Abstract
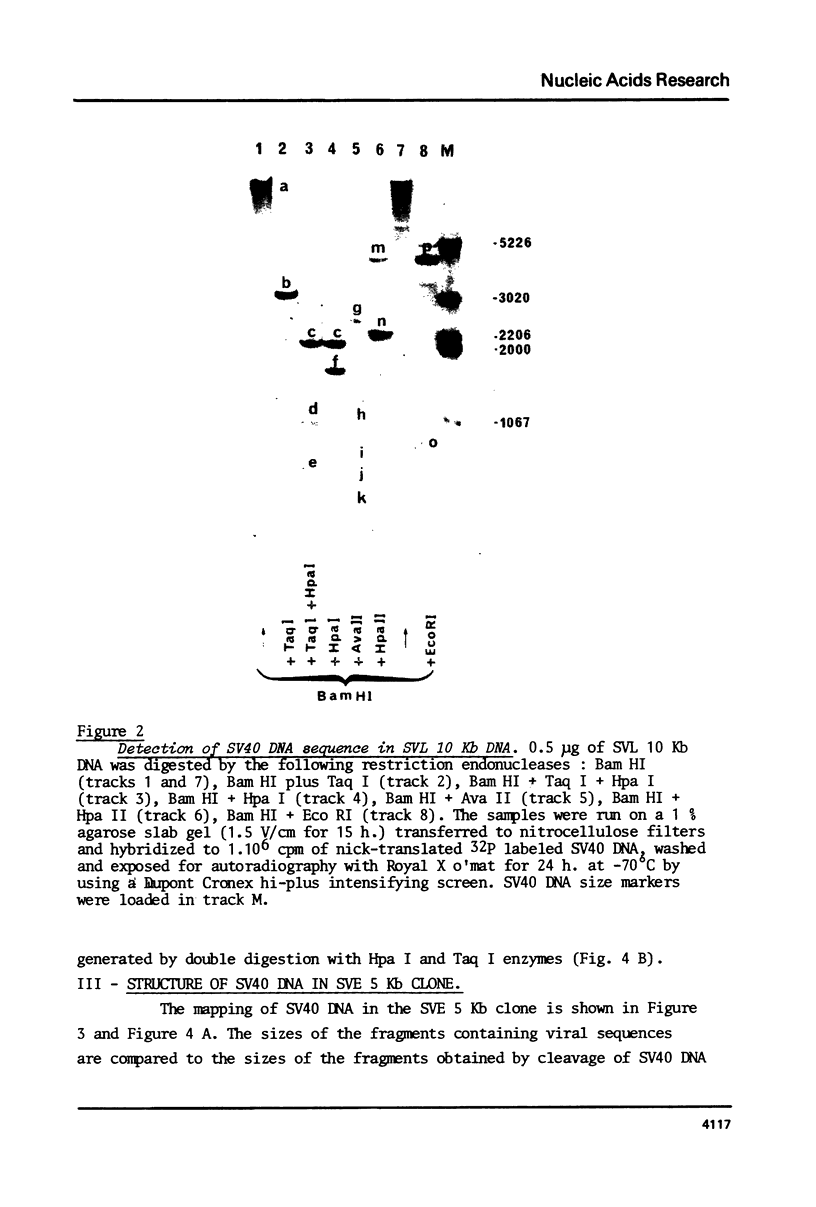
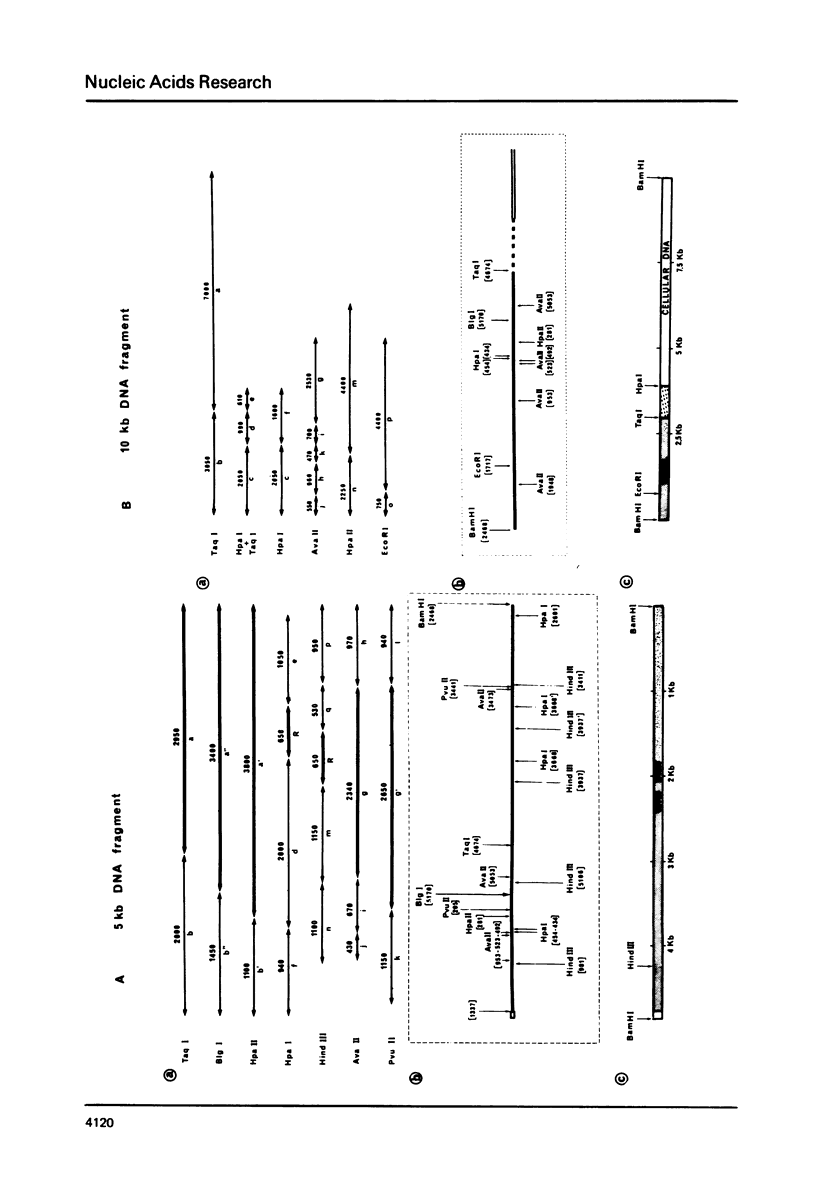
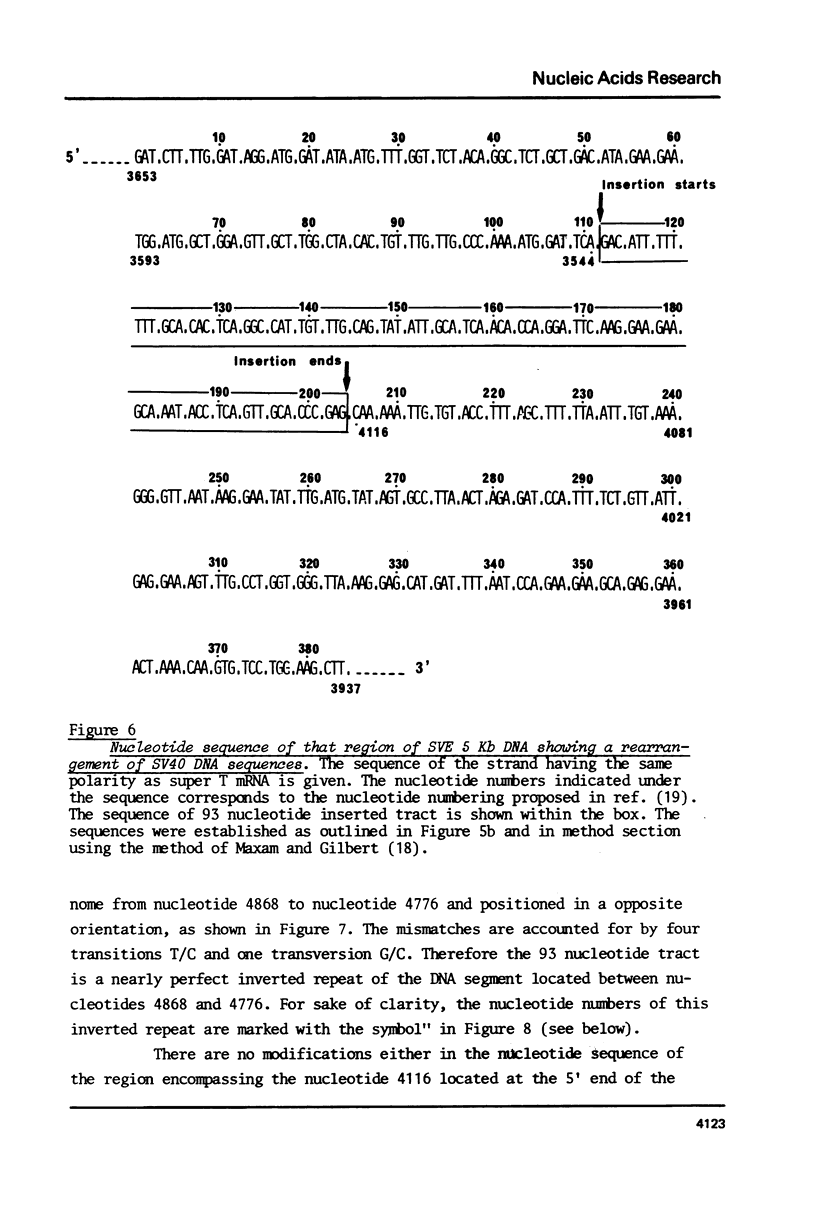
It has been reported that SV40-transformed V 11 F 1 clone 1 subclone 7 rat cells (subclone 7) produce a super T antigen of 115,000 M. This super T antigen is entirely SV40 coded and is synthesized by translation of an elongated form of SV40 early mRNA (May, E., Kress, M. Daya-Grosjean, L., Monier, R. and May, P. (1981) J. Virol., 37, 24-35). The results reported here show that there is only one independent insertion of viral DNA in the cellular genome of subclone 7 cells. When DNA from subclone 7 cells was cleaved with Bam HI endonuclease two distinct SV40 sequence containing fragments were generated with sizes of 5 Kb and 10 Kb, respectively. Two recombinant cosmids were constructed by insertion of the 5 Kb and 10 Kb fragments, respectively, into cosmid pHC 79. Using restriction map analysis and nucleotide sequencing, we showed that the 5 Kb fragment actually contained the complete sequence of a gene encoding super T antigen. As compared to the normal SV40 early gene, the sequence of super T gene showed the following rearrangements: (i) The segment between nucleotides 4116 - 3544 was duplicated in a direct order and (ii) these two copies of 573 nucleotide sequence were separated by a 93 nucleotide tract which was a nearly perfect inverted repeat of the segment located between nucleotides 4868 and 4776 (nucleotide numbering used here = Weissmann number +17).
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birg F., Dulbecco R., Fried M., Kamen R. State and organization of polyoma virus DNA sequences in transformed rat cell lines. J Virol. 1979 Feb;29(2):633–648. doi: 10.1128/jvi.29.2.633-648.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. doi: 10.1016/0092-8674(76)90118-5. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Mandel J. L., Chambon P. Ovalbumin gene is split in chicken DNA. Nature. 1977 Nov 24;270(5635):314–319. doi: 10.1038/270314a0. [DOI] [PubMed] [Google Scholar]
- Campo M. S., Cameron I. R., Rogers M. E. Tandem integration of complete and defective SV40 genomes in mouse-human somatic cell hybrids. Cell. 1978 Dec;15(4):1411–1426. doi: 10.1016/0092-8674(78)90065-x. [DOI] [PubMed] [Google Scholar]
- Chang C., Simmons D. T., Martin M. A., Mora P. T. Identification and partial characterization of new antigens from simian virus 40-transformed mouse cells. J Virol. 1979 Aug;31(2):463–471. doi: 10.1128/jvi.31.2.463-471.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni V., Dailey L., Basilico C. Amplification of integrated viral DNA sequences in polyoma virus-transformed cells. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3850–3854. doi: 10.1073/pnas.77.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna K. J., Sack G. H., Jr, Nathans D. Studies of simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol. 1973 Aug 5;78(2):363–376. doi: 10.1016/0022-2836(73)90122-8. [DOI] [PubMed] [Google Scholar]
- Gannon F., Jeltsch J. M., Perrin F. A detailed comparison of the 5'-end of the ovalbumin gene cloned from chicken oviduct and erythrocyte DNA. Nucleic Acids Res. 1980 Oct 10;8(19):4405–4421. doi: 10.1093/nar/8.19.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garapin A. C., Lepennec J. P., Roskam W., Perrin F., Cami B., Krust A., Breathnach R., Chambon P., Kourilsky P. Isolation by molecular cloning of a fragment in the split ovalbumin gene. Nature. 1978 Jun 1;273(5661):349–354. doi: 10.1038/273349a0. [DOI] [PubMed] [Google Scholar]
- Hiscott J., Murphy D., Defendi V. Amplification and rearrangement of integrated SV40 DNA sequences accompany the selection of anchorage-independent transformed mouse cells. Cell. 1980 Nov;22(2 Pt 2):535–543. doi: 10.1016/0092-8674(80)90363-3. [DOI] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Hohn B. In vitro packaging of lambda and cosmid DNA. Methods Enzymol. 1979;68:299–309. doi: 10.1016/0076-6879(79)68021-7. [DOI] [PubMed] [Google Scholar]
- Kress M., May E., Cassingena R., May P. Simian virus 40-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. J Virol. 1979 Aug;31(2):472–483. doi: 10.1128/jvi.31.2.472-483.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. A map of temperature-sensitive mutants of simian virus 40. Virology. 1975 Jul;66(1):70–81. doi: 10.1016/0042-6822(75)90179-8. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- May E., Kress M., Daya-Grosjean L., Monier R., May P. Mapping of the viral mRNA encoding a super-T antigen of 115,000 daltons expressed in simian virus 40-transformed rat cell lines. J Virol. 1981 Jan;37(1):24–35. doi: 10.1128/jvi.37.1.24-35.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P., Kress M., Lange M., May E. New genetic information expressed in SV40-transformed cells: characterization of the 55K proteins and evidence for unusual SV40 mRNAs. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):189–200. doi: 10.1101/sqb.1980.044.01.022. [DOI] [PubMed] [Google Scholar]
- McCormick F., Chaudry F., Harvey R., Smith R., Rigby P. W., Paucha E., Smith A. E. T antigens of SV40-transformed cells. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):171–178. doi: 10.1101/sqb.1980.044.01.020. [DOI] [PubMed] [Google Scholar]
- Mougneau E., Birg F., Rassoulzadegan M., Cuzin F. Integration sites and sequence arrangement of SV40 DNA in a homogeneous series of transformed rat fibroblast lines. Cell. 1980 Dec;22(3):917–927. doi: 10.1016/0092-8674(80)90569-3. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohmori H., Ohtsubo E. Nucleotide-sequence analysis of Tn3 (ap): implications for insertion and deletion. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1269–1277. doi: 10.1101/sqb.1979.043.01.144. [DOI] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sager R., Anisowicz A., Howell N. Genomic rearrangements in a mouse cell line containing integrated SV40 DNA. Cell. 1981 Jan;23(1):41–50. doi: 10.1016/0092-8674(81)90268-3. [DOI] [PubMed] [Google Scholar]
- Smith A. E., Smith R., Paucha E. Characterization of different tumor antigens present in cells transformed by simian virus 40. Cell. 1979 Oct;18(2):335–346. doi: 10.1016/0092-8674(79)90053-9. [DOI] [PubMed] [Google Scholar]
- Van Heuverswyn H., Fiers W. Nucleotide sequence of the Hind-C fragment of simian virus 40 DNA. Comparison of the 5'-untranslated region of wild-type virus and of some deletion Mutants. Eur J Biochem. 1979 Oct;100(1):51–60. doi: 10.1111/j.1432-1033.1979.tb02032.x. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]






