity are separable from TIA.
Figure 2. Enforced H9M Expression Permits Surface Marker Plasticity in TIC Resulting in a Dynamic Tumorigenic Hierarchy.
(A) Flow cytometry of representative marrow from secondary mice transplanted with indicated cell number from specified compartment. Shaded region indicates the population transplanted. (B) (i) Single cells from primary AML were expanded into colonies and after one week transplanted into secondary mice. (ii) Flow cytometric analysis of the GFP+ marrow compartment of secondary mice transplanted with single-cell derived Lym+kit+ colony (top) or Gr1+kit+ colony (bottom). (C–F) Survival curves from mice transplanted with 100 primary or secondary (C) Lin-kit+, (D) Lym+kit+, (E) Gr1+kit+, or (F) Gr1+kitlo cells. See also Figure S3.
Dynamic Hierarchy in Secondary AML
To determine whether secondary tumors had the same hierarchical tumorigenic capacity as the primary tumors, 100 Lin−kit+, Lym+kit+, Gr1+kit+, or Gr1+kitlo cells from secondary AML were transplanted into tertiary recipients. As in primary AML, all animals transplanted with 100 Lin−kit+ or Lym+kit+ cells from secondary AML succumbed rapidly to disease (Fig 2C,D). Strikingly, although no animals transplanted with 100 Gr1+kit+ cells from primary AML developed leukemia, all animals transplanted with 100 Gr1+kit+ cells from secondary AML developed fatal AML indistinguishable (except longer latency, 38 v. 23 days, p=0.002, Log-Rank Test) from tumors arising from Lin−kit+ or Lym+kit+ subsets (Fig 2E). As before, Gr1+ kitlo cells failed to reconstitute tumors (Fig 2F) demonstrating a continued lack of TIA in this fraction. Thus, while TIA does not remain fixed within immunophenotypic compartments as tumors progress, H9M AML remains hierarchically organized in secondary disease as TIC give rise to terminal blasts without TIA.
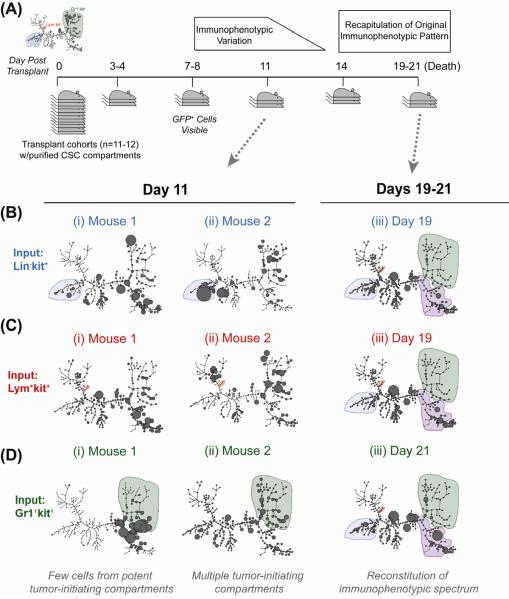
To determine whether H9M AML, like normal hematopoiesis, has a directional developmental progression, cohorts of mice were transplanted with each primary TIC and groups of mice were sacrificed sequentially to assess the immunophenotypes of leukemic progeny and capture individual variability of leukemic expansion (Fig 3A). GFP+ cells were first detectable on day 7-post transplantation. SPADE analysis showed substantial heterogeneity in immunophenotypic reconstitution prior to day 14, after which nearly all mice (11/12) consolidated to a uniform immunophenotypic equilibrium that was nearly identical to primary AML (Fig 3B-Diii and Fig S4). Thus, while each TIC ultimately recapitulated the immunophenotypic spectrum of primary AML, unlike in normal development, the in vivo developmental program that did not appear to follow a strict directional progression. Whether the regeneration occurs by a transdifferentiation-like mechanism, or dedifferentiation followed by forward differentiation remains to be determined, as it rapid and beyond the current level of resolution.
Figure 3. In Vivo Tumorigenic Differentiation Does Not Proceed Along a Strictly Defined Pathway.
(A) Experimental scheme. Defined numbers of each TIC in primary AML were transplanted into cohorts of secondary recipients. Animals were sacrificed every 3–4 four days thereafter to assess for frequency and immunophenotype of progeny. (B–D). SPADE analysis of bone marrow of individual mice transplanted with (B) 500 Lin-kit+ cells, (C) 500 Lym+kit+ cells, or (D) 2500 Gr1+kit+ cells on (i, ii) day 11 post transplant or (iii) 19–21 days post transplant. Shaded regions in panels i and ii are the compartment transplanted, and in panel iii are compartments present in primary leukemia. See also Figure S4.
TICs Share Genetic, Signaling and Survival Pathways
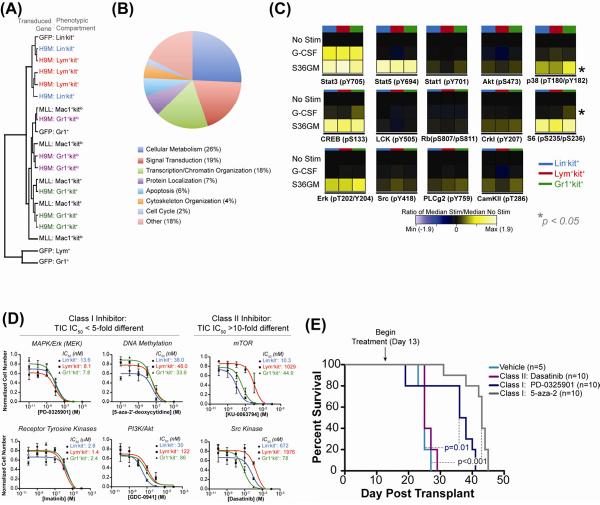
We hypothesized these phenotypically distinct TIC, which shared biological and developmental potential, would utilize overlapping genetic, signaling and survival pathways, and targeting such shared pathways could affect TIC survival. Microarray analysis and unsupervised hierarchal clustering was performed on TICs from primary H9M AML, populations from MLL-AF9 AML with high or low TIA (Mac1+kit+ or Mac1+kitlo, respectively) (Somervaille and Cleary, 2006), and analogous populations from WT-GFP marrow (Lin−kit+, Lym+, and Gr1+) (Fig 4A). This analysis showed H9M Lin−kit+ and Lym+kit+ compartments clustered with normal HSPC (GFP:Lin−kit+), H9M Gr1+kit+ cells clustered with the Mac1+kit+ TIC from MLL-AF9 AML, and terminal blasts clustered together (H9M:Gr1+kitlo and MLL-AF9:Mac1+kitlo). This suggests that TICs in primary H9M AML (Lin−kit+ and Lym+kit+), share a genetic signature resembling normal HSPC, as occurs in human AML (Eppert et al., 2011), and is distinct from the program utilized in MLL-AF9 TICs.
Figure 4. Phenotypically Distinct TICs Have Conserved Signaling and Survival Networks & Targeting Conserved Nodes in vivo Increases Survival.
(A) Hierarchical clustering of phenotypic compartments from primary H9M AML (H9M: Lin-kit+, Lym+kit+, Gr1+kit+, Gr1+kitlo), populations from MLL-AF9 AML with high or low TIA (Mac1+kit+ or Mac1+kitlo, respectively), and analogous populations from GFP marrow (Lin-kit+, Lym+, and Gr1+). Each line represents a unique biological replicate. (B) GO analysis of genes upregulated in primary H9M TICs (Lin-kit+ and Lym+kit+) relative to the Gr1+kit+ compartment based on biological process annotations. See also Figure S5 & Table S1. (C) Heatmap of the phospho-response in TICs (colored bar indicates compartment) isolated from secondary AML, measured by mass cytometry. ANOVA was used to test for statistically significant differences in fold-change values between TIC in all mice tested (n=3). Asterisks indicate statistically significant differences (p<0.05). See also Figure S6. (D) Defined numbers of cells from each TIC (Lin-kit+, blue; Lym+kit+, red; Gr1+kit+, green) were sorted into liquid medium with S36GM and concentrations of inhibitors for the pathways indicated above the plots. Plots represent numbers after 7 days measured quadruplicate ± SEM. (E) Survival curves of secondary mice transplanted with 104 total GFP+ primary AML marrow cells treated with dasatinib, PD-0325901, and 5-aza-2'-deoxycytidine. Survival differences between mice treated vehicle and PD-0325901 (p=0.01, Log-Rank Test) or 5-aza-2'-deoxycytidine (p<0.0001, Log-Rank Test) are statistically significant.
To determine if primary H9M TICs shared enrichment for leukemogenesis-associated genes, the top 2500 differentially expressed probes between Lin−kit+ and Lym+kit+ cells relative to primary Gr1+kit+ cells (which lacked TIA) were assessed and common genes were identified (Table S1). GO-term enrichment analysis of the resultant 151 genes (94 with annotations) showed the major biological functions were cellular metabolism (26%), signal transduction regulation (19%), and transcription/chromatin organization (18%) (Fig 4B). Specifically, this list included targets whose improper regulation has been previously implicated in AML such as Satb1, DNMT3A, and Msi2 (Kharas et al., 2010; Ley et al., 2010; Steidl et al., 2007). Therefore, primary H9M TICs shared expression of genes implicated in leukemogenesis, suggesting a core cellular program can be induced by H9M expression—although the roles of these genes as drivers or passengers remains to be elucidated. Despite this shared program, these populations are unique as 12 of the top 20 genes differentially expressed between them have roles in B-cell development, Lym+kit+ cells express B220, and have rearranged the D-J gene segments on the immunoglobulin heavy chain locus (Fig S5). Therefore, the Lym+kit+ progenitor represents a lymphoid-specified AML TIC, potentially analogous to other lymphoid-primed AML TICs (Deshpande et al., 2006; Goardon et al., 2011).
To determine if TICs shared signaling networks, mass cytometry was utilized providing a single-cell view of TIC signaling architecture (Bendall et al., 2011). After measuring potentiated phosphorylation of 14 intracellular nodes in each TIC after cytokine stimulation, analysis of variance (ANOVA) was utilized to test for statistically significant differences in the fold-change values (FCV) between TICs for each stimulation-phospho node pair (Fig 4B). This analysis showed no statistically significant differences in the FCV for 26 of 28 conditions tested (Fig S6A), demonstrating these distinct TICs have largely conserved signaling networks (unlike WT samples where FCV between compartments were significantly different for 3 of 5 conditions (Fig S6B)).
To determine which nodes had conserved roles in TIC proliferation, each compartment was sorted into liquid media supplemented with S36GM—which selectively expands TIC but not terminal blasts in vitro—and proliferation was assessed after 7 days in the presence of pharmacological inhibitors targeting a suite of cellular pathways implicated in TIC-maintenance (Fig 4D). For inhibitors targeting the MAP kinase pathway (Mek inhibitor PD-0325901), DNA methyltransferase (5-aza-2'-deoxycytidine), receptor tyrosine kinases (imatinib), and PI3K/Akt (GDC-0941), termed class I inhibitors, the IC50 values for each TIC were overlapping or within 5-fold, suggesting these nodes play conserved roles in TIC survival and proliferation. In contrast, inhibitors targeting Src-family kinases (dasatinib) and mTOR (KU-0063794), class II inhibitors, had IC50 values greater than 10-fold different in the TICs, suggesting disparate requirements for these pathways across TICs. Collectively, these proliferation and signaling data show that overlapping survival and signaling nodes govern phenotypically distinct TICs.
Targeting Conserved Nodes in vivo Increases Survival
We hypothesized that pathways with conserved roles in TIC survival could serve as effective therapeutic targets in vivo while those with disparate roles would not. To determine if conserved signaling pathways were essential for AML maintenance in vivo, secondary recipients transplanted with total GFP+ H9M marrow were treated with either class I (PD-0325901, 5-aza-2'-deoxycytidine) or class II (Dasatinib) inhibitors starting two weeks after transplant to allow for immunophenotypic reconstitution. All mice treated with vehicle or the class II inhibitor dasatinib (with 10-fold differential selectivity across the TICs) succumbed rapidly to AML (median survival 25 days; Fig 4D). Strikingly, mice treated with the class I inhibitors PD-0325901 and 5-aza-2'-deoxcytidine had statistically significant increases in overall survival (median survival 37 days, p=0.01 and 42 days, p<0.001, respectively, Log-Rank Test). These results show that in vivo pharmacologic targeting of pathways with conserved roles in TIC cell survival in vitro significantly increased survival, suggesting phenotypically distinct TICs rely on conserved survival nodes in vivo and targeting nodes only important for certain classes of TIC in the tumor allows for other TICs to “escape” and for disease to rapidly progress.
Discussion
Our data support the central tenet of the CSC model, i.e. hierarchical tumor organization. In H9M AML, self-renewing TICs with multilineage potential gave rise to terminal blasts (Gr1+kitlo) unable to regenerate the tumor. However, this hierarchy existed outside the strict phenotypic and unidirectional developmental bounds characterizing normal hematopoiesis and other leukemic systems (Goardon et al., 2011; Sarry et al., 2010) such that TIA was decoupled from stable immunophenotype or stage specificity. Notably, TICs had overlapping enrichment of leukemic- and HSPC-associated gene signatures, signaling networks, and survival determinants, and pharmacological targeting these shared pathways in vivo significantly improved host survival. Therefore, the ability to initiate disease (or “cancer stemness”) can represent a coordinately regulated cellular state as opposed to a defined cell phenotype.
This view provides context to the disparate results regarding the nature of AML CSCs (Bonnet and Dick, 1997; Goardon et al., 2011; Sarry et al., 2010; Somervaille and Cleary, 2006; Taussig et al., 2010). Much of the controversy regarding CSCs comes from the notion that the CSC model's implications are mutually exclusive from other views. That is, cancer either represents caricatured developmental process with a strictly defined CSC population, or that stochastic disequilibrium rules, with every cell being an equally bad agent. In this retroviral, LTR-driven model with a single oncogenic initiating event in a genetically homogenous background with syngeneic recipients, the frequency and phenotype of cells with TIA in the resultant oligoclonal disease are dynamic. It is therefore not surprising that in human AML, driven by numerous genetic lesions in genetically diverse backgrounds, and where CSC activity is assayed across a xeno-barrier, disparate interpretations could arise. While it remains to be seen whether phenotypic interconversion and a tumor-initiating “stem cell state” that appears to transit between different cellular phenotypes is a general CSC mechanism or is confined to rare oncogene sets, these results add a new layer of complexity to the heterogeneity underlying normal and leukemic stem cell systems (Anderson et al., 2011; Gibbs et al., 2011; Notta et al., 2011). While stable hematopoietic marker expression proved unreliable in isolating H9M TICs, we believe identifying CSC surface markers does have important mechanistic and therapeutic value (Jin et al., 2009; Majeti et al., 2009). Future work must elucidate molecular modules endowing TIA, identify consistent CSC markers or pathways, and unravel targetable signaling events conserved in-but not necessarily exclusive to-CSC survival.
Methods
Vectors, Generation of H9M AML & Colony Formation Assay
H9M AML was generated and colony-forming assay were done as previously described (Majeti et al., 2009; Somervaille and Cleary, 2006). Animal studies were done in compliance with the Stanford Administrative Panel on Laboratory Animal Care Protocol 15986.
Gene Expression Profiling
Total RNA from 5×103–105 TICs was purified by RNAqueous kit (Ambion). Microarray hybridization probes were synthesized from total RNA; RNA was amplified using the Ovation Pico WTA kit and biotin labeled using the Encore Biotin Module (NuGEN). Probes were hybridized to Affymetrix Mouse Genome 430 2.0 arrays. The primary data was quantile normalized and analyzed with RMAexpress software. Unweighted hierarchal clustering was performed using Spotfire.
Mass Cytometry and SPADE Analysis
Stimulation, antibody preparation cell staining, and data analysis was performed based on procedures previously described (Bendall et al., 2011). Briefly, H9M AML cells were rested in culture media for 90 minutes at 37°C, stimulated with S36GM (100 ng/ml SCF and 20 ng/ml IL-3, IL-6, and GM-CSF) for 15 minutes, fixed with 1.6% paraformaldehyde for 10 minutes at 25°C, stained with surface markers for 30 minutes at 25°C, permeabilized with ice-cold, 100% methanol at −80°C overnight, and stained with intracellular anti bodies 30 min at 25°C. Mass cytometry antibodies are described in Table S2.
In Vitro Drug Screen
Culture medium (IMDM with 10% FCS + penicillin/streptomycin/gluatmine + 25 mM HEPES, and non-essential amino acids (Gibco)) supplemented with S36GM cocktail and inhibitor were added to 96-well plates. Defined cell numbers were clone sorted into wells as 4-fold replicates, and cultured for 7 days at 37°C. Cell number was calculated by flow cytometry (BDTruCount beads), and normalized to the highest individual replicate for a specific cell population and inhibitor. Statistical analysis, least-squares regression and IC50 values were calculated using GraphPad Prism. PD-0325901, dasatinib, imatinib, KU-0063794 and GDC-0941 purchased from Selleck Chemicals; 5-aza-2'-deoxycytidine purchased from Sigma-Aldrich.
In Vivo Pharmacological Inhibition
10-week-old, C57BL/6 mice were sublethally irradiated (4.8 Gy) and transplanted with 104 total GFP+ marrow cells from primary H9M mice and 5×104 helper marrow cells. Treatment began 13 days after transplantation. Vehicle (0.5% HPMC/0.05% Tween-80 solution; daily) and Dasatinib (10 mg/kg d13–23) were administered via oral gavage until mice were moribund. PD-0325901 was administered via oral gavage (25 mg/kg on d13–18 and 15 mg/kg on d23–26 post transplantation). 5-Aza-2'-deoxycytidine (0.25 mg/kg in PBS) was administered three times per week (d14, 17, 20, 22, 24, 27) via intraperitoneal injection.
Supplementary Material
Acknowledgments
We thank E Simonds, G Behbehani and W Fantl for critical review of the manuscript, and S Bendall, E Zunder and R Majeti for helpful conversations. KG is funded by NSF GRFP and Stanford DARE Fellowship. This work was supported by NIH/NCI grants 1R01CA130826, U54CA149145, 5U54CA143907 as well as CIRM grant DR1-01477. GPN is the Rachford and Carlotta A. Harris Endowed Professor, is a member of the scientific advisory board at DVS Sciences, and is a paid consultant for BD Biosciences and Nodality, Inc.
Abbreviations
- H9M
HoxA9-Meis1
- CSC
Cancer stem cells
- TIC
Tumor-initiating cells
- TIA
Tumor-initiating activity
- CFA
Colony forming activity
- HSPC
Hematopoietic stem-progenitor cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions KG designed the study, performed experiments, analyzed results and wrote the manuscript. AJ, OC, YG, AT, and CER performed experiments. GPN designed experiments and wrote the manuscript.
References
- Abramovich C, Humphries RK. Hox regulation of normal and leukemic hematopoietic stem cells. Current opinion in hematology. 2005;12:210–216. doi: 10.1097/01.moh.0000160737.52349.aa. [DOI] [PubMed] [Google Scholar]
- Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature medicine. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer Stem Cells--Perspectives on Current Status and Future Directions: AACR Workshop on Cancer Stem Cells. Cancer research. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- Clevers H. The cancer stem cell: premises, promises and challenges. Nature medicine. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- Deshpande AJ, Cusan M, Rawat VP, Reuter H, Krause A, Pott C, Quintanilla-Martinez L, Kakadia P, Kuchenbauer F, Ahmed F, et al. Acute myeloid leukemia is propagated by a leukemic stem cell with lymphoid characteristics in a mouse model of CALM/AF10-positive leukemia. Cancer Cell. 2006;10:363–374. doi: 10.1016/j.ccr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, Metzeler KH, Poeppl A, Ling V, Beyene J, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nature medicine. 2011 doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- Gibbs KD, Jr., Gilbert PM, Sachs K, Zhao F, Blau HM, Weissman IL, Nolan GP, Majeti R. Single-cell phospho-specific flow cytometric analysis demonstrates biochemical and functional heterogeneity in human hematopoietic stem and progenitor compartments. Blood. 2011;117:4226–4233. doi: 10.1182/blood-2010-07-298232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, Soneji S, Woll P, Mead A, Alford KA, Rout R, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Jin L, Lee EM, Ramshaw HS, Busfield SJ, Peoppl AG, Wilkinson L, Guthridge MA, Thomas D, Barry EF, Boyd A, et al. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell. 2009;5:31–42. doi: 10.1016/j.stem.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Jordan CT. Cancer stem cells: controversial or just misunderstood? Cell Stem Cell. 2009;4:203–205. doi: 10.1016/j.stem.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, Morgan K, Tam W, Paktinat M, Okabe R, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nature medicine. 2010;16:903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD, Jr., van Rooijen N, Weissman IL. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, Phillips LA, Ma J, Minden MD, Downing JR, Dick JE. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr., Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011 doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Sarry JE, Murphy K, Perry R, Sanchez PV, Secreto A, Keefer C, Swider CR, Strzelecki AC, Cavelier C, Recher C, et al. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in NOD/SCID/IL2Rgammac-deficient mice. J Clin Invest. 2010;121:384–395. doi: 10.1172/JCI41495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Steidl U, Steidl C, Ebralidze A, Chapuy B, Han HJ, Will B, Rosenbauer F, Becker A, Wagner K, Koschmieder S, et al. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J Clin Invest. 2007;117:2611–2620. doi: 10.1172/JCI30525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taussig DC, Vargaftig J, Miraki-Moud F, Griessinger E, Sharrock K, Luke T, Lillington D, Oakervee HE, Cavenagh J, Agrawal SG, et al. Leukemia initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34- fraction. Blood. 2010 doi: 10.1182/blood-2009-02-206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI, Armstrong SA. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.