Abstract
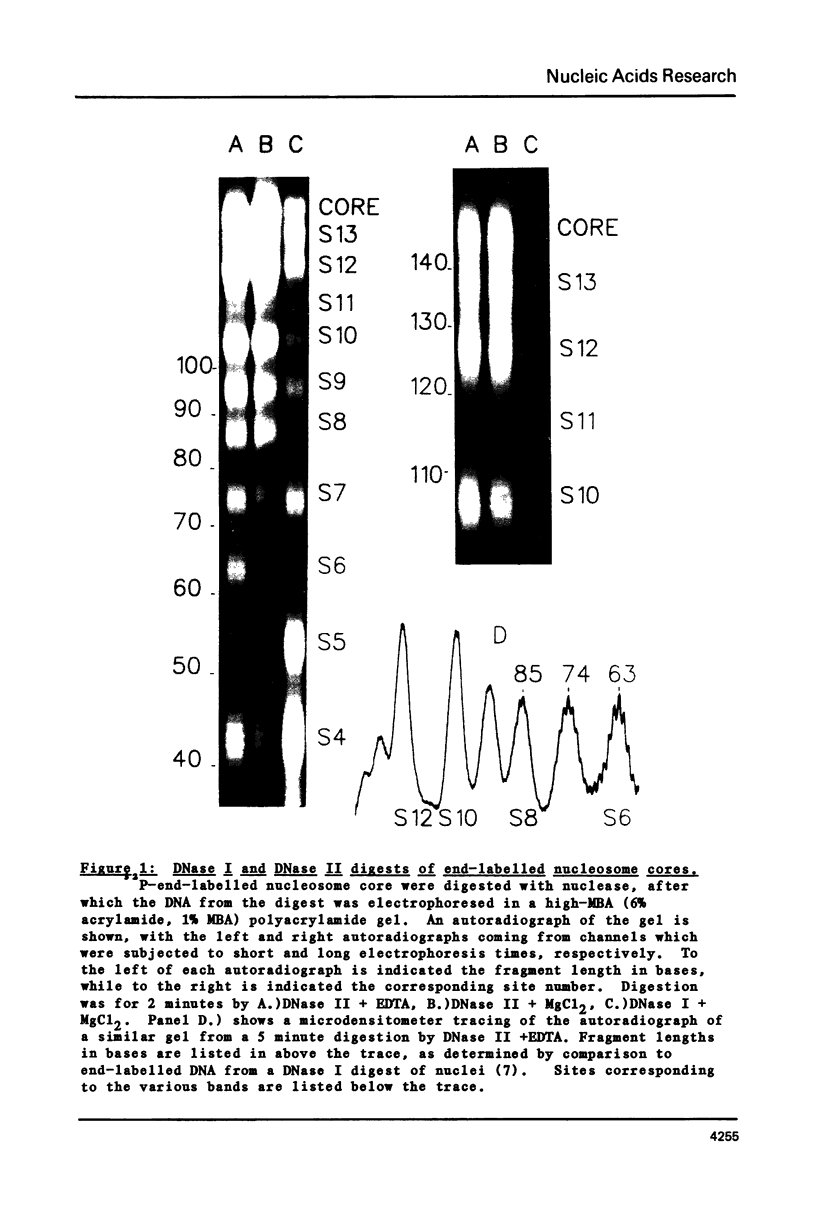
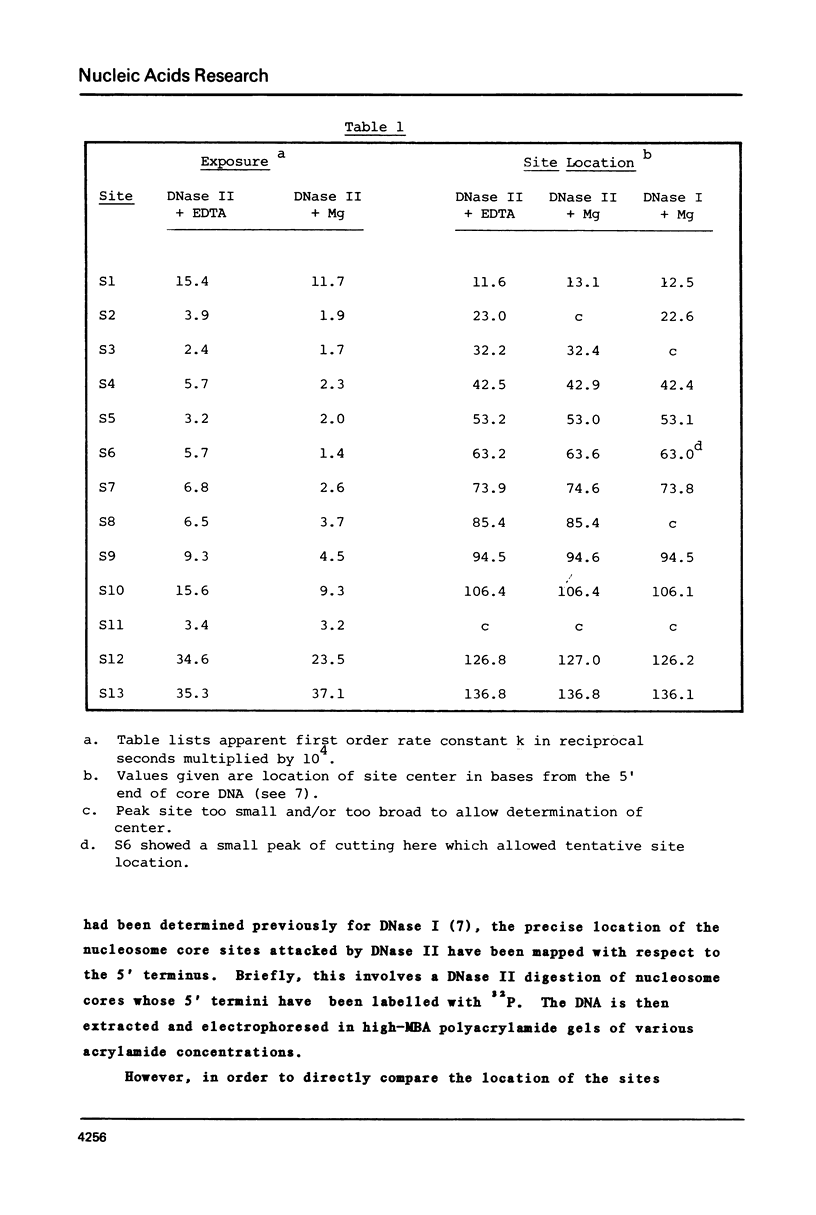
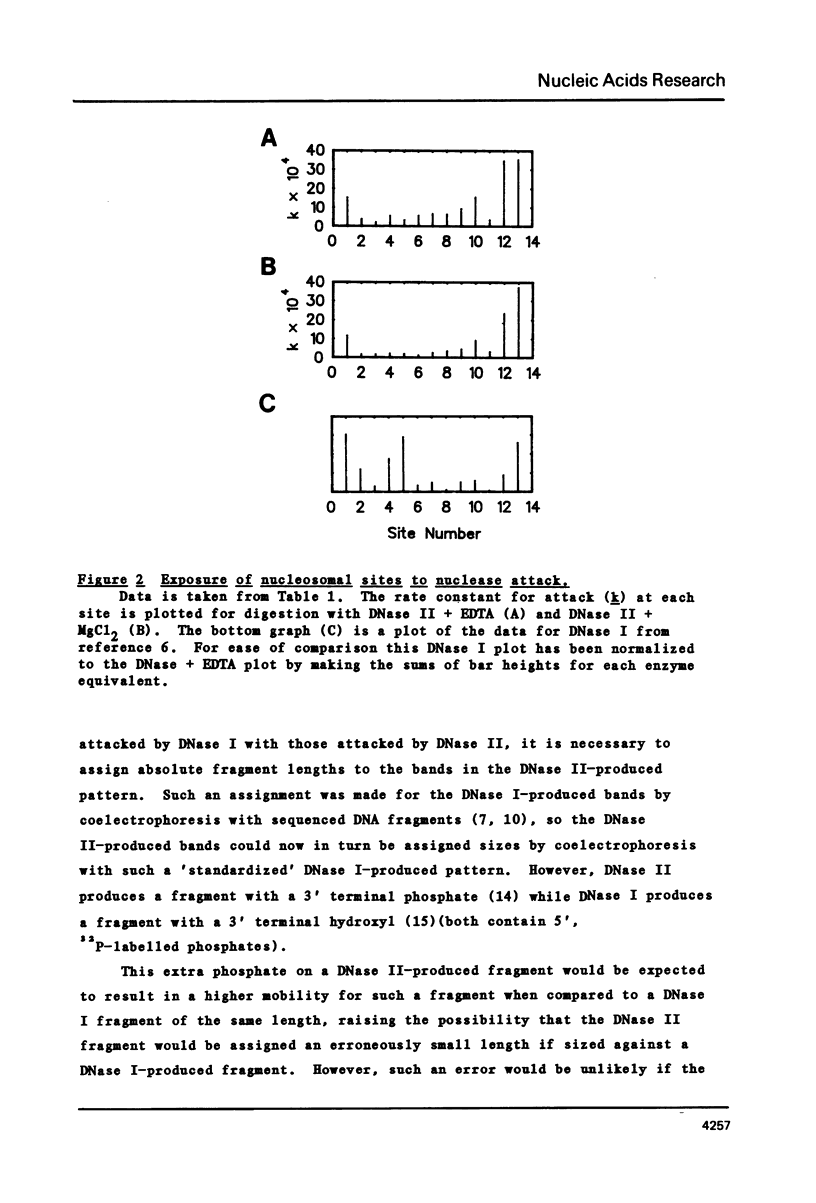
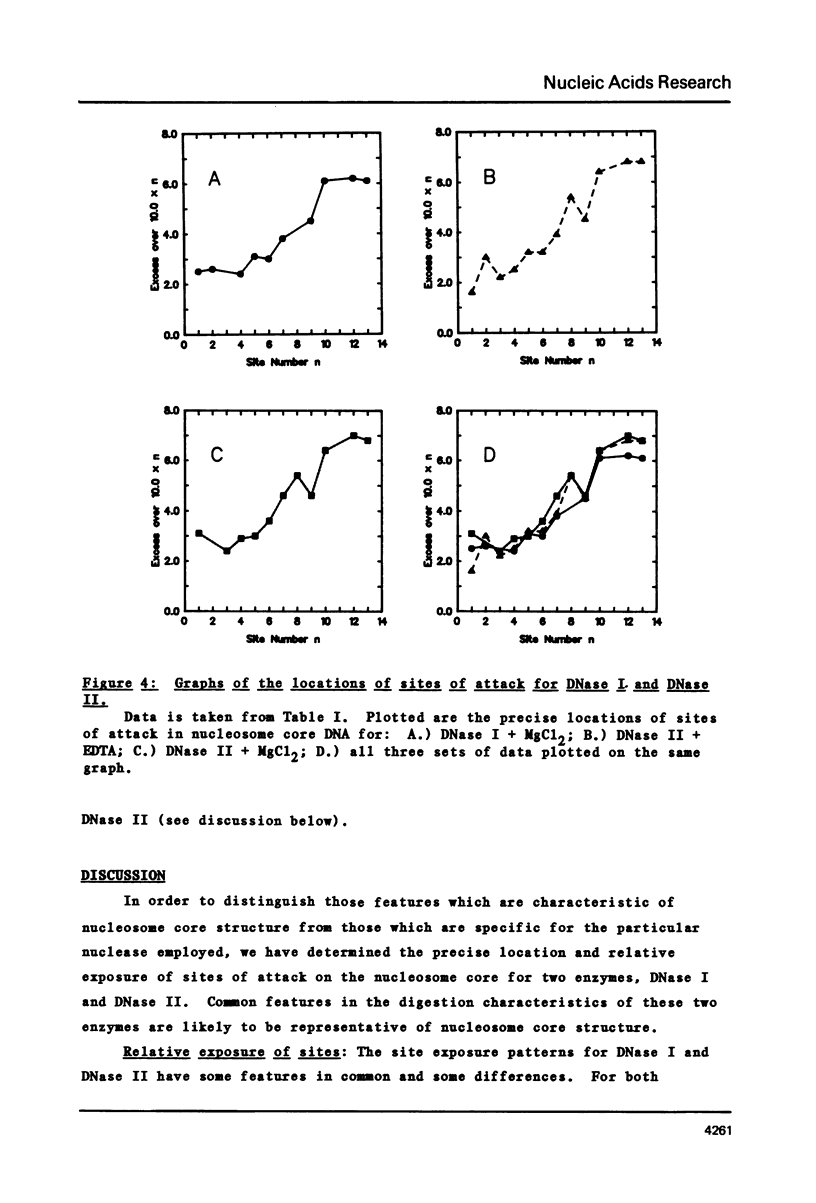
The precise locations and relative exposures of the DNase II-accessible sites in the nucleosome core DNA are determined using techniques previously employed for the enzyme DNase I. It is found that there are a number of similarities between the site exposure patterns for the two enzymes but that in general the DNase II seems to discriminate less among adjacent sites' accessibilities than does DNase I. The two enzymes attack essentially the same positions in the DNA, the average difference between the precise location of the site being less than one-half base for the two enzymes. Such close similarities in the digestion patterns of two enzymes with such different mechanisms of scission show that the patterns reflect the structure of the nucleosome core and not merely the properties of the particular enzyme used.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bryan P. N., Wright E. B., Olins D. E. Core nucleosomes by digestion of reconstructed histone-DNA complexes. Nucleic Acids Res. 1979 Apr;6(4):1449–1465. doi: 10.1093/nar/6.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P. Summary: the molecular biology of the eukaryotic genome is coming of age. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1209–1234. doi: 10.1101/sqb.1978.042.01.122. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Brown R. S., Richmond T., Rushton B., Lutter L. C., Klug A. X-ray diffraction study of a new crystal form of the nucleosome core showing higher resolution. J Mol Biol. 1981 Feb 5;145(4):757–769. doi: 10.1016/0022-2836(81)90313-2. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Pardon J. F. Structure and function of chromatin. Annu Rev Genet. 1979;13:197–233. doi: 10.1146/annurev.ge.13.120179.001213. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Characterization of DNase-I cleavage sites in the nucleosome. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):137–147. doi: 10.1101/sqb.1978.042.01.015. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Deoxyribonuclease I produces staggered cuts in the DNA of chromatin. J Mol Biol. 1977 Nov 25;117(1):53–69. doi: 10.1016/0022-2836(77)90022-5. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Precise location of DNase I cutting sites in the nucleosome core determined by high resolution gel electrophoresis. Nucleic Acids Res. 1979 Jan;6(1):41–56. doi: 10.1093/nar/6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell D. J., Searcy D. G., Sutcliffe J. G. A restriction enzyme Tha I from the thermophilic mycoplasma Thermoplasma acidophilum. Nucleic Acids Res. 1978 Jun;5(6):1729–1739. doi: 10.1093/nar/5.6.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D., Lutter L., Klug A., Levitt M., Crick F. H. Periodicity of deoxyribonuclease I digestion of chromatin. Science. 1979 May 25;204(4395):855–858. doi: 10.1126/science.441739. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Melchior W., Jr, Felsenfeld G. DNAase I, DNAase II and staphylococcal nuclease cut at different, yet symmetrically located, sites in the nucleosome core. Cell. 1978 Jul;14(3):611–627. doi: 10.1016/0092-8674(78)90246-5. [DOI] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Rushizky G. W., Simpson R. T. DNase-sensitive sites in nucleosomes. Their relative suspectibilities depend on nuclease used. J Biol Chem. 1977 May 10;252(9):3003–3006. [PubMed] [Google Scholar]