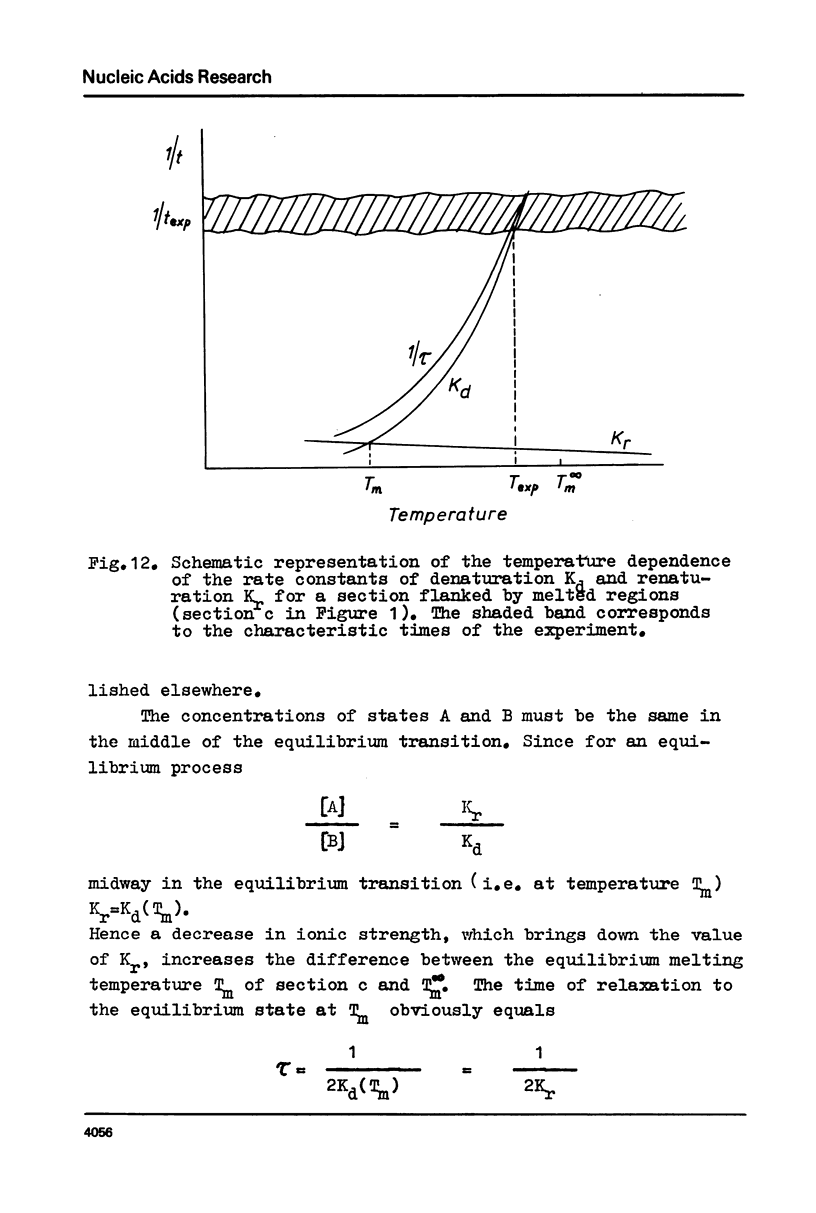
Abstract
The reversibility of DNA melting has been thoroughly investigated at different ionic strengths. We concentrated on those stages of the process that do not involve a complete separation of the strands of the double helix. The differential melting curves of pBR 322 DNA and a fragment of T7 phage DNA in a buffer containing 0.02M Na+ have been shown to differ substantially from the differential curves of renaturation. Electron-microscopic mapping of pBR 322 DNA at different degrees of unwinding (by a previously elaborated technique) has shown that the irreversibility of melting under real experimental conditions is connected with the stage of forming new helical regions during renaturation. In a buffer containing 0.2M Na+ the melting curves of the DNAs used (pBR322, a fragment of T7 phage DNA, a fragment of phage Lambda DNA, a fragment of phiX174 phage DNA) coincide with the renaturation curves, i.e. the process is equilibrium. We have carried out a semi-quantitative analysis of the emergence of irreversibility in the melting of a double helix. The problem of comparing theoretical and experimental melting curves is discussed.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allington W. B., Cordry A. L., McCullough G. A., Mitchell D. E., Nelson J. W. Electrophoretic concentration of macromolecules. Anal Biochem. 1978 Mar;85(1):188–196. doi: 10.1016/0003-2697(78)90289-0. [DOI] [PubMed] [Google Scholar]
- Benight A. S., Wartell R. M., Howell D. K. Theory agrees with experimental thermal denaturation of short DNA restriction fragments. Nature. 1981 Jan 15;289(5794):203–205. doi: 10.1038/289203a0. [DOI] [PubMed] [Google Scholar]
- Blake R. D., Fresco J. R. Polynucleotides. VII. Spectrophotometric study of the kinetics of formation of the two-stranded helical complex resulting from the interaction of polyriboadenylate and polyribouridylate. J Mol Biol. 1966 Aug;19(1):145–160. doi: 10.1016/s0022-2836(66)80057-8. [DOI] [PubMed] [Google Scholar]
- Borovik A. S., Kalambet Y. A., Lyubchenko Y. L., Shitov V. T., Golovanov E. I. Equilibrium melting of plasmid ColE1 DNA: electron-microscopic visualization. Nucleic Acids Res. 1980 Sep 25;8(18):4165–4184. doi: 10.1093/nar/8.18.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grachev M. A., Perelroyzen M. P. Measurement of the differential melting profile of a promoter-containing fragment of T7 DNA by means of a microspectrophotometer. Nucleic Acids Res. 1978 Jul;5(7):2557–2564. doi: 10.1093/nar/5.7.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravchev M. A., Zaichikov E. F., Kravchenko V. V., Pletnev A. G. Vydelenie promotornykh i terminatornogo fragmentov DNK faga T7 is gidrolizata, poluchennogo deivistviem éndonukleazy restrikstii BsuR. Dokl Akad Nauk SSSR. 1978;239(2):475–478. [PubMed] [Google Scholar]
- Hoff A. J., Roos A. L. Hysteresis of denaturation of DNA in the melting range. Biopolymers. 1972;11(6):1289–1294. doi: 10.1002/bip.1972.360110612. [DOI] [PubMed] [Google Scholar]
- Michel F. Hysteresis and partial irreversibility of denaturation of DNA as a means of investigating the topology of base distribution constraints: application to a yeast rho- (petite) mitochondrial DNA. J Mol Biol. 1974 Oct 25;89(2):305–326. doi: 10.1016/0022-2836(74)90521-x. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Guryev S. O., Krayev A. S., Monastyrskaya G. S., Skryabin K. G., Sverdlov E. D., Zakharyev V. M., Bayev A. A. Primary structure of an EcoRI fragment of lambda imm434 DNA containing regions cI-cro of phage 434 and cII-o of phage lambda. Gene. 1979 Jul;6(3):235–249. doi: 10.1016/0378-1119(79)90060-x. [DOI] [PubMed] [Google Scholar]
- Pörschke D., Eigen M. Co-operative non-enzymic base recognition. 3. Kinetics of the helix-coil transition of the oligoribouridylic--oligoriboadenylic acid system and of oligoriboadenylic acid alone at acidic pH. J Mol Biol. 1971 Dec 14;62(2):361–381. doi: 10.1016/0022-2836(71)90433-5. [DOI] [PubMed] [Google Scholar]
- Spatz H. C., Crothers D. M. The rate of DNA unwinding. J Mol Biol. 1969 Jun 14;42(2):191–219. doi: 10.1016/0022-2836(69)90038-2. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Effects of the conformation of single-stranded DNA on renaturation and aggregation. J Mol Biol. 1969 Apr;41(2):199–209. doi: 10.1016/0022-2836(69)90385-4. [DOI] [PubMed] [Google Scholar]
- Vizard D. L., White R. A., Ansevin A. T. Comparison of theory to experiment for DNA thermal denaturation. Nature. 1978 Sep 21;275(5677):250–251. doi: 10.1038/275250a0. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. V., Frank-Kamenetskii M. D. Theoretical melting profiles and denaturation maps of DNA with known sequence: fdDNA. Nucleic Acids Res. 1978 Jul;5(7):2547–2556. doi: 10.1093/nar/5.7.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki S., Fuke M., Wada A. The fine structures in melting curves of deoxyribonucleic acids of bacteriophage lambda. I. J Biochem. 1971 Jan;69(1):191–207. doi: 10.1093/oxfordjournals.jbchem.a129447. [DOI] [PubMed] [Google Scholar]


