Abstract
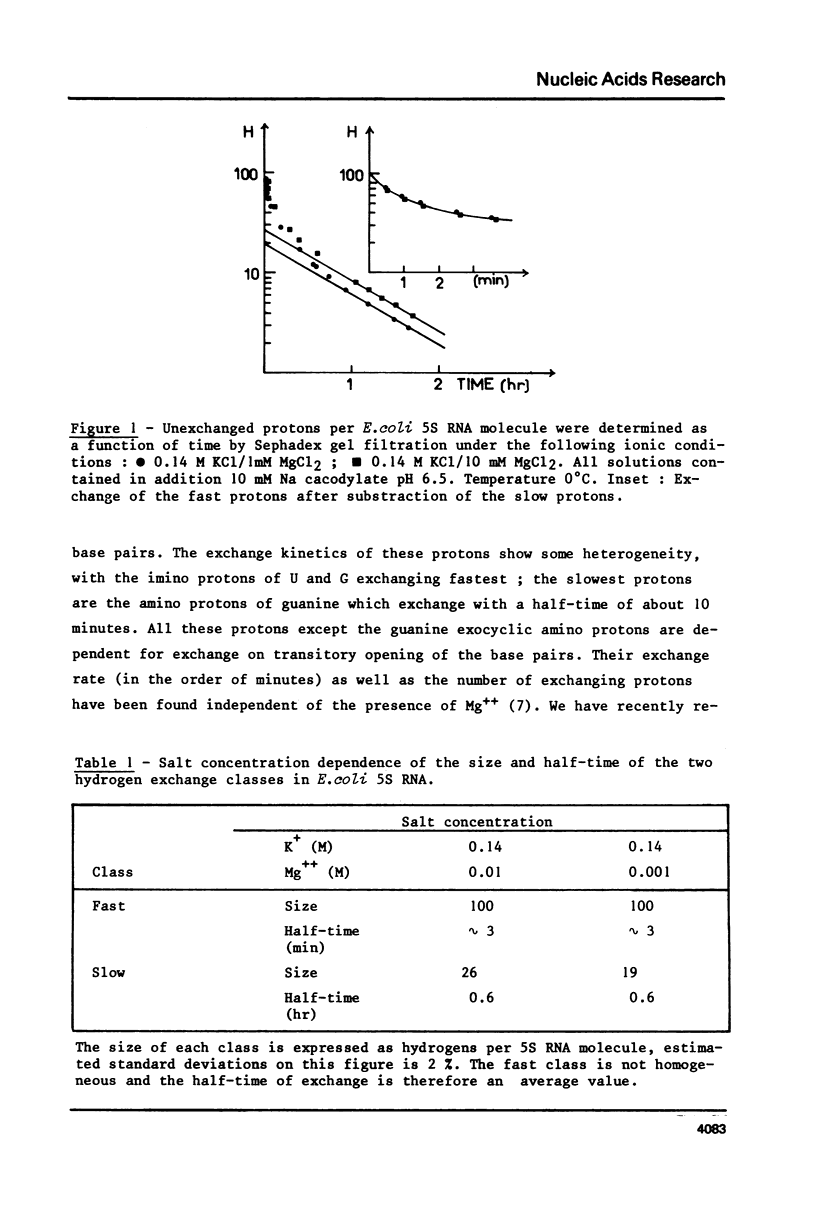
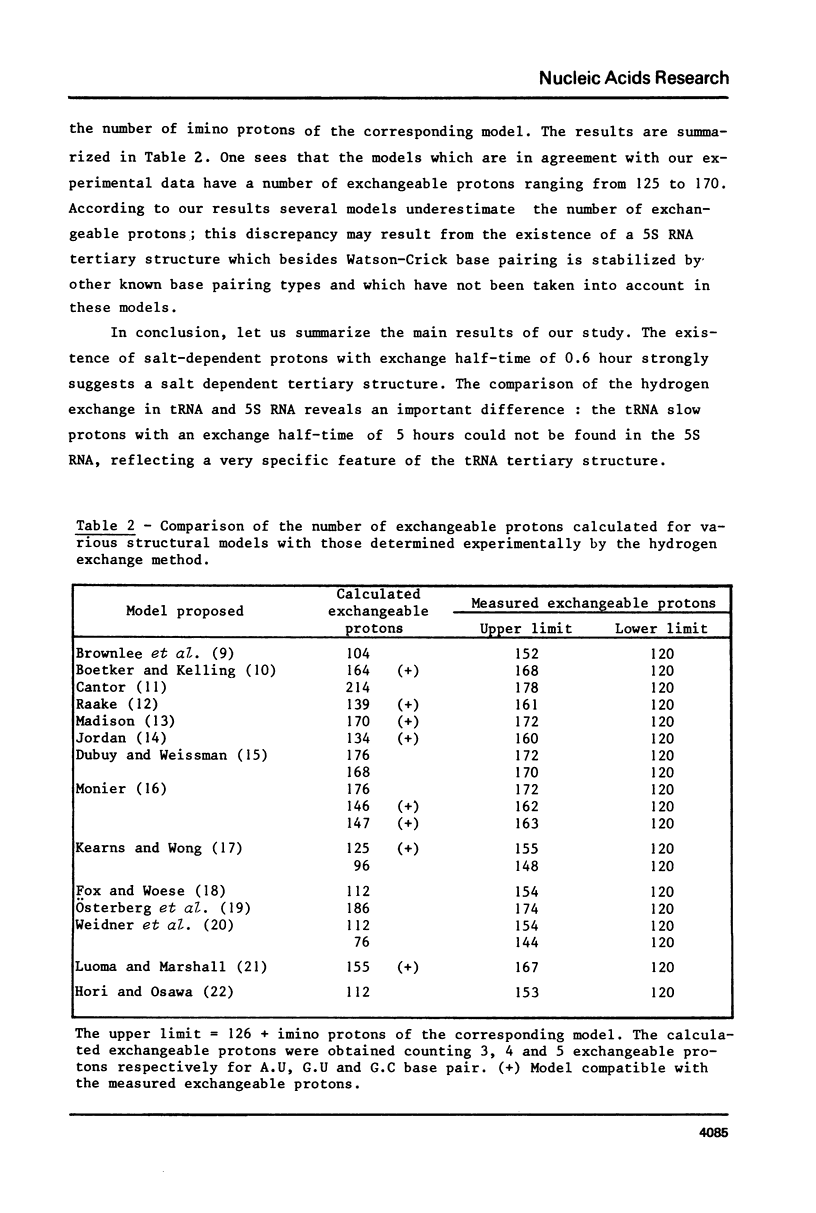
Using the tritium Sephadex method, the number of exchangeable protons in E. coli 5S RNA and the kinetics of their exchange reactions have been measured at two different Mg++ concentrations (10(-2) M and 10(-3) M). A quantitative analysis of these results indicates the presence of two classes of protons exchanging with very different rates. The protons of the slow class, not seen in linear molecules of double helical RNA, exchange with a half-time of 0.6 hour and their exchange kinetics are independent of Mg++ concentration. Reduction of the Mg++ concentration from 10(-2) ot 10(-3) M, however, results in a decrease in the number of these exchangeable protons form 26 to 19. Neither the total number, nor the exchange kinetics of the fast protons are affected by this change in Mg++ concentration. Comparison of these results with those previously obtained with tRNA, suggests the presence of a Mg++ dependent tertiary structure in 5S RNA. The number of exchangeable protons obtained from extrapolation of the exchange curves (120 and 126 respectively for 10(-3) and 10(-2) M Mg++ concentration) are compared to the calculated number of exchangeable protons predicted by previous proposed structural models for E.Coli 5S RNA.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appel B., Erdmann V. A., Stulz J., Ackerman T. Determination of base pairing in Escherichia coli and Bacillus stearothermophilus 5S RNAs by infrared spectroscopy. Nucleic Acids Res. 1979 Oct 25;7(4):1043–1057. doi: 10.1093/nar/7.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedtker H., Kelling D. G. The ordered structure of 5S RNA. Biochem Biophys Res Commun. 1967 Dec 15;29(5):758–766. doi: 10.1016/0006-291x(67)90283-5. [DOI] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F., Barrell B. G. Nucleotide sequence of 5S-ribosomal RNA from Escherichia coli. Nature. 1967 Aug 12;215(5102):735–736. doi: 10.1038/215735a0. [DOI] [PubMed] [Google Scholar]
- Cantor C. R. Possible conformations of 5-S ribosomal RNA. Nature. 1967 Nov 4;216(5114):513–514. doi: 10.1038/216513a0. [DOI] [PubMed] [Google Scholar]
- DuBuy B., Weissman S. M. Nucleotide sequence of Pseudomonas fluorescens 5 S ribonucleic acid. J Biol Chem. 1971 Feb 10;246(3):747–761. [PubMed] [Google Scholar]
- Englander J. J., Kallenbach N. R., Englander S. W. Hydrogen exchange study of some polynucleotides and transfer RNA. J Mol Biol. 1972 Jan 14;63(1):153–169. doi: 10.1016/0022-2836(72)90527-x. [DOI] [PubMed] [Google Scholar]
- Englander S. W., Englander J. J. Hydrogen-tritium exchange. Methods Enzymol. 1972;26:406–413. doi: 10.1016/s0076-6879(72)26021-9. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A., Doberer H. G. Structure and function of 5S RNA: the role of the 3' terminus in 5S RNA function. Mol Gen Genet. 1972;114(2):89–94. doi: 10.1007/BF00332779. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A. Structure and function of 5S and 5.8 S RNA. Prog Nucleic Acid Res Mol Biol. 1976;18:45–90. [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. The architecture of 5S rRNA and its relation to function. J Mol Evol. 1975 Oct 3;6(1):61–76. doi: 10.1007/BF01732674. [DOI] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B. R. Computer generation of pairing schemes for RNA molecules. J Theor Biol. 1972 Feb;34(2):363–378. doi: 10.1016/0022-5193(72)90168-3. [DOI] [PubMed] [Google Scholar]
- Kearns D. R., Wong Y. P. Investigation of the secondary structure of Escherichia coli 5 S RNA by high-resolution nuclear magnetic resonance. J Mol Biol. 1974 Aug 25;87(4):755–774. doi: 10.1016/0022-2836(74)90083-7. [DOI] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Laser Raman evidence for new cloverleaf secondary structures for eukaryotic 5.8S RNA and prokaryotic 5S RNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4901–4905. doi: 10.1073/pnas.75.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison J. T. Primary structure of RNA. Annu Rev Biochem. 1968;37:131–148. doi: 10.1146/annurev.bi.37.070168.001023. [DOI] [PubMed] [Google Scholar]
- Osterberg R., Sjöberg B., Garrett R. A. Molecular model for 5-S RNA. A small-angle x-ray scattering study of native, denatured and aggregated 5-S RNA from Escherichia coli ribosomes. Eur J Biochem. 1976 Sep 15;68(2):481–487. doi: 10.1111/j.1432-1033.1976.tb10835.x. [DOI] [PubMed] [Google Scholar]
- Raacke I. D. "Cloverleaf" conformation for 5S RNAs. Biochem Biophys Res Commun. 1968 May 23;31(4):528–533. doi: 10.1016/0006-291x(68)90509-3. [DOI] [PubMed] [Google Scholar]
- Ramstein J., Buckingham R. H. Tritium exchange on transfer RNA: slowly exchanging protons sensitive to a change in the dihydrouridine stem. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1567–1571. doi: 10.1073/pnas.78.3.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum H., Englander S. W. Open states in native polynucleotides. I. Hydrogen-exchange study of adenine-containing double helices. J Mol Biol. 1975 Feb 15;92(1):55–78. doi: 10.1016/0022-2836(75)90091-1. [DOI] [PubMed] [Google Scholar]
- Teitelbaum H., Englander S. W. Open states in native polynucleotides. II. Hydrogen-exchange study of cytosine-containing double helices. J Mol Biol. 1975 Feb 15;92(1):79–92. doi: 10.1016/0022-2836(75)90092-3. [DOI] [PubMed] [Google Scholar]
- Weidner H., Yuan R., Crothers D. M. Does 5S RNA function by a switch between two secondary structures? Nature. 1977 Mar 10;266(5598):193–194. doi: 10.1038/266193a0. [DOI] [PubMed] [Google Scholar]


