Abstract
Study Objectives:
EEG slow waves are the hallmark of deep NREM sleep and may reflect the restorative functions of sleep. Evidence suggests that increased sleep slow waves after sleep deprivation reflect plastic synaptic processes, and that brain-derived neurotrophic factor (BDNF) is causally involved in their homeostatic regulation. The functional Val66Met polymorphism of the gene encoding pro-BDNF causes impaired activity-dependent secretion of mature BDNF protein. We investigated whether this polymorphism contributes to the pronounced inter-individual variation in sleep slow wave activity (SWA) in humans.
Setting:
Sleep laboratory in temporal isolation unit.
Participants:
Eleven heterozygous Met allele carriers and 11 individually sex- and age-matched Val/Val homozygotes.
Interventions:
Forty hours prolonged wakefulness.
Measurements and Results:
Cognitive performance, subjective state, and waking and sleep EEG in baseline and after sleep deprivation were studied. Val/Val homozygotes showed better response accuracy than Met allele carriers on a verbal 2-back working memory task. This difference did not reflect genotype-dependent differences in sleepiness, well-being, or sustained attention. In baseline and recovery nights, deep stage 4 sleep and NREM sleep intensity as quantified by EEG SWA (0.75-4.5 Hz) were higher in Val/Val compared to Val/Met genotype. Similar to sleep deprivation, the difference was most pronounced in the first NREM sleep episode. By contrast, increased activity in higher EEG frequencies (> 6 Hz) in wakefulness and REM sleep was distinct from the effects of prolonged wakefulness.
Conclusion:
BDNF contributes to the regulation of sleep slow wave oscillations, suggesting that genetically determined variation in neuronal plasticity modulates NREM sleep intensity in humans.
Citation:
Bachmann V; Klein C; Bodenmann S; Schäfer N; Berger W; Brugger P; Landolt HP. The BDNF Val66Met polymorphism modulates sleep intensity: EEG frequency- and state-specificity. SLEEP 2012;35(3):335-344.
Keywords: Brain derived neurotrophic factor, electroencephalogram, sleep homeostasis, slow wave sleep, synaptic plasticity
INTRODUCTION
Deep NREM sleep is characterized by slow brain oscillations which are homeostatically regulated and can be reliably quantified by electroencephalographic (EEG) slow wave activity (SWA; 0.75-4.5 Hz range).1 A current hypothesis on the function of sleep posits that neuronal plastic changes occurring during wakefulness are causally related to sleep SWA.2 Brain-derived neurotrophic factor (BDNF) is an established mediator of activity-dependent synaptic plasticity.3,4 Studies in vitro and in genetically modified mice suggest that this neurotrophin is important for hippocampal long-term potentiation and rearrangements of cortical circuits due to sensory input.5,6 With respect to the sleep-wake cycle, cortical BDNF in rats is higher after wakefulness than after sleep, and increases after sleep deprivation.7–9 Developmental studies indicate that expression of cortical Bdnf reflects the onset of adult sleep homeostasis.8 Moreover, in adult rats, the degree of exploratory behavior predicts the level of BDNF as well as of SWA during subsequent sleep.10 Finally, pharmacological experiments such as injections of exogenous BDNF, an anti-BDNF antibody, and an inhibitor of tyrosine kinase B receptor, the high-affinity BDNF receptor, consistently suggest that this relationship is causal.11
Based on this evidence obtained in animals, we hypothesized that functional genetic variation of BDNF in humans affects slow brain oscillations during sleep. The gene encoding BDNF is located on chromosome 11q13 and contains a common functional single nucleotide polymorphism (SNP; SNP-ID: rs6265) which causes a valine (Val) to methionine (Met) amino acid substitution at codon 66 of pro-BDNF (Val66Met). This precursor peptide is proteolytically cleaved to mature BDNF protein. Presence of the Met allele modifies the intracellular distribution, processing and secretion of BDNF in cultured rat hippocampal neurons when compared to cells expressing the Val isoform.12 More specifically, met-BDNF bound to green fluorescence protein (GFP) exhibits diffuse localization mainly in cell bodies, whereas val-BDNF-GFP is found in secretory granules mostly localized in dendrites. Moreover, the activity-dependent secretion of BDNF is markedly reduced in neurons transfected with met-BDNF-GFP. Studies employing fMRI in human subjects performing the N-back working memory task and MRI spectroscopy to measure N-acetyl aspartate as an intracellular marker of neuronal function corroborated that the Met allele is associated with altered hippocampal response and impaired neuronal integrity/synaptic activity.12,13 On the behavioral level, homozygous Val/Val allele carriers often perform better than Val/Met heterozygotes on cognitive tasks, in particular on tasks probing working memory, and show reduced stress-induced anxiety-like behavior.12–15
We investigated whether the Val66Met polymorphism of BDNF modulates working memory, subjective sleepiness, personal well-being, and sustained vigilant attention during sleep deprivation. Moreover, we quantified the waking and sleep EEG in baseline and after one night without sleep. In accordance with previous work, the data confirm that BDNF genotype affects performance on a 2-back working memory task. Furthermore, they demonstrate for the first time that functional genetic variation of BDNF in healthy humans contributes to the pronounced inter-individual differences in slow wave oscillations in NREM sleep. Similar to the effects of sleep deprivation, the genotype-dependent differences were frequency-specific and most pronounced in the beginning of baseline and recovery nights. On the contrary, EEG alterations in wakefulness and REM sleep were not reminiscent of the effects of prolonged wakefulness. These findings are consistent with the notion that BDNF contributes to the regulation of SWA in NREMS and that sleep slow oscillations may reflect plastic synaptic processes.
MATERIALS AND METHODS
Subject Recruitment and Genotyping
The local ethics committees for research on human subjects reviewed and approved study protocol and all experimental procedures. They were conducted according to the principles of the Declaration of Helsinki and all participants provided written informed consent.
Forty-three young adults were genotyped for the 196G > A polymorphism of the coding DNA sequence (aka Val66Met variant of the protein sequence) of BDNF (SNP identification number: rs6265). Genomic DNA was extracted from 3 mL fresh EDTA blood and genotypes were determined with a TaqMan SNP Genotyping Assay (Applied Biosystems, Rotkreuz, Switzerland; Assay ID: C_11592758_10). Allele-specific polymerase chain reaction (PCR) on a MJ Research PTC-225 thermal cycler (MJ Research/Bio-Rad, Reno, NV, USA) was performed. The reaction volume contained 20 ng genomic DNA, 4 μL TaqMan Universal Master Mix (Applied Biosystems, Rotkreuz, Switzerland), 0.4 μL 20X SNP Genotyping Assay Mix and 1.6 μL H2O. Annealing temperature was set to 60°C. After running the PCR, an end-point fluorescence measurement with the SDS 2.2 software package (Applied Biosystems, Rotkreuz, Switzerland) was obtained, to examine the samples and discriminate between the specific alleles. All PCRs were independently replicated for data confirmation. Allele and genotype frequencies (Val/Val: 65.1% [28/43], Val/Met: 32.6% [14/43], Met/Met: 2.3% [1/43]) were as expected in healthy Caucasian populations12 and did not deviate from Hardy-Weinberg equilibrium (χ2 = 0.24, P > 0.6).
Among the 14 Val/Met genotypes, 11 fulfilled all inclusion criteria (20-30 years old, right-handed, nonsmoker, body mass index [BMI]: 18-25 kg/m2, no extreme diurnal preference, Epworth Sleepiness Scale [ESS] score < 10, moderate alcohol and caffeine consumption) and agreed to participate in a sleep deprivation study. They were individually matched in case-control fashion to 11 Val/Val allele carriers based on age, sex, BMI, diurnal preference, ESS score, trait anxiety,16 and habitual consumption of alcohol and caffeine (Table 1). Women were matched with respect to their phase of the menstrual cycle (follicular [n = 2] or luteal [n = 2] phase). All subjects reported to have no medical history of neurologic and psychiatric disease and not taking any medication. They were polysomnographically screened prior to the study, to exclude poor sleep efficiency and unrecognized sleep disorders.
Table 1.
Subject characteristics
| Val/Val | Val/Met | P-value | |
|---|---|---|---|
| Sex (female / male) | 4 / 7 | 4 / 7 | n/a |
| Age (years) | 23.7 ± 0.6 | 24.0 ± 0.8 | 0.3 |
| Alcohol consumption (drinks/week) | 2.9 ± 0.6 | 2.9 ± 0.9 | 0.9 |
| Caffeine consumption (mg/day) | 124.1 ± 27.1 | 116.4 ± 30.2 | 0.8 |
| Body mass index (kg/m2) | 21.6 ± 0.4 | 22.2 ± 0.4 | 0.3 |
| Trait anxiety | 38.7 ± 2.2 | 36.8 ± 2.4 | 0.4 |
| Diurnal preference | 2.9 ± 0.3 | 3.2 ± 0.1 | 0.3 |
| Daytime sleepiness | 6.6 ± 0.5 | 7.5 ± 0.6 | 0.3 |
Values represent means ± SEM (n = 11 per group). Caffeine consumption was estimated based on the following average caffeine contents per serving: Coffee: 100 mg; Ceylon or green tea: 30 mg; Cola drink: 40 mg (2 dL); Energy drink: 80 mg (2 dL); Chocolate: 50 mg (100 g). Trait anxiety,16 diurnal preference,50 and daytime sleepiness51 values were based on validated questionnaires. P-values refer to 2-tailed, paired t-tests.
Study Design
For 2 weeks prior to the experiment, study volunteers were asked to abstain from all sources of caffeine (coffee, tea, coke, chocolate, and energy drinks), to wear a wrist-activity monitor on the non-dominant arm, and to keep a sleep-wake diary. For 3 days before and during the study, they abstained from alcohol and maintained regular 8-h sleep/16-h wake cycles, with bed times scheduled from 24:00 to 08:00. When subjects did not obey these instructions, they were excluded from the study. After arrival in the sleep laboratory, breath ethanol concentration was determined and saliva samples for the measurement of caffeine were collected.
All subjects spent 4 nights (time in bed: 24:00-08:00) and 2 days in the sleep laboratory. The first night served for adaptation to the laboratory environment, the second night provided the baseline. The following 2 days and 1 night, subjects were not allowed to sleep; thus were kept awake for 40 hours. All subjects were under constant supervision by members of the research team. They were free to read, study, play games, watch movies, and occasionally take a walk outside the laboratory. Three times a day, they either prepared a meal by themselves and ate in the kitchen adjacent to the sleep laboratory or they were served in the University cafeteria. The sleep laboratory was controlled for constant temperature and light intensity, which was kept below 150 lux. The last night of the study served as recovery night from prolonged wakefulness.
Working Memory, Subjective Sleepiness, Sustained Vigilant Attention, and Personal Well-Being
Working memory was assessed with a visual verbal 2-back task, administered at 6-h intervals, starting at 08:45 on day one of prolonged wakefulness. On a computer screen, subjects were asked to compare a single consonant with a consonant presented 2 trials earlier.17 When the target letter was the same as the letter 2 trials before (e.g., f-M-F), subjects had to respond with their right index finger. By contrast, when these 2 consonants were different, subjects had to respond with their right middle finger. They were instructed to respond as fast and accurately as possible. The task consisted of 24 targets and 56 non-targets, which were presented in random order. In addition to a training session on the evening before study start, each session was preceded by a short practice block with feedback, including 3 targets and 7 non-targets. The actual task lasted for roughly 10 minutes. Mean response accuracy (percentage of correct responses) and speed (reciprocal values of reaction time) were calculated for each individual and session. The data of the first session after awakening from sleep were not analyzed because of possible sleep inertia.
To estimate the evolution of subjective sleepiness and sustained vigilant attention during sleep deprivation, participants completed at 3-h intervals the Stanford Sleepiness Scale (SSS)18 and a 10-min psychomotor vigilance task (PVT), for the first time at 08:15 after wakening. The PVT is one of the most often used measures of sustained vigilant attention in human sleep and chronobiology research.19 Here, the task was implemented on a PC using the software e-Prime (Psychology Software Tools Inc., Pittsburgh, PA, USA). Subjects were instructed to press a button on a response box as quickly as possible, to stop a digital millisecond counter appearing in the middle of the computer screen. They conducted a training session on the evening prior the adaptation night. In each session, a total of 100 stimuli were presented at variable (2-10 s) inter-stimulus intervals (task duration: ∼ 10 min). All reaction times below 100 ms (“errors of commission”) were excluded from analyses. Here we report the outcome variable 90th - 10th inter-percentile range of speed. During 20 min before each PVT session, all participants stayed in the non-social environment of their bedroom and were engaged in filling in questionnaires and recording of the waking EEG.
Subjective well-being was assessed with von Zerssens's “Befindlichkeits-Skala (Bf-S),”20 administered at 10:45 and 22:45 on day 1 (baseline) and 2 (sleep deprivation) of prolonged wakefulness.
Waking EEG Recordings
The EEG, mental electromyogram (EMG), bipolar electrooculogram (EOG) and electrocardiogram (ECG) were recorded in standardized manner in 14 sessions at 3-h intervals, starting 15 min after lights-on from the baseline night. To record the waking EEG, participants were instructed to relax comfortably on a chair and to place their chin on an individually adjusted head-rest. A 3-min period with eyes closed was followed by a 5-min period with eyes open, while subjects were fixating a black dot attached at a distance of 3 m to the wall. When individuals tended to fall asleep (i.e., reduced EEG alpha activity, rolling eyes), they were alerted by addressing them over the intercom. One hour before each waking EEG recording, subjects had to stay in the laboratory, and 15 min before data collection they were by themselves in their bedroom.
All bioelectric signals were recorded with Rembrandt Datalab (Version 8; Embla Systems, Broomfield, CO, USA) and the polygraphic amplifier Artisan (Micromed, Mogliano Veneto, Italy). Analogue signals were conditioned by a high-pass filter (EEG: —3 dB at 0.15 Hz; EMG: 10 Hz; ECG: 1 Hz) and an antialiasing low-pass filter (—3 dB at 67.2 Hz), digitized, and transmitted via fiberoptic cables to a personal computer. Data were sampled with a frequency of 256 Hz. The EEG signal was recorded from 1 referential (C3A2) and 3 bipolar (fronto-central [FC], centro-parietal [CP], parieto-occipital [PO]) derivations along the left and right antero-posterior axes. Artifacts in all derivations were visually identified. The power spectra (fast Fourier transform, Hanning window) of artifact-free, 50%-overlapping 2-s (waking EEG) or 4-s (sleep EEG) epochs were computed with MATLAB (The MathWorks Inc, Natick, MA, USA). Analyses were performed for referential (C3A2) and bipolar derivations along the antero-posterior axes. Because SWA did not differ between left and right hemispheres, mean values over left and right FC, CP, and PO derivations are reported. Absolute and/or relative EEG power spectra between 0 and 20 Hz (0.5-Hz resolution) of the 5-min recording periods with eyes open were computed. The data of the C3A2 derivation are reported.
Sleep EEG Recordings
During all experimental nights, all-night continuous recordings of EEG, EOG, EMG, and ECG were obtained in the same way as during wakefulness. Standard sleep stages were visually scored for 20-s epochs (C3A2 derivation).21 Consecutive 4-s EEG spectra were averaged over 5 consecutive epochs and matched with the sleep scores. Twenty-sec epochs with movement- and arousal-related artifacts were visually identified and eliminated. To compute all-night power spectra in NREM (stages 2, 3, and 4) and REM sleep, all artifact-free 20-s values were averaged. In recovery nights, data analyses were restricted to the first 8 h of the 10-h sleep opportunity. Absolute and/or relative power values between 0 and 20 Hz of C3A2, FC, CP, and PO EEG derivations were computed for consecutive 0.25-Hz bins. The evolution during the nights of SWA (0.75-4.5 Hz) in NREM sleep and alpha activity (8.5-10.5 Hz) in REM sleep were also quantified.
Data Analysis and Statistics
Working memory, subjective sleepiness and well-being, sustained vigilant attention, sleep architecture, and waking and sleep EEG in baseline, during/after prolonged wakefulness, and during recovery were compared between individually matched Val/Val and Val/Met genotype subjects of BDNF. EEG power was computed for consecutive 0.5 Hz (waking) and 0.25 Hz (sleep) bins and for specific frequency bands. The frequency bins and bands were indicated by the encompassing frequency range (i.e., the 1.0 Hz bin denotes 0.75-1.25 Hz in waking and 0.875-1.125 in NREM sleep). All statistical analyses were performed with SAS 9.1.3 software (SAS Institute, Cary, NC). If variables were not normally distributed, they were either log-transformed (absolute data of EEG) or analyzed by nonparametric tests. Mixed-effect analysis of variance (ANOVA) models included the factors “genotype” (Val/Val, Val/Met), “session” (6 [2-back task], 11 [SSS] or 4 [Bf-S]), “condition” (baseline, deprivation/recovery), “NREM sleep episode” (1-4), “2-min epoch” (1-15), “derivation” (FC, CP, PO), as well as their interactions. Tukey HSD tests were conducted to localize significant differences. The significance level for statistical tests was set at α < 0.05. If not stated otherwise, only significant results are reported.
RESULTS
Better Working Memory in Val/Val Genotype of BDNF
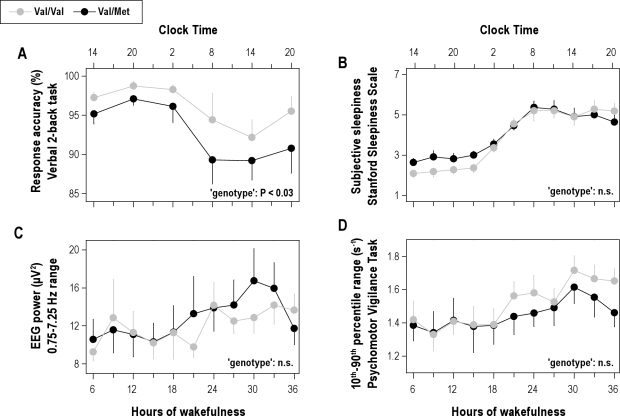
Corroborating the functional importance of the Val66Met polymorphism of BDNF, Val/Val homozygotes showed better response accuracy on a 2-back working memory task than Val/Met allele carriers (Figure 1A). Performance in both genotypes exhibited a diurnal modulation and was reduced by roughly 10% after sleep deprivation. The genotype-dependent difference between the groups persisted throughout prolonged wakefulness. Response speed was similar in both genotypes (data not shown).
Figure 1.
Better working memory in Val/Val genotype of BDNF. The data were analyzed with 2-way, mixed-model ANOVA with the factors “genotype” (Val/Val, Val/Met) and “session” (6 [2-back] or 11 [sleepiness and PVT] consecutive assessments). (A) Time course of response accuracy (percentage of correct responses) on a visual verbal 2-back task (means ± SEM). Starting after 6.75 h of wakefulness (at 14:45), each subject completed 6 test sessions at 6-h intervals. Tick marks on the x-axes were rounded down to the nearest hour. Val/Val allele carriers (gray dots, n = 11) performed better than Val/Met allele carriers (black dots, n = 11) throughout prolonged wakefulness (genotype: F1,40 = 5.1, P < 0.03; session: F5,41 = 11.8, P < 0.0001; genotype × session: F5,59 = 0.2, P > 0.9). (B) Time course of subjective sleepiness as quantified with the Stanford Sleepiness Scale. Mean scores (± SEM) for consecutive 3-h intervals, starting at 6.25 h of wakefulness (at 14:15) are plotted. Tick marks on the x-axes were rounded down to the nearest hour. Sleep deprivation similarly increased subjective sleepiness in both genotypes (genotype: F1,65 = 1.8, P = 0.1; session: F10,120 = 38.7, P < 0.0001; genotype × session: F10,131 = 0.94, P > 0.4). (C) Time course of EEG 0.75-7.25 Hz power in wakefulness. Mean absolute power values (± SEM) for consecutive 3-h intervals, starting at 6.25 h of wakefulness (at 14:15) are plotted. Tick marks on the x-axes were rounded down to the nearest hour. Sleep deprivation similarly affected EEG power in both genotypes (genotype: F1,45 = 0.4, P > 0.5; session: F10,167 = 15.9, P < 0.001; genotype × session: F10,117 = 0.6, P > 0.8). (D) Time course of global alertness as quantified with the 10th-to-90th inter-percentile range of reaction times on the psychomotor vigilance task. Mean scores (± SEM) for consecutive 3-h intervals, starting at 6.25 h of wakefulness (at 14:15) are plotted. Tick marks on the x-axes were rounded down to the nearest hour. Sleep deprivation induced similar response instability in Val/Val and Val/Met allele carriers of BDNF (genotype: F1,46 = 2.0, P > 0.1; session: F10,160 = 7.1, P < 0.0001; genotype × session: F10,114 = 0.3, P > 0.9).
To investigate whether the difference in working memory was caused by reduced vigilance, the evolution of subjective sleepiness, sustained vigilant attention and personal well-being were quantified during prolonged wakefulness. Subjective sleepiness (Figure 1B), absolute EEG 0.75-7.25 Hz activity (Figure 1C), global alertness (10th-90th inter-percentile range on the PVT; Figure 1D) and current strain (Val/Val: baseline: 20.6 ± 6.3, deprivation: 41.0 ± 5.8; Val/Met: baseline: 23.3 ± 4.1, deprivation: 48.1 ± 5.7; condition: F1,30 = 25.5, P < 0.0001) evolved similarly in both genotypes and did not differ between the groups.
Genotype-dependent difference in EEG activity is vigilance/sleep state-specific
To examine whether prolonged wakefulness and the Val66Met polymorphism of BDNF similarly affect brain oscillations, the EEG before and after sleep deprivation, as well as between Val/Val and Val/Met allele carriers were compared.
Effects of sleep deprivation
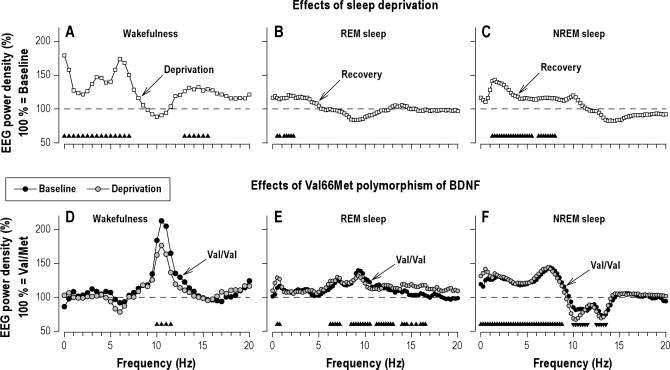
Prolonged wakefulness increased EEG power in delta and theta frequencies in wakefulness (0.25-7.25 Hz, Figure 2A) and NREM sleep (0.875-5.625 and 6.125-8.125 Hz, Figure 2C),and in low-delta frequencies in REM sleep (0.375-2.375 Hz, Figure 2B). The changes in wakefulness also included enhanced power in the 12.75-15.75 Hz band.
Figure 2.
Sleep deprivation and BDNF genotype modulate EEG oscillations in vigilance/sleep state-specific manner. Effects of sleep deprivation (A-C): To quantify the effects of sleep deprivation on the waking EEG (C3A2 derivation, eyes open), absolute EEG power values were averaged over 4 recording sessions (08:00, 11:00, 14:00, 17:00) after sleep loss (day 2, deprivation) and compared to the corresponding values in baseline (day 1). In REM sleep and NREM sleep, EEG activity in the recovery night was expressed as a percentage of the corresponding values in the baseline night (horizontal dashed lines at 100%). Geometric mean values were plotted for each 0.5-Hz bin in wakefulness, and for each 0.25-Hz bin in REM sleep and NREM sleep (n = 22). Triangles at the bottom of the panels indicate frequency bins, which differed significantly from baseline (condition: F1,30 ≥ 4.2, P < 0.05). Effects of the Val66Met polymorphism of BDNF (D-F): Absolute power values in individuals with Val/Val genotype (n = 11) were expressed as a percentage of the corresponding value of individuals with Val/Met genotype (n = 11; horizontal dashed lines at 100%). Wakefulness: Relative EEG power spectra in recording sessions at 8:00, 11:00, 14:00, and 17:00 on day 1 (baseline) and day 2 (deprivation). REM sleep and NREM sleep: Relative EEG power spectra in baseline and recovery nights. Geometric mean values were plotted for each 0.5-Hz bin in wakefulness, and for each 0.25-Hz bin in REM sleep and NREM sleep. Triangles at the bottom of the panels indicate frequency bins, in which power differed significantly between Val/Val and Val/Met genotypes (genotype: F1,30 ≥ 4.3, P < 0.05).
Effects of Val66Met polymorphism of BDNF
In rested (baseline) and sleep-deprived state, the Val66Met polymorphism of BDNF induced frequency-specific alterations in EEG activity in wakefulness and sleep. EEG alpha power (9.75-11.75 Hz) in wakefulness was roughly doubled in Val/Val homozygotes compared to Val/Met genotype subjects (Figure 2D). Also in REM sleep, alpha (8.375-10.625 and 11.125-13.125 Hz), but also theta (6.125-7.375 Hz) and sigma (13.875-16.625 Hz) activity were higher in Val/Val than in Met allele carriers (Figure 2E). Thus, except for 2 bins in REM sleep (0.375-0.875 Hz), the genotype-dependent differences in wakefulness and REM sleep were distinct from the EEG signature of sleep deprivation.
A different picture emerged in NREM sleep. EEG activity in the entire delta/theta range (0.125- 8.875 Hz) was consistently enhanced in the Val/Val genotype compared to Val/Met allele carriers (Figure 2F). By contrast, power was reduced in alpha (9.875-11.625 Hz) and sigma (12.375-13.625 Hz) frequencies. These genotype-dependent differences were reminiscent in shape and magnitude of the EEG changes in NREM sleep, which are reliably induced after 1 night of sleep deprivation (Figure 2C).
Of note, all genotype-dependent EEG differences in wakefulness, REM sleep and NREM sleep were remarkably similar in rested and sleep deprived state (“condition” × “genotype” interaction: Pall > 0.1).
More Slow Wave Sleep in Val/Val Genotype of BDNF
All study participants showed healthy sleep, including short sleep latency (< 12 min) and high sleep efficiency (> 93%). Nevertheless, the sleep deprivation- and genotype-dependent differences were also reflected in sleep architecture. Sleep deprivation reduced sleep latency, stage 1 sleep, and wakefulness after sleep onset, while slow wave sleep, total sleep time, and sleep efficiency were increased compared to baseline (Table 2). The effects were similar in both genotypes. In both baseline and recovery nights, however, Val/Val allele carriers spent roughly 20 min more in deep stage 4 sleep than Val/Met allele carriers. By contrast, superficial stage 2 sleep was reduced (Table 2).
Table 2.
Visually scored sleep variables in Val/Val and Val/Met genotypes of BDNF
| Variable | Val/Val (n = 11) |
Val/Met (n = 11) |
“Condition” | “Genotype” | “Condition’ × ‘genotype” | ||
|---|---|---|---|---|---|---|---|
| Baseline | Recovery | Baseline | Recovery | F1,30 (P) | F1,30 (P) | F1,30 (P) | |
| Episode | 469.8 ± 1.3 | 476.7 ± 0.8 | 467.9 ± 1.8 | 477.4 ± 0.6 | 68.7 (0.001) | 1.6 (0.21) | 0.4 (0.54) |
| TST | 448.8 ± 5.1 | 467.1 ± 1.7 | 454.4 ± 2.7 | 466.7 ± 1.7 | 38.0 (0.001) | 1.1 (0.30) | 1.4 (0.24) |
| Efficiency | 93.6 ± 1.1 | 97.3 ± 0.4 | 94.7 ± 0.6 | 97.2 ± 0.4 | 35.6 (0.001) | 1.1 (0.31) | 1.4 (0.24) |
| SL | 11.6 ± 1.8 | 3.2 ± 0.8 | 9.8 ± 1.3 | 2.6 ± 0.6 | 62.0 (0.001) | 1.5 (0.24) | 0.4 (0.54) |
| RL | 68.0 ± 8.5 | 79.2 ± 12.2 | 62.6 ± 3.7 | 79.5 ± 14.3 | 2.0 (0.17) | 0.8 (0.80) | 0.1 (0.77) |
| WASO | 10.2 ± 3.7 | 0.4 ± 0.2 | 4.3 ± 1.3 | 0.7 ± 0.2 | 12.2 (0.02) | 2.1 (0.15) | 2.5 (0.12) |
| Stage 1 | 32.2 ± 4.2 | 20.3 ± 4.0 | 31.2 ± 3.6 | 13.2 ± 2.0 | 40.9 (0.001) | 3.0 (0.09) | 1.7 (0.20) |
| Stage 2 | 201.8 ± 6.7 | 189.7 ± 9.8 | 220.3 ± 13.2 | 209.8 ± 12.0 | 1.7 (0.21) | 4.9 (0.04) | 0.0 (0.92) |
| Stage 3 | 39.1 ± 4.3 | 41.8 ± 5.9 | 41.2 ± 4.0 | 46.3 ± 2.9 | 1.0 (0.32) | 0.7 (0.40) | 0.1 (0.76) |
| Stage 4 | 73.6 ± 7.9 | 122.0 ± 10.5 | 56.6 ± 7.9 | 101.0 ± 11.3 | 34.4 (0.001) | 5.8 (0.02) | 0.1 (0.81) |
| SWS | 112.8 ± 9.8 | 163.8 ± 11.2 | 97.8 ± 10.5 | 147.3 ± 10.5 | 36.4 (0.001) | 3.6 (0.07) | 0.0 (0.93) |
| REM sleep | 102.0 ± 4.4 | 93.3 ± 8.5 | 105.1 ± 5.9 | 96.4 ± 7.4 | 3.0 (0.10) | 0.4 (0.54) | 0.0 (0.99) |
| MT | 9.0 ± 1.3 | 9.2 ± 1.3 | 11.2 ± 1.8 | 10.0 ± 1.4 | 0.2 (0.64) | 2.2 (0.15) | 0.5 (0.49) |
Mean values ± SEM in minutes (except sleep efficiency in %) for the first 480 min from lights-off.
Baseline, baseline night; Recovery, recovery night after 40 h of wakefulness; Episode, sleep episode, time after sleep onset until final awakening; TST, total sleep time; Efficiency, sleep efficiency, percentage of TST per 480 min; SL, sleep latency, time from lights-off to first occurrence of stage 2; RL, REM sleep latency, time from sleep onset to first occurrence of REM sleep; WASO, waking after sleep onset; SWS, slow wave sleep; MT, movement time. F- and P-values: 2-way mixed-model ANOVA with the factors “condition” (baseline, recovery) and “genotype” (Val/Val, Val/Met).
Elevated EEG Slow Wave Activity in NREM Sleep in Val/Val Genotype of BDNF
To further investigate whether the Val66Met polymorphism of BDNF modulates sleep pressure and NREM sleep intensity, the dynamics of SWA, the best documented physiological marker of sleep homeostasis, were analyzed in detail.
In baseline (374.3 ± 32.6 vs. 296.1 ± 29.7 μV2) and recovery (494.8 ± 55.6 vs. 379.0 ± 40.6 μV2) nights, Val/Val homozygotes produced more SWA than Val/Met allele carriers (condition: F1,30 = 13.0, P < 0.002; genotype: F1,30 = 12.6, P < 0.002).
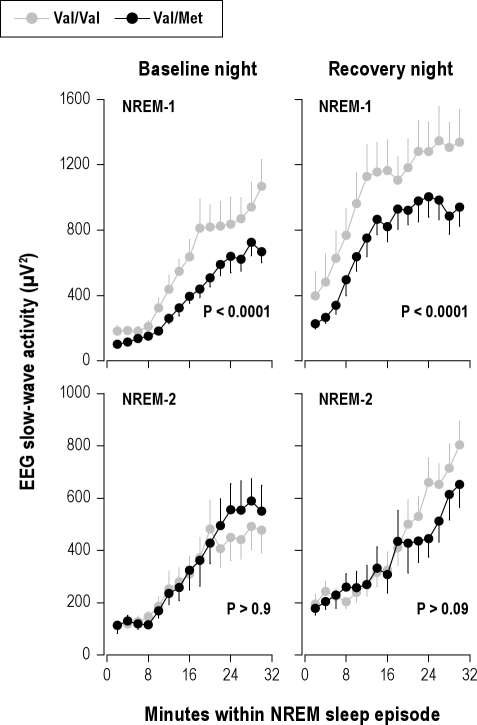
The evolution of absolute SWA values during the first 30 min of NREM sleep episodes 1 and 2 are plotted in Figure 3. Sleep deprivation increased SWA in both genotypes (condition: Pall < 0.0001). Additionally, in NREM sleep episode 1, SWA was significantly higher in Val/Val allele carriers than in Val/Met allele carriers (genotype: F1,109 = 55.8, P < 0.0001).
Figure 3.
Steeper build-up of initial slow-wave activity (SWA) in NREM sleep in Val/Val genotype of BDNF. Absolute EEG SWA (0.75-4.5 Hz; means ± SEM) values of consecutive 2-min epochs during the first 30 min of NREM sleep episodes 1 and 2 in baseline and recovery nights were plotted for Val/Val (gray dots, n = 11) and Val/Met (black dots, n = 11) genotypes. Data were analyzed with 4-way, mixed-model ANOVA with the within-subjects factors “genotype” (Val/Val, Val/Met), “condition” (baseline, recovery), “NREM sleep episode” (1, 2), and “2-min epoch” (1-15). SWA was significantly higher in Val/Val allele carriers than in Val/Met allele carriers (genotype: F1,91 = 9.3, P = 0.003), and the difference between the genotypes was modulated by NREM sleep episode (genotype × sleep episode: F1,148 = 6.1, P = 0.01). Separate analyses of NREM sleep episodes 1 and 2 revealed that the difference was restricted to NREM-1 (genotype: baseline, F1,56.4 = 37.7, P < 0.0001; recovery, F1,47.4 = 19.2, P < 0.0001) and not present in NREM-2 (baseline, F1,50 = 0.0, P > 0.9; recovery, F1,52 = 2.9, P > 0.09). To estimate the rise rates of SWA in Val/Val and Val/Met genotypes, the median slopes of adjacent 2-min intervals in the first 30 minutes of NREM sleep episodes 1 and 2 were calculated in baseline and recovery nights. The data were analyzed with 3-way, mixed-model ANOVA with the within-subjects factors “genotype” (Val/Val, Val/Met), “NREM sleep episode” (1, 2), and “condition” (baseline, recovery). The build-up rate of SWA was faster in Val/Val genotype than in Val/Met genotype (NREM-1, baseline: 64.3 ± 13.1 vs. 27.2 ± 5.4 μV2/min; NREM-1, recovery: 76.3 ± 14.8 vs. 57.0 ± 14.5 μV2/min; NREM-2, baseline: 29.8 ± 5.4 vs. 17.6 ± 10.0 μV2/min; NREM-2, recovery: 37.5 ± 7.3 vs. 29.3 ± 9.7 μV2/min) (genotype: F1,70 = 3.8, P < 0.06; NREM sleep episode: F1,70 = 13.8, P < 0.001; condition: F1,70 = 3.8, P < 0.06; genotype × NREM sleep episode: F1,70 = 5.2, P < 0.03). Separate analyses of NREM sleep episodes 1 and 2 showed that the difference was present in NREM-1 (genotype: F1,30 = 7.4, P < 0.02), but not in NREM-2 (genotype: F1,30 = 0.1, P > 0.7).
The build-up rate of EEG SWA in NREM sleep faithfully reflects the level of homeostatic sleep pressure.22,23 To investigate whether it differed between the genotypes, the rise rates of SWA in NREM sleep episodes 1 and 2 were estimated from the median slopes of adjacent 2-min intervals. In both genotypes and in baseline and recovery nights, SWA rose faster in the first when compared to the second NREM sleep episode (Figure 3). Moreover, SWA accumulated more rapidly in Val/Val genotype than in Val/Met genotype. Also this difference was restricted to NREM sleep episode 1.
Faster Dissipation of EEG Slow Wave Activity in NREM Sleep in Val/Val Genotype of BDNF
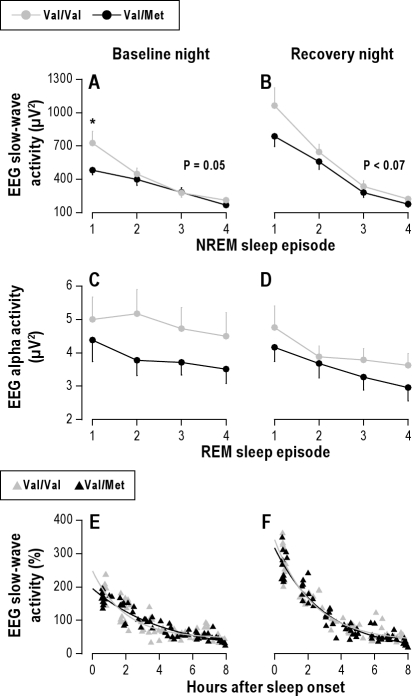
In both nights and genotypes, SWA was highest in the first NREM sleep episode and declined in the course of night. Suggesting elevated sleep pressure in Val/Val homozygotes at sleep onset, the difference between the groups was most pronounced in the first NREM sleep episode (Figure 4). The decline in absolute SWA from the first to the second NREM sleep episode was increased after sleep deprivation (condition: F1,30 = 7.0, P < 0.02), and differed between BDNF genotypes (genotype: F1,30 = 13.0, P = 0.01). Thus, SWA tended to decline more in Val/Val genotype than in Val/Met genotype in baseline (278.1 ± 100.0 vs. 82.8 ± 41.3 μV2, P = 0.05, Tukey HSD test) as well as in recovery nights (416.6 ± 97.4 vs. 227.6 ± 43.3 μV2, P < 0.07) (Figures 4A & 4B).
Figure 4.
Elevated level and faster dissipation of slow-wave activity (SWA) in Val/Val genotype of BDNF. Mean absolute (± SEM) and individual relative SWA values were plotted for the first 4 NREM sleep episodes (stages 2-4) in baseline and recovery nights (C3A2 derivation). (A & B) Gray dots: Val/Val allele carriers (n = 11). Black dots: Val/Met allele carriers (n = 11). The data were analyzed with 3-way, mixed-model ANOVA with the within-subject factors “genotype” (Val/Val, Val/Met), “NREM sleep episode” (1-4), and “condition” (baseline, recovery). Val/Val genotype subjects exhibited higher SWA than Val/Met genotype subjects (genotype: F1,30 = 7.7, P < 0.01). The difference was largest in NREM sleep episode 1 (genotype × NREM sleep episode × condition: F6,133 = 4.1, P < 0.001). The asterisk indicates the significant difference between Val/Val and Val/Met genotypes (P = 0.01, Tukey HSD test). In both genotypes, SWA decreased across consecutive NREM sleep episodes (NREM sleep episode: F3,124 = 391.2, P < 0.0001) and was enhanced after sleep deprivation (condition: F1,115 = 39.3, P < 0.0001). The P-values refer to the difference between the genotypes in the decline in SWA from the first to the second NREM sleep episode (Tukey HSD tests). (C & D) Time course of EEG alpha activity (8.375-10.625 Hz) in REM sleep. Gray dots: Val/Val allele carriers (n = 11). Black dots: Val/Met allele carriers (n = 11). Three-way mixed model ANOVA with the factors genotype (Val/Val, Val/Met), REM sleep episode (1-4) and condition (baseline, recovery) revealed significant main effects (genotype: F1,27 = 7.0, P < 0.02; REM sleep episode: F3,121 = 13.4, P < 0.0001; night: F1,104 = 16.3, P = 0.0001), yet no significant interactions. (E & F) Individual SWA values per NREM sleep episode in baseline and recovery nights were plotted at episode midpoint times relative to sleep onset. The lines represent exponential decay fits SWAt = SWA∞ + SWAo × e-r*t in Val/Val (gray triangles, gray line) and Val/Met (black triangles, black line) allele carriers (t = time, τ = time constant, SWAo = initial value, and SWA∞ = lower asymptote). The decay rate r of the exponential decay of SWA was estimated across consecutive NREM sleep episodes. Two-way, mixed-model ANOVA showed significant effects of genotype (F1,30 = 8.1, P < 0.01) and genotype × condition interaction (genotype × condition: F1,30 = 4.6, P < 0.05).
To compare the dissipation of NREM sleep pressure throughout the nights, individual episodic SWA values were expressed as a percentage of the mean all-night value in baseline (Figures 4E & 4F). Employing nonlinear regression analysis, an exponential function was fitted to the data. The decay rate r was estimated from the decay of SWA across consecutive NREM sleep episodes. In baseline, SWA declined faster in Val/Val homozygotes (r = 239.7 ± 25.4 min) than in Val/Met allele carriers (r = 139 ± 25.6 min; P < 0.02, Tukey HSD test). This difference was not significant in recovery sleep (171.4 ± 12.4 vs. 157.4 ± 20.7 min, P > 0.5).
Genotype-Dependent Difference in Slow Wave Activity is Independent of EEG Location
To investigate whether the genotype-dependent difference in SWA between Val/Val and Val/Met allele carriers varied with scalp location, the time course of SWA in NREM sleep was analyzed in FC, CP, and PO bipolar derivations. Highest SWA was found in the first NREM sleep episode and a significant anterior predominance in SWA was present, regardless of night and genotype (Figure 5). Moreover, the increase in SWA after sleep deprivation was identical in both genotypes. Nevertheless, corroborating the findings derived from the C3A2 derivation, the Val/Val allele carriers exhibited elevated SWA when compared to Val/Met allele carriers (genotype: F1,122 = 27.8, P < 0.0001). The difference was exclusively present in the first NREM sleep episode.
Figure 5.
Genotype-dependent difference in initial slow-wave activity (SWA) is independent of EEG location. Mean absolute SWA (0.75-4.5 Hz) values (± SEM) in fronto-central (FC), centro-parietal (CP), and parieto-occipital (PO) EEG derivations are plotted for the first 4 NREM sleep episodes in baseline and recovery nights. Gray dots: Val/Val allele carriers (n = 11). Black dots: Val/Met allele carriers (n = 11). Data were analyzed with 4-way, mixed-model ANOVA with the factors genotype (Val/Val, Val/Met), condition (baseline, recovery), NREM sleep episode (1-4) and derivation (FC, CP, PO). Val/Val genotype subjects exhibited higher SWA than Val/Met genotype subjects (genotype: F1,122 = 27.8, P < 0.0001). No interactions involving the factor genotype were found (main effects: condition: F1,209 = 39.0, P < 0.0001; NREM sleep episode: F3,257 = 403.2, P < 0.0001; derivation: F2,377 = 45.7, P < 0.0001). Except for the FC derivation in the recovery night, post hoc Tukey HSD tests indicated a difference between the genotypes in all derivation in NREM sleep episode 1 (P-values).
DISCUSSION
This study shows that the non-synonymous Val66Met polymorphism of BDNF affects functional brain oscillations in healthy adults in frequency- and vigilance/sleep state-specific manner. Following normal as well as prolonged wakefulness, EEG SWA, an established physiological marker of sleep need and intensity, was specifically higher in the first NREM sleep episode in Val/Val compared to individually matched Val/Met genotype subjects. Furthermore, a genotype-dependent reduction in high-alpha/low-sigma frequencies (∼ 10-13.5 Hz range) was also reminiscent of the effects of sleep deprivation (see Achermann and Borbély1 for recent overview). In REM sleep and wakefulness, genotype-dependent differences in the alpha/sigma range were virtually opposite to those reflecting elevated sleep pressure. The findings, thus, support an important role for BDNF in modulating established EEG markers of NREM sleep intensity.
Accumulating evidence indicates a detrimental effect of the Met allele on working memory in healthy adults.12,13,24 Accordingly, we found on a 2-back working memory task higher response accuracy in Val/Val homozygotes than in Met allele carriers. By contrast, response speed was not affected. A similar observation was previously made with the same task in schizophrenia patients,25 indicating that this polymorphism affects visuospatial working memory and executive control without affecting vigilance and attention. This conclusion is supported by the similar evolution in both genotype groups of subjective sleepiness, EEG low-frequency activity, sustained vigilant attention and well-being during sleep deprivation.
The neurotrophin BDNF is a key signaling molecule in regulating neurogenesis, cell survival, synaptic strength, and neural circuit activity throughout the brain.3,4 Given this role in synaptic transmission, the Val66Met polymorphism was hypothesized to cause an endophenotype of the waking EEG.26 Nevertheless, investigation of over 300 individuals revealed no genotype-related differences in absolute EEG activity, except for reduced frontal beta (14.5-30 Hz) power in Met/Met compared to Val/Met genotype. On the other hand, standardized alpha (8-13 Hz) power was reduced in Met/Met vs. Val/Met and Val/Val genotype subjects across all cortical regions. In contrast to these previous results, we found significantly higher absolute alpha (9.75-11.75 Hz) activity in Val/Val compared to Val/Met allele carriers in rested and sleep deprived state. This inconsistency may be explained by different experimental (e.g., single vs. multiple recordings) and analytical (e.g., broad bands vs. 0.5-Hz bins) procedures.
The principal aim of our investigation was to examine a possible role for BDNF in regulating SWA in NREM sleep. The study was inspired by previous findings in rats, suggesting a causal relationship between BDNF and SWA.10,11 More specifically, blockade of BDNF by unilateral injection of anti-BDNF during wakefulness reduced sleep SWA in the injected hemisphere when compared to the contralateral side. By contrast, local infusion of BDNF during wakefulness increased SWA in the initial 2 hours of subsequent NREM sleep without affecting sleep duration. The authors postulated that BDNF triggers SWA by promoting synaptic potentiation.11 Highly reminiscent of these data obtained in rats, we found higher and more rapidly accumulating SWA in initial NREM sleep in Val/Val compared to Val/Met genotype subjects. Cell culture and animal models suggest that the Met allele impairs the activity-dependent secretion of BDNF in the brain.12,14 The question, however, whether the Val66Met polymorphism influences circulating BDNF levels is unanswered.27 To clarify the roles for BDNF in sleep-wake regulation, levels of BDNF in body fluids should be quantified in the future.
Spectral sleep EEG variables are among the most heritable traits in humans.28–30 Their heritability estimates are only matched by brain architecture such as the distribution of cortical gray matter.31 Thus, the trait-like inter-individual differences in the sleep EEG may reflect differences in functional brain anatomy.32–34 The Val66Met polymorphism of BDNF was indeed associated with pronounced neuroanatomical phenotypes. The differences may reflect reduced gray matter volume in particular in hippocampus and prefrontal areas,35–37 but possibly also involve more widespread brain structures.38–40 Taken together, complementary lines of research suggest that genotype-dependent differences in brain anatomy could underlie trait-like inter-individual differences in SWA in NREM sleep between Val/Val and Val/Met genotype subjects of BDNF. Consistent with this notion, a recent report showed that developmental changes in sleep slow waves correlate positively with changes in cortical thickness of widespread brain areas.41
The inter-individual differences in the sleep EEG are highly stable and robust against external influences. A recent study suggested that trait-like, inter-individual differences in SWA are more than 10 times larger than the average SWA rebound after sleep deprivation.42 Moreover, the genetic influence on higher EEG frequencies (8-16 Hz) in monozygotic twins was independent of elevated sleep pressure after prolonged wakefulness.30 These findings indicate that genotype-dependent differences in sleep EEG variables are equally present in baseline and recovery sleep. Our data are consistent with this notion. The enhanced SWA and reduced high-alpha/low-sigma activity in NREM sleep in baseline and recovery nights suggest that Val/Val genotypes exhibit overall higher NREM sleep pressure than Val/Met genotypes. By contrast, the rebound in SWA after sleep deprivation and the dissipation of SWA throughout the nights were not or only subtly affected by BDNF genotype.
In conclusion, our study adds a new gene to the growing list of allelic variants of candidate genes (PER3, ADA, ADORA2A, COMT, DQB1*0602), which determine functional aspects of sleep EEG profiles in healthy humans.43–47 Most of these polymorphisms were determined in the current sample and similarly distributed in Val/Val and Val/Met allele carriers (Table 3). Nevertheless, all three individuals with PER35/5 genotype were Val/Val homozygote of BDNF. Given that the PER35/5 genotype was previously shown to have higher SWA in early sleep when compared to the PER34/4 genotype,43 we performed a subgroup analysis without the three pairs of subjects including PER35/5 individuals. The genotype-dependent differences in NREM sleep between Val/Val and Val/Met allele carriers of BDNF were virtually the same as in the entire study sample. These analyses suggest that the frequency-specific differences within the slow wave and alpha/sigma ranges in NREM sleep were specific for BDNF. Future studies are warranted to investigate whether the Val66Met polymorphism of BDNF modifies the effects of light on endogenous circadian time, alters EEG topography when using a referential setup rather than bipolar derivations, and whether it is associated with sleep disorders. Such an association was recently suggested for adult sleepwalking and the 22G > A polymorphism of ADA,48 which causes a sleep EEG phenotype similar to the effects of the gene variant presented here.44,45 In vitro studies show that the facilitatory action of BDNF on long-term potentiation is prevented in hippocampal slices when adenosine is removed with adenosine deaminase.49 This may be the reason why the two polymorphisms have similar consequences on sleep intensity.
Table 3.
Distribution of polymorphisms of candidate genes
|
BDNF rs6265 (G/A) |
PER3 rs57875989 del(3031-3084 nt) |
ADA rs73598374 (G/A) |
ADORA2A rs5751876 (T/C) |
COMT rs4680 (G/A) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4/4 | 4/5 | 5/5 | G/G (Asp/Asp) | G/A (Asp/Asn) | T/T | T/C | C/C | G/G (Val/Val) | G/A (Val/Met) | A/A (Met/Met) | |
| Val/Val | 4 | 4 | 3 | 9 | 2 | 3 | 4 | 4 | 5 | 2 | 4 |
| Val/Met | 7 | 4 | 0 | 9 | 2 | 3 | 5 | 3 | 6 | 2 | 3 |
NCBI gene symbols indicate genes encoding brain-derived neurotrophic factor (BDNF), PERIOD3 (PER3), adenosine deaminase (ADA), adenosine A2A receptor (ADORA2A), and catechol-O-methyltransferase (COMT). Analyses employing χ2-tests revealed no differences in the frequency of PER3, ADA, ADORA2A, and COMT genotypes between Val/Val and Val/Met allele carriers of BDNF.
DISCLOSURE STATEMENT
This was not an industry supported study. The authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
This work was supported by the University Research Priority Program Integrative Human Physiology at the University of Zürich, the Swiss National Science Foundation grant # 310000-120377, the Schüller Stiftung, and the OPO Foundation (to HPL). We thank K. Hefti, Dr. R. Wehrle, Dr. R. Dürr, and Prof. P. Achermann for their help with data collection and analyses.
Footnotes
A commentary on this article appears in this issue on page 309.
REFERENCES
- 1.Achermann P, Borbély AA. Sleep homeostasis and models of sleep regulation. In: Kryger MH, Roth T, Dement WC, editors. Principles and practice of sleep medicine. 5th ed. St. Louis, MI: Elsevier Saunders; 2011. pp. 431–44. [Google Scholar]
- 2.Tononi G, Cirelli C. Sleep function and synaptic homeostasis. Sleep Med Rev. 2006;10:49–62. doi: 10.1016/j.smrv.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Lu B. BDNF and activity-dependent synaptic modulation. Learn Mem. 2003;10:86–98. doi: 10.1101/lm.54603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waterhouse EG, Xu BJ. New insights into the role of brain-derived neurotrophic factor in synaptic plasticity. Mol Cell Neurosci. 2009;42:81–9. doi: 10.1016/j.mcn.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korte M, Carroll P, Wolf E, Brem G, Thoenen H, Bonhoeffer T. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc Natl Acad Sci U S A. 1995;92:8856–60. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genoud C, Knott GW, Sakata K, Lu B, Welker E. Altered synapse formation in the adult somatosensory cortex of brain-derived neurotrophic factor heterozygote mice. J Neurosci. 2004;24:2394–400. doi: 10.1523/JNEUROSCI.4040-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cirelli C, Tononi G. Differential expression of plasticity-related genes in waking and sleep and their regulation by the noradrenergic system. J Neurosci. 2000;20:9187–94. doi: 10.1523/JNEUROSCI.20-24-09187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hairston IS, Peyron C, Denning DP, et al. Sleep deprivation effects on growth factor expression in neonatal rats: a potential role for BDNF in the mediation of delta power. J Neurophysiol. 2004;91:1586–95. doi: 10.1152/jn.00894.2003. [DOI] [PubMed] [Google Scholar]
- 9.Conti B, Maier R, Barr AM, et al. Region-specific transcriptional changes following the three antidepressant treatments electro convulsive therapy, sleep deprivation and fluoxetine. Mol Psychiatry. 2007;12:167–89. doi: 10.1038/sj.mp.4001897. [DOI] [PubMed] [Google Scholar]
- 10.Huber R, Tononi G, Cirelli C. Exploratory behavior, cortical BDNF expression, and sleep homeostasis. Sleep. 2007;30:129–39. doi: 10.1093/sleep/30.2.129. [DOI] [PubMed] [Google Scholar]
- 11.Faraguna U, Vyazovskiy VV, Nelson AB, Tononi G, Cirelli C. A causal role for brain-derived neurotrophic factor in the homeostatic regulation of sleep. J Neurosci. 2008;28:4088–95. doi: 10.1523/JNEUROSCI.5510-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egan MF, Kojima M, Callicott JH, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–69. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 13.Hariri AR, Goldberg TE, Mattay VS, et al. Brain-derived neurotrophic factor val(66)met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci. 2003;23:6690–4. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen ZY, Jing DQ, Bath KG, et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–3. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soliman F, Glatt CE, Bath KG, et al. A genetic variant BDNF polymorphism alters extinction learning in both mouse and human. Science. 2010;327:863–6. doi: 10.1126/science.1181886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spielberger CD, Gorsuch RL, Lushene RE. Palo Alto, CA: Consulting Psychologists Press; 1970. Manual for the state-trait anxiety inventory. [Google Scholar]
- 17.Kirchner WK. Age-differences in short-term retention of rapidly changing information. J Exp Psychol. 1958;55:352–8. doi: 10.1037/h0043688. [DOI] [PubMed] [Google Scholar]
- 18.Sturm A, Clarenbach P. Stuttgart, New York: Thieme Verlag; 1997. Schlafstörungen. [Google Scholar]
- 19.Durmer JS, Dinges DF. Neurocognitive consequences of sleep deprivation. Semin Neurol. 2005;25:117–29. doi: 10.1055/s-2005-867080. [DOI] [PubMed] [Google Scholar]
- 20.von Zerssen D, Doerr P, Emrich HM, Lund R, Pirke KM. Diurnal variation of mood and the cortisol rhythm in depression and normal states of mind. Eur Arch Psychiatry Neurol Sci. 1987;237:36–45. doi: 10.1007/BF00385665. [DOI] [PubMed] [Google Scholar]
- 21.Rechtschaffen A, Kales A. Bethesda, MD: National Institutes of Health; 1968. A manual of standardized terminology, techniques and scoring system for sleep stages of human subjects. [Google Scholar]
- 22.Brunner DP, Dijk DJ, Borbély AA. Repeated partial sleep deprivation progressively changes the EEG during sleep and wakefulness. Sleep. 1993;16:100–13. doi: 10.1093/sleep/16.2.100. [DOI] [PubMed] [Google Scholar]
- 23.Dijk DJ, Brunner DP, Borbély AA. Time course of EEG power density during long sleep in humans. Am J Physiol. 1990;258:R650–R661. doi: 10.1152/ajpregu.1990.258.3.R650. [DOI] [PubMed] [Google Scholar]
- 24.Richter-Schmidinger T, Alexopoulos P, Horn M, et al. Influence of brain-derived neurotrophic-factor and apolipoprotein E genetic variants on hippocampal volume and memory performance in healthy young adults. J Neur Transm. 2011;118:249–57. doi: 10.1007/s00702-010-0539-8. [DOI] [PubMed] [Google Scholar]
- 25.Rybakowski JK, Borkowska A, Skibinska M, et al. Prefrontal cognition in schizophrenia and bipolar illness in relation to Val66Met polymorphism of the brain-derived neurotrophic factor gene. Psychiatry Clin Neurosci. 2006;60:70–6. doi: 10.1111/j.1440-1819.2006.01462.x. [DOI] [PubMed] [Google Scholar]
- 26.Gatt JM, Kuan SA, Dobson-Stone C, et al. Association between BDNF Val66Met polymorphism and trait depression is mediated via resting EEG alpha band activity. Biol Psychol. 2008;79:275–84. doi: 10.1016/j.biopsycho.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Terracciano A, Martin B, Ansari D, et al. Plasma BDNF concentration, Val66Met genetic variant and depression-related personality traits. Genes Brain Behav. 2010;9:512–8. doi: 10.1111/j.1601-183X.2010.00579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buckelmüller J, Landolt HP, Stassen HH, Achermann P. Trait-like individual differences in the human sleep electroencephalogram. Neuroscience. 2006;138:351–6. doi: 10.1016/j.neuroscience.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Ambrosius U, Lietzenmaier S, Wehrle R, et al. Heritability of sleep electroencephalogram. Biol Psychiatry. 2008;64:344–8. doi: 10.1016/j.biopsych.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 30.De Gennaro L, Marzano C, Fratello F, et al. The EEG fingerprint of sleep is genetically determined: a twin study. Ann Neurol. 2008;64:455–60. doi: 10.1002/ana.21434. [DOI] [PubMed] [Google Scholar]
- 31.Andretic R, Franken P, Tafti M. Genetics of sleep. Annu Rev Genet. 2008;42:361–88. doi: 10.1146/annurev.genet.42.110807.091541. [DOI] [PubMed] [Google Scholar]
- 32.Finelli LA, Borbély AA, Achermann P. Functional topography of the human nonREM sleep electroencephalogram. Eur J Neurosci. 2001;13:2282–90. doi: 10.1046/j.0953-816x.2001.01597.x. [DOI] [PubMed] [Google Scholar]
- 33.De Gennaro L, Ferrara M, Vecchio F, Curcio G, Bertini M. An electroencephalographic fingerprint of human sleep. NeuroImage. 2005;26:114–122. doi: 10.1016/j.neuroimage.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 34.Tinguely G, Finelli LA, Landolt HP, Borbely AA, Achermann P. Functional EEG topography in sleep and waking: state-dependent and state-independent features. NeuroImage. 2006;32:283–92. doi: 10.1016/j.neuroimage.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 35.Pezawas L, Verchinski BA, Mattay VS, et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bueller JA, Aftab M, Sen S, Gomez-Hassan D, Burmeister M, Zubieta JK. BDNF val(66) met allele is associated with reduced hippocampal volume in healthy subjects. Biol Psychiatry. 2006;59:812–5. doi: 10.1016/j.biopsych.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 37.Frodl T, Schuele C, Schmitt G, et al. Association of the brain-derived neurotrophic factor Val66Met polymorphism with reduced hippocampal volumes in major depression. Arch Gen Psychiatry. 2007;64:410–6. doi: 10.1001/archpsyc.64.4.410. [DOI] [PubMed] [Google Scholar]
- 38.Toro R, Chupin M, Garnero L, et al. Brain volumes and Val66Met polymorphism of the BDNF gene: local or global effects? Brain Struct Funct. 2009;213:501–9. doi: 10.1007/s00429-009-0203-y. [DOI] [PubMed] [Google Scholar]
- 39.Montag C, Weber B, Jentgens E, Elger C, Reuter M. An epistasis effect of functional variants on the BDNF and DRD2 genes modulates gray matter volume of the anterior cingulate cortex in healthy humans. Neuropsychologia. 2010;48:1016–21. doi: 10.1016/j.neuropsychologia.2009.11.027. [DOI] [PubMed] [Google Scholar]
- 40.Montag C, Weber B, Fliessbach K, Elger C, Reuter M. The BDNF Val66Met polymorphism impacts parahippocampal and amygdala volume in healthy humans: incremental support for a genetic risk factor for depression. Psychol Med. 2009;39:1831–9. doi: 10.1017/S0033291709005509. [DOI] [PubMed] [Google Scholar]
- 41.Buchmann A, Ringli M, Kurth S, et al. EEG sleep slow-wave activity as a mirror of cortical maturation. Cereb Cortex. 2011;21:607–15. doi: 10.1093/cercor/bhq129. [DOI] [PubMed] [Google Scholar]
- 42.Tucker AM, Dinges DF, Van Dongen HPA. Trait interindividual differences in the sleep physiology of healthy young adults. J Sleep Res. 2007;16:170–80. doi: 10.1111/j.1365-2869.2007.00594.x. [DOI] [PubMed] [Google Scholar]
- 43.Viola AU, Archer SN, James LM, et al. PER3 polymorphism predicts sleep structure and waking performance. Curr Biol. 2007;17:613–8. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 44.Rétey JV, Adam M, Honegger E, et al. A functional genetic variation of adenosine deaminase affects the duration and intensity of deep sleep in humans. Proc Natl Acad Sci U S A. 2005;102:15676–81. doi: 10.1073/pnas.0505414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bachmann V, Klaus F, Bodenmann S, et al. Functional ADA polymorphism increases sleep depth and reduces vigilant attention in humans. Cereb Cortex. 2011 doi: 10.1093/cercor/bhr173. in press. doi: 10.1093/cercor/bhr173. [DOI] [PubMed] [Google Scholar]
- 46.Bodenmann S, Rusterholz T, Durr R, et al. The Functional Val158Met polymorphism of COMT predicts interindividual differences in brain alpha oscillations in young men. J Neurosci. 2009;29:10855–62. doi: 10.1523/JNEUROSCI.1427-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goel N, Banks S, Mignot E, Dinges DF. DQB1*0602 predicts interindividual differences in physiologic sleep, sleepiness, and fatigue. Neurology. 2010;75:1509–19. doi: 10.1212/WNL.0b013e3181f9615d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Licis AK, Desruisseau DM, Yamada KA, Duntley SP, Gurnett CA. Novel genetic findings in an extended family pedigree with sleepwalking. Neurology. 2011;76:49–52. doi: 10.1212/WNL.0b013e318203e964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fontinha BM, Diogenes MJ, Ribeiro JA, Sebastiao AM. Enhancement of long-term potentiation by brain-derived neurotrophic factor requires adenosine A(2A) receptor activation by endogenous adenosine. Neuropharmacology. 2008;54:924–33. doi: 10.1016/j.neuropharm.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Horne JA, Östberg O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int J Chronobiol. 1976;4:97–110. [PubMed] [Google Scholar]
- 51.Johns MW. A new method for measuring daytime sleepiness - the Epworth Sleepiness Scale. Sleep. 1991;14:540–5. doi: 10.1093/sleep/14.6.540. [DOI] [PubMed] [Google Scholar]