Abstract
Understanding human pre-implantation development has important implications for assisted reproductive technology (ART) and for human embryonic stem cell (hESC)-based therapies. Owing to limited resources, the cellular and molecular mechanisms governing this early stage of human development are poorly understood. Nonetheless, recent advances in non-invasive imaging techniques and molecular and genomic technologies have helped to increase our understanding of this fascinating stage of human development. Here, we summarize what is currently known about human pre-implantation embryo development and highlight how further studies of human pre-implantation embryos can be used to improve ART and to fully harness the potential of hESCs for therapeutic goals.
Keywords: Human embryo, Oocyte to embryo transition, Zygote, Cleavage division, Blastocyst, Human embryonic stem cells, Aneuploidy
Introduction
Studies of mammalian embryo development, especially in the mouse, have provided key insights into early mammalian developmental pathways. However, species-specific differences, for example in the timing of the major wave of genome activation, the patterns of gene expression, the frequency of chromosome missegregation and the patterns of epigenetic modifications, may limit the extrapolation of some findings to human embryo development. To date, studies of human pre-implantation development have utilized spare human pre-implantation embryos derived from in vitro fertilization (IVF; see Glossary, Box 1) to obtain insights into aspects of development specific to humans. Historically, these studies have focused on the morphological examination of embryos and the identification of factors that can improve in vitro culture, such as the conditions required to fertilize human oocytes in vitro, to cryopreserve and thaw human embryos and to promote human blastocyst formation. More recently, with the advent of advanced imaging techniques and sensitive gene expression profiling technologies, these studies are beginning to provide a clearer understanding of human pre-implantation development at cellular and molecular resolution. In addition, spare human pre-implantation embryos have enabled the derivation of human embryonic stem cells (hESCs), leading to the establishment of novel tools for human developmental biology and the emergence of a new field of research, namely hESC-based regenerative medicine. Thus, further investigations into the fundamental aspects of human pre-implantation development might provide not only insights into human developmental biology and common birth defects but also potential benefits for reproductive health and improvements in regenerative medicine.
Box 1. Glossary
Aneuploidy. Chromosomal abnormality characterized by an abnormal chromosome number.
Assisted reproductive technology (ART). ART encompasses clinical procedures including stimulation of ovulation via hormonal induction, intrauterine insemination (IUI), IVF and intracytoplasmic sperm injection (ICSI), a variation of IVF in which the sperm is injected directly into the oocyte cytoplasm.
Cleavage divisions. A series of cell divisions after fertilization in which the net size of the embryo remains the same, but following DNA synthesis mitosis results in cells of approximately equal, decreased size. In humans, there are three cleavage divisions from 1 cell to 2 cells, 2 cells to 4 cells and 4 cells to 8 cells.
Compaction. A process during early embryo development, when blastomeres adhere to each other to form a cluster of cells (the morula).
Embryonic genome activation (EGA). The process during which the embryonic genome is activated, i.e. when transcription is evident (day 3 of human embryo development, at the 4- to 8-cell stage).
Epiblast. The part of the embryo containing pluripotent cells that are able to give rise to all the tissues of the fetus.
Germinal vesicle (GV) oocyte. An immature oocyte that has a visible nucleus (the germinal vesicle) and is arrested in metaphase I (of meiosis I), prior to ovulation.
Embryo transfer. The process of transferring embryos from in vitro culture to the uterus. This is often done at day 3 (at the 4- to 8-cell stage), but is now increasingly performed at day 5 (blastocyst stage).
Inner cell mass (ICM). Comprises pluripotent cells that are able to give rise to all cells of the fetus.
In vitro fertilization (IVF). The fertilization of the oocyte by sperm in a Petri dish.
In vitro maturation (IVM). This involves removing immature, germinal vesicle stage oocytes from the ovaries, then culturing and maturing them in vitro.
Metaphase II (MII) oocyte. A mature human oocyte that is capable of being fertilized and of reprogramming gametic (sperm and egg) pronuclei.
Oocyte to embryo transition. The stage of development following fertilization in which the molecular programs of the oocyte are degraded and those of the embryo are activated (days 0-3).
Primitive endoderm. Extra-embryonic cells that do not contribute to the fetus; instead, they give rise to extra-embryonic endoderm cells that will form the yolk sac.
Reprogramming. The reversal of cell fate from a differentiated state to an embryonic state. In vivo, this occurs during embryogenesis with the innate reprogramming of the germ cell pronuclei to an embryonic fate. Differentiated somatic cells can also be reprogrammed by somatic cell nuclear transfer (SCNT) to an oocyte, or in vitro by transgenically expressing a set of pluripotency-associated transcription factors (induced pluripotency).
Trophectoderm (TE). Extra-embryonic cells that surround the ICM and, upon implantation, give rise to the placental cytotrophoblast, syncytiotrophoblast and extravillous trophoblast.
Vitrification. A cryopreservation process that involves the addition of a cryoprotectant followed by rapid freezing, allowing embryos (and oocytes) to be frozen free of damaging ice crystal formation (Fujioka et al., 2004; Hunt and Timmons, 2007; Reubinoff et al., 2001).
In this Primer, we delve further into the specifics of human pre-implantation embryo development and discuss its relationship to that of other species. We also provide a summary of our current understanding of the molecular pathways of early human embryo development. Finally, we discuss how studies of hESCs can be used to further our understanding of early human development and, vice versa, how studies of human pre-implantation development might impact the field of stem cell biology.
Assisted reproductive technology
An historical perspective
In the most severe cases of infertility, especially when germ cell quantity and quality are most compromised, assisted reproductive technology (ART; see Glossary, Box 1) may be used to increase the chances of conception (Cedars, 2005). These technologies range from administration of therapeutics to induce ovulation, to artificial insemination (also termed intrauterine insemination or IUI), to methods of IVF. Although often considered to be a modern development, ART in the most simple form can be traced back to the end of the 18th century (see Box 2), when it began with the use of artificial insemination in animals (dogs) (Spallanzani, 1785). In spite of continual use, even in the mid-1900s, human artificial insemination, however, has remained controversial (Clarke, 2006). By 1950-1970, research in animals had progressed such that oocyte and sperm retrieval were routine and the culture of embryos was optimized for some species (Jones, 2003; Inge et al., 2005; Clarke, 2006). Against this backdrop, Robert Edwards and Patrick Steptoe made advances on two fronts that set the stage for human IVF: first, in 1969, the early stages of IVF with human eggs were described; and second, the culture of human cleavage stage embryos was reported in 1970 (Edwards et al., 1969; Edwards et al., 1970). Subsequently, the researchers continued to pursue studies of human reproduction in basic and clinical settings and, in 1978, they announced the birth of Louise Brown, the first child conceived via IVF (Steptoe and Edwards, 1978; Edwards et al., 1980).
Box 2. A history of ART
1785. Conception by artificial insemination in dogs by Lazzaro Spallanzani (University of Pavia, Italy) (Spallanzani, 1785).
1900s. Artificial insemination techniques for use in horses, cattle and sheep developed by Ilya Ivanov (State Veterinary Institute, Russia).
1940s. Techniques to freeze and store animal spermatozoa developed by Chris Polge (University of Cambridge, UK).
1934. IVF of rabbit oocytes followed by transfer into the fallopian tubes by George Pincus and Ernst Enzmann (Harvard University, USA).
1954. First characterization of human pre-implantation embryos (at the 2-cell and later stages) by Arthur Hertig and John Rock (Free Hospital for Women in Brookline, USA).
1957. Development of superovulation in mice using gonadotrophins by Robert Edwards and Ruth Fowler (University of Cambridge, UK).
1959. In vitro fertilized rabbit oocytes capable of proceeding to live birth were demonstrated by Min Chueh Chang (Worcester Foundation, USA).
1969. Early stages of IVF of human eggs by Robert Edwards and colleagues (University of Cambridge, UK).
1970. Successful culture of human cleavage stage embryos by Robert Edwards and colleagues (University of Cambridge, UK).
1972. Cryopreservation techniques for the long-term storage of pre-implantation mammalian embryos by David Whittingham (Oak Ridge National Laboratory, USA).
1978. Birth of the first child conceived via IVF reported by Robert Edwards and Patrick Steptoe (Oldham General Hospital and Bourn Hall Clinic).
1992. Development and successful use of intracytoplasmic sperm injection (ICSI) to assist with infertility by Gianpiero Palermo, Paul Devroey and Andre Van Steirteghem (Vrije Universiteit Brussel, Belgium).
1998. Development of serum-free culture methods for human blastocysts in vitro by David Gardner and colleagues (University of Oxford, UK).
Advances in ART have continued over the years. There have been improvements in the conditions used for the culture of human pre-implantation embryos, including the development of one- and two-step protocols that mimic the factors present as an embryo travels through the maternal fallopian environment (Bongso and Tan, 2005; Mercader et al., 2006; Ilic et al., 2007; Biggers and Summers, 2008; Sathananthan and Osianlis, 2010). Other advances include in vitro maturation (IVM; see Glossary, Box 1), which may allow conception by women who are susceptible to developing ovarian hyperstimulation syndrome as a result of negative reactions to fertility drugs to receive infertility treatment (Hardy et al., 2000). Major advances in cryopreservation, such as vitrification (see Glossary, Box 1), have also improved post-thaw survival and live birth rates. Finally, intracytoplasmic sperm injection (ICSI) has revolutionized the treatment of male infertility (Palermo et al., 1992).
Yet, some historical problems with IVF have remained unsolved. In particular, in the absence of sufficient knowledge of human pre-implantation embryo development, success rates of IVF have remained relatively low; even though apparently healthy-looking embryos are selected for embryo transfer (see Glossary, Box 1) back to the mother, they frequently fail to implant (see Box 3). To improve the odds of pregnancy given the uncertainty regarding embryo developmental potential, multiple embryos are transferred in some clinics, resulting in multiple births and the associated complications of low-birth weight, prematurity and, in some cases, the need to reduce fetal number for the health of the mother or siblings (Racowsky, 2002). Generally, the best IVF clinics attempt to optimize the pregnancy rate while minimizing adverse outcomes. In the UK and USA, single embryo transfer is recommended and is most frequently balanced by consideration of maternal age and other factors that might impact pregnancy as outlined (www.cdc.gov/art; www.oneatatime.org.uk). In the UK, for example, many IVF clinics now routinely transfer single embryos and the UK Human Fertilisation and Embryology Authority (HFEA) discourages multiple embryo transfer. In the USA, single embryo transfers comprise the minority of transfers and consideration of the number of embryos to transfer can be complicated. A better understanding of human pre-implantation embryo development and the parameters that can predict developmental outcome is therefore needed to achieve better methods for identifying those embryos resulting from ART that are most likely to lead to a successful pregnancy.
Box 3. Embryo implantation: a barrier to ART?
Implantation in humans occurs on approximately day 7 of development and depends on steroid hormones, such as estradiol-17β and progesterone, to induce changes in the expression of cytokines and growth factors that will facilitate uterine receptivity to the blastocyst (Norwitz et al., 2001). Implantation of the human embryo into the uterine wall is required for further development of the embryo proper; unsuccessful implantation is a major limitation to ART with likely etiologies linked to both poor embryo development and poor uterine receptivity (Troncoso et al., 2003). A better understanding of the molecular mechanisms involved in peri- and post-implantation human development will undoubtedly improve infertility treatment and disorders related to early pregnancy loss. The establishment of useful ex vivo models of early human implantation and post-implantation events (within ethical boundaries for culture durations) would be of significant benefit to ART and for understanding the mechanisms of early human development. Until now, our understanding of early human post-implantation development has depended on studying implantation events in other organisms (Enders and Lopata, 1999), with the caveat that not all events may be analogous. In the future, improved in vitro models for human implantation and gastrulation using embryos or stem cells might well supersede these limitations.
A source of pre-implantation embryos
With advances in ART, including ovarian stimulation and improved handling, culturing and storing of embryos, ART now generates many more embryos than are typically needed to alleviate infertility; in the USA, ∼1.5 million embryos are produced annually with ∼500,000 discarded each year (www.cdc.gov/sart). In addition, although worldwide estimates are difficult to ascertain, a survey of American clinics suggested that 400,000 embryos were in cryostorage as of 2002, with the majority (88.2%) intended for family building (Hoffman et al., 2003). Only a small minority (2.8%) were donated for research, and of these the majority were not of the highest quality (i.e. were unable to grow at normal rates) (Hoffman et al., 2003). However, in the USA there is currently just a single embryo bank that accepts embryos for research (the RENEW BioBank at Stanford University) and, in fact, many more couples seek to donate embryos than are accepted for research (Kalista et al., 2011). Some sources of optimal embryos for research include those that are derived from fertile couples who seek pre-implantation genetic diagnosis (PGD) or those that are cryopreserved at the one-cell (zygote) stage prior to assessment of quality as described (Wong et al., 2010). Still, many human embryos consented for research will not reach an optimal stage for hESC derivation and will provide limited information for developmental studies.
Significant ethical considerations surround the donation of human embryos for research and special procedures generally exist, as outlined recently (Kalista et al., 2011) (see Box 4). In consideration of ethical issues, the Court of Justice of the European Union recently passed a ruling banning patents from being issued for hESC research, although research related to therapeutic or diagnostic purposes, as applied to the embryo (i.e. improving embryo viability and reducing malformation), are excluded from the ban. This ruling might place European scientists in the unfortunate position of justifying the translational goals of embryonic stem cell research without the backing of patents to develop the therapies that would result from this research. This ruling might also jeopardize translational goals of hESC research by disincentivizing collaborations between academic scientists and biotechnology and pharmaceutical companies.
Box 4. Ethical considerations
Only embryos donated with ethics committee approval and patient informed consent can be used for research (Kalista et al., 2011). Human pre-implantation embryos used for research are generally obtained from two sources: (1) embryos destined for discard during the course of ART that are deemed to be of too poor quality for transfer to the uterus or cryopreservation; (2) embryos that are in excess of reproductive need; couples may complete child-bearing and excess embryos remain under cryopreservation. Couples can have three choices for these embryos: they can continue to keep them cryopreserved, they can have them discarded, or they can donate them, either for reproductive purposes to other couples or for research purposes (provided that an embryo protocol or bank is willing to accept the embryos). The creation of embryos for research rarely occurs and is not consistent with policy in most countries. In summary, the majority of human embryos used for research are surplus to the treatment of infertility and would otherwise be discarded.
During the process of hESC derivation, pre-implantation embryos (or their isolated ICM) attach to the surface of culture dishes, which results in the loss of their normal three-dimensional structure. Consequently, such embryos are no longer surrounded by an outer TE layer, precluding them from further intrauterine development. In addition, rules governing the culture of IVF embryos include specific limitations to the number of days that a human embryo may be cultured in vitro. In the UK, the Human Fertilisation and Embryology Authority has limited the in vitro culture of human embryos to 14 days following fertilization; the Academy of Sciences has implemented similar recommendations in the USA. This ensures that there is no possibility of growing a viable human embryo in vitro to the stage of primitive streak formation, when the primary tissue layers form and when axial organization of the fetus begins.
Early human embryo development
The stages of human embryo pre-implantation development
Arthur Hertig and John Rock first characterized human pre-implantation embryos from volunteers undergoing elective hysterectomy (Hertig et al., 1954; Hertig et al., 1956). The earliest embryos obtained were at the 2-cell stage and the latest were undergoing gastrulation. Hertig and Rock made several key observations of early human development, including the first estimate of fertilization dynamics. They also hypothesized that up to half of in vivo human embryos are developmentally abnormal (Hertig et al., 1954; Hertig et al., 1956). Since then, our understanding of human pre-implantation embryo development has primarily emerged from studies of human embryo development in an IVF context.
Human embryo development begins in relative transcriptional silence with an oocyte to embryo transition (see Glossary, Box 1) that lasts for ∼3 days and encompasses fusion of the egg and sperm, migration and fusion of the germ cell pronuclei, genetic and epigenetic reprogramming (see Glossary, Box 1) and a series of cleavage divisions (see Glossary, Box 1) that culminate with a major wave of embryonic genome activation (EGA; see Glossary, Box 1) between the 4- and 8-cell stages (Fig. 1). In 1988, Peter Braude and colleagues determined the timing of EGA in humans and found that distinct aspects of protein synthesis, linked to transcriptional activation, were first evident following the 4-cell stage at ∼8 cells (Braude et al., 1988). Moreover, cleavage was not sensitive to inhibition of transcription by α-amanitin until after the 4-cell stage, an observation consistent with the uridine radiolabel experiments of Tesarík and colleagues, suggesting that the EGA activation occurs between the 4- and 8-cell stages in human embryos (Tesarík et al., 1987; Braude et al., 1988). Later, other studies by Taylor et al. reported the initial detection of paternal transcripts at the 3- to 4-cell stage (Taylor et al., 1997). Subsequently, Dobson and colleagues characterized the transcriptome of pre-implantation human embryos; these authors demonstrated that ∼1800 mRNAs were modulated in expression through the first 3 days of development, with the majority being downregulated or targeted for destruction, a small group being upregulated on days 1 and 2, and a large group of mRNAs increasing in abundance on day 3 (Dobson et al., 2004). The authors suggested that increases in mRNAs prior to day 3 represent minor transcriptional activity or preferentially stable mRNAs in the pool of rapidly degrading maternal mRNAs. In addition, they demonstrated that the major wave of EGA was independent of cell number, occurring at day 3 even in embryos that arrested with fewer than 8 cells (Dobson et al., 2004). These findings appeared to differ from those made in other species, such as the mouse, in which zygotic gene activation (ZGA) is initiated during the 1- to 2-cell stage (Flach et al., 1982; Wang et al., 2004). Although debate has been re-ignited on occasion regarding when and how transcription is initiated, recent evidence (see below) indicates that, regardless of the initial detection of changes, the major wave of EGA occurs on day 3 of human embryo development and this wave corresponds to the first wave of ZGA in mice at 26-29 hours post-fertilization (Vassena et al., 2011).
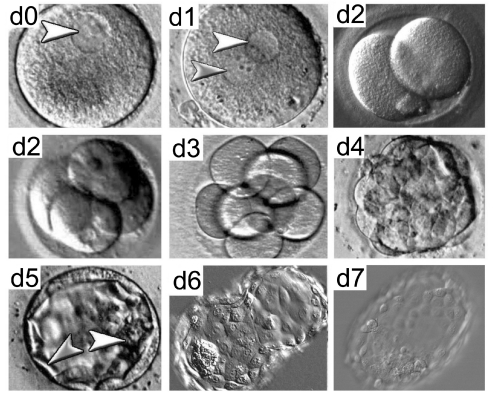
Fig. 1.
Stages of human pre-implantation embryo development. Phase-contrast images of human embryo development from day (d) 0 to day 7. Following fertilization, embryos undergo a series of mitotic cell divisions. Arrowheads in d0 and d1 indicate pronuclei. On or around day 4, the embryo compacts, resulting in the formation of a morula that consists of cells (or blastomeres) in a compact cluster contained within the zona pellucida (the glycoprotein layer that surrounds the embryo). The blastocyst, which forms on day 5, is a fluid-filled structure composed of an inner cell mass (white arrowhead) and trophectoderm (gray arrowhead). On day 6, the blastocyst ‘hatches’ from the zona pellucida and it is ready to implant into the uterine wall on day 7.
Following EGA, the embryo subsequently undergoes compaction (see Glossary, Box 1) to form a morula that marks the first morphological indication of a break in radial symmetry. As human embryos continue to progress despite lysis or fragmentation of one or more blastomeres, morphological changes during human pre-implantation development, such as compaction or cavitation, are a function of the timing of development (i.e. days post-fertilization), rather than cell number (Fig. 1). Subsequent cell divisions lead to the development of a blastocyst that comprises a fluid-filled blastocyst cavity and an inner cell mass (ICM; see Glossary, Box 1), surrounded by trophectoderm (TE; see Glossary, Box 1) cells (Fig. 1). Just before the blastocyst implants into the uterine wall, the ICM further diverges into early epiblast (see Glossary, Box 1) and primitive endoderm (see Glossary, Box 1) cells. Implantation, which in humans occurs at approximately day 7 of development, is required for further development of the embryo proper.
Recent, time-lapse imaging studies have provided observations of the dynamic behaviors of human embryos during their first week of in vitro development (Wong et al., 2010) (Fig. 2). These studies show that successful development to the blastocyst stage can be predicted as early as the 4-cell stage, based on parameters of the first three mitotic divisions (Wong et al., 2010). Human embryos that successfully develop to the blastocyst stage undergo cytokinesis within 14.3±6.0 minutes, complete the second mitotic division (to 3 cells) within 11.1±2.2 hours of the conclusion of the first cytokinesis, and have synchronized divisions to the 3-cell and 4-cell stages to within 1.0±1.6 hours (Wong et al., 2010). As successful pre-implantation development can be predicted prior to the major wave of EGA, it is likely that human embryo development is in large part influenced by the inheritance of maternal and/or paternal factors by the nascent zygote (Fig. 3). Potential factors that might be involved in this process include those that mediate diverse aspects of RNA metabolism/translation and cytokinesis (Wong et al., 2010). Other factors might include epigenetic and genetic factors, especially the status of ploidy.
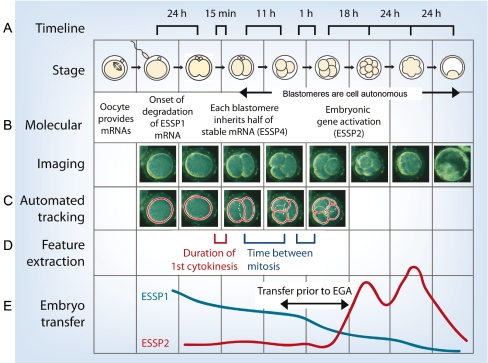
Fig. 2.
Imaging human embryo development and predicting developmental potential. (A) Timeline of human embryo development from the zygote to the blastocyst stage, highlighting critical times between stages that predict successful development. (B) Molecular events and time-lapse images of human embryo development. ESSP1 describes maternally inherited oocyte mRNAs destined for degradation, ESSP4 genes are stably expressed, and ESSP2 refers to a set of embryonic activated genes. (C) Automated tracking demonstrates modeling via computer algorithms to predict success or failure. (D) Critical features that influence developmental success include the duration of first cytokinesis, the time between the first and second mitotic division, and the synchronicity of appearance of the third and fourth blastomeres. (E) Potential application of prediction of embryo developmental potential in ART. Figure reproduced with permission from Wong et al. (Wong et al., 2010).
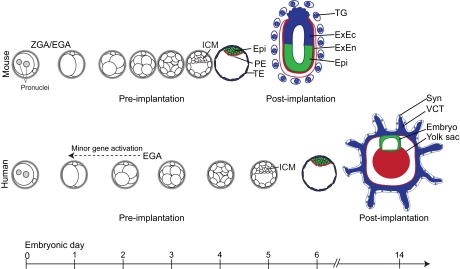
Fig. 3.
Cell fate decisions and their timing in mouse versus human early embryo development. Prior to implantation, both human and mouse embryos similarly undergo cell divisions culminating in the development of a blastocyst comprising a discernible ICM and TE. Mouse zygotic/embryo genome activation (ZGA/EGA) begins at the 2-cell stage (Flach et al., 1982), whereas human EGA begins at ∼4- to 8-cell stage on day 3, although minor human EGA may occur as early as the 2-cell stage (Taylor et al., 1997; Vassena et al., 2011). The timing of compaction and blastocyst formation also differs significantly, with human embryos showing delayed development compared with mouse embryos; the mouse blastocyst forms between days 3 and 4, whereas human blastocysts form between days 5 and 6. Both human and mouse pre-implantation blastocysts comprise an outer layer of trophectoderm (TE) cells, which form the trophoblast lineage of the placenta, and an inner cell mass (ICM) that segregates into epiblast (Epi) and primitive endoderm (PE) layers. Epiblast cells eventually give rise to all the tissues of the future fetus, whereas the PE gives rise to extra-embryonic endoderm (ExEn) cells that will form the yolk sac. In the mouse, the TE gives rise to a proliferative stem cell pool of extra-embryonic ectoderm (ExEc) cells that bud off differentiated polyploid trophoblast giant (TG) cells. By contrast, human TE gives rise to villous cytotrophoblast (VCT) cells, a multinucleated syncytium (Syn) and extravillous trophoblast cells (not shown). The dashed arrow indicates the possibility of earlier minor gene activation.
The effects of growth factors on human embryo development
It has been suggested that the viability and growth of human embryos are inversely correlated with the metabolism of growth factors and thus metabolically ‘quiet’ embryos are more viable than those that are ‘active’ (Leese, 2002). Although this has never been confirmed for human pre-implantation embryos, the ‘quiet embryo hypothesis’ predicts that hyperactive metabolism arising in embryos as a result of culture stresses would be accompanied by the increased expression of genes involved in glycolysis and glucose transport (Lazzari et al., 2002) as well as lactate (anion efflux) transport. Principally, this suggestion is based on radiolabeling assays that determine the rate of protein synthesis during bovine embryo development and from experiments assaying the embryo culture medium for the gain and loss of amino acids (Leese, 2002; Lazzari et al., 2002). In addition, cell survival and the allocation of cells to the ICM and TE have been suggested to be regulated by specific growth factors, such as insulin-like growth factor 1 (IGF1) and granulocyte-macrophage colony-stimulating factor (GM-CSF, or CSF2) (Sjöblom et al., 1999; Brison, 2000; Spanos et al., 2000; Robertson et al., 2001; Sjöblom et al., 2002; Lin et al., 2003). However, it remains unclear whether there are cell-specific or general survival mechanisms that correlate with unique growth factor receptor expression in human ICM or TE cells. Understanding the influence of growth factors on human embryo development will be important for improving embryo viability and might also benefit efforts to derive in vitro stem cells.
Parallels with mouse pre-implantation development
Human and mouse embryos appear morphologically similar during pre-implantation development; however, there are key molecular differences that might underlie later significant differences in developmental timing (Fig. 3). These differences include unique gene expression patterns (Dobson et al., 2004; Wang et al., 2004; Hamatani et al., 2004; Zeng and Schultz, 2005; Bell et al., 2008), programs of epigenetic modification (Fulka et al., 2004; Beaujean et al., 2004), susceptibility to genetic instability (Vanneste et al., 2009; Vanneste et al., 2011), and a protracted period of transcriptional silence in the human embryo relative to the mouse (Flach et al., 1982; Braude et al., 1988; Memili and First, 2000; Duranthon et al., 2008; Rother et al., 2011). As development progresses, human embryos lag behind mouse embryos in the timing of compaction and blastocyst formation (Fig. 3). Moreover, human embryos are likely to undergo at least one additional round of cell division before implantation (to the ∼256-cell stage in human blastocysts compared with 164 cells in mouse blastocysts) (Fig. 3, day 6).
Notably, human embryo development is inefficient, with 50-70% of embryos routinely failing to reach blastocyst stage in vitro (Gardner et al., 2000; French et al., 2010) and up to 50% similarly failing in vivo (Hertig et al., 1954). These high failure rates are likely to be reflected in vivo in the low fecundity rates of humans relative to many other species, which is largely a result of pre- and post-implantation embryo loss (Evers, 2002; Macklon et al., 2002). The processes of implantation and placental development in humans are also distinct from those observed in the mouse (Fig. 3). In humans, TE cells give rise to placental cytotrophoblast cells that are initially largely invasive. Cytotrophoblast cells in the placental villi proliferate, giving rise to differentiated cells, such as the multinucleated syncytial cells that are generated from the fusion of cytotrophoblast cells, and to extravillous trophoblast cells that invade the maternal decidualised uterus (Norwitz et al., 2001; Moffett and Loke, 2006) (Fig. 3). By contrast, in the mouse there is minimal early invasion and TE cells initially give rise to extra-embryonic ectoderm as early proliferative cells adjacent to the epiblast that will bud off polyploid trophoblast giant cells through a process of endoreduplication (Simmons et al., 2007) (Fig. 3).
Molecular control of human embryo development
Historically, it has been difficult to obtain reliable human embryo gene expression data owing to: (1) limitations of the techniques for analyzing single oocyte, embryo or blastomere expression; and (2) the variable quality of available human embryos, which can compromise data reliability. Nonetheless, recent findings from several groups have provided a framework for our understanding of gene expression in pre-implantation human development.
Lineage commitment
The molecular mechanisms that underlie lineage decisions during human pre-implantation development are important for understanding how the ICM and TE form. During mouse development, cells become committed to either the ICM or TE by the 32-cell stage, although molecular differences and lineage bias in early blastomeres can be observed earlier (Gardner, 2001; Khosla et al., 2001; Piotrowska et al., 2001; Torres-Padilla et al., 2007; Jedrusik et al., 2008; Plachta et al., 2011) (Fig. 2). Alternatively, it has been proposed that mouse pre-implantation patterning may be a result of stochastic processes and that, although some cells have begun to express markers of different lineages, cell fate is not determined until the early blastocyst stage (Motosugi et al., 2005; Dietrich and Hiiragi, 2007; Ralston and Rossant, 2008; Tarkowski et al., 2010).
Whether molecular differences are a cause or consequence of lineage commitment is incompletely understood and is an ongoing point of discussion in the field. It has been suggested that cell fate in human embryos may be determined as early as the 4-cell stage, even in the absence of transcription, based on the localization of TE-associated gene expression (Hansis et al., 2002; Edwards and Hansis, 2005). However, other studies have not confirmed this expression of TE-associated genes prior to the 8-cell to morula stage in single embryo or single blastomere analysis (Galan et al., 2010; Wong et al., 2010). In studies by Galan and colleagues, human blastomeres were dissected up to the 8-cell stage and global gene expression in single cells was investigated (Galan et al., 2010). The results suggested that human blastomeres are transcriptionally similar up to the precompaction 8-cell stage and therefore are of undefined lineage (Galan et al., 2010). Notably, however, one cannot exclude the possibility that blastomeres at the 8-cell stage are poised to differentially express lineage-associated genes. Notably, in the study by Wong et al. (Wong et al., 2010), blastomeres from 8-cell embryos (day 3) were characterized by different molecular programs that were either maternal or embryonic, suggesting that each blastomere may develop in a cell-autonomous manner; however, expression of markers of the TE was not observed.
These findings contrast with evidence for earlier lineage definition in the mouse embryo (Jedrusik et al., 2008). These differences might be linked to the distinct timing of embryonic (zygotic) genome activation in the mouse, differences in the localization of lineage-defining proteins that have not yet been fully characterized, or alternative mechanisms for lineage commitment. Species differences have been noted for the expression of the lineage-defining transcription factors OCT4 (POU5F1), a POU homeodomain protein, and CDX2, a caudal-related homeodomain protein, during mouse, rhesus monkey, human and bovine pre-implantation development, with human and bovine embryos having protracted OCT4 and CDX2 colocalization in the TE (Chen et al., 2009; Berg et al., 2011) compared with mouse and rhesus monkey embryos (Mitalipov et al., 2003; Dietrich and Hiiragi, 2007; Ralston and Rossant, 2008). It will be important to determine whether such differences are unique to OCT4 and CDX2 and whether species differences in the interaction of these proteins indeed reflect a lack of evolutionary conservation in the formation of ICM and TE. Differences in the signaling response of rodent and bovine versus human embryos have recently been noted (Roode et al., 2012; Kuijk et al., 2012) (see note added in proof). Moreover, whereas the zinc-finger proteins GATA4 and GATA6 and the SOX protein SOX17 are expressed in the primitive endoderm of both mouse and human embryos (Plusa et al., 2008; Morris et al., 2010; Niakan et al., 2010; Roode et al., 2012; Kuijk et al., 2012), comprehensive temporal and spatial analysis of protein localization throughout human pre-implantation development is still lacking. Detailed analysis of the molecular mechanisms of lineage restriction in the early human embryo will undoubtedly lead to improvements in stem cell biology.
Embryonic genome activation
Genome-wide transcriptional analysis of human pre-implantation embryos has uncovered several interesting patterns: (1) few genes are up- or downregulated during maturation of human oocytes from the immature germinal vesicle oocyte (GV oocyte; see Glossary, Box 1) stage to the metaphase II oocyte (MII oocyte; see Glossary, Box 1) stage; (2) maternally expressed transcripts inherited from the MII oocyte are predominantly downregulated or degraded during progression to the 4-cell stage; (3) many genes are upregulated after the 4-cell stage, reflecting the major wave of EGA; (4) genes involved in lineage commitment are upregulated later in pre-implantation development; and (5) many dynamically expressed genes encode transcription factors, epigenetic modifiers and chromatin remodeling factors (Fig. 4) (Dobson et al., 2004; Hamatani et al., 2004; Hamatani et al., 2006; Zhang et al., 2009; Galan et al., 2010). However, more extensive transcriptional analyses of individual embryos and blastomeres throughout human pre-implantation development are still needed, together with confirmation at the protein level, to understand the extent of variation in each of these stereotypic patterns of gene expression.
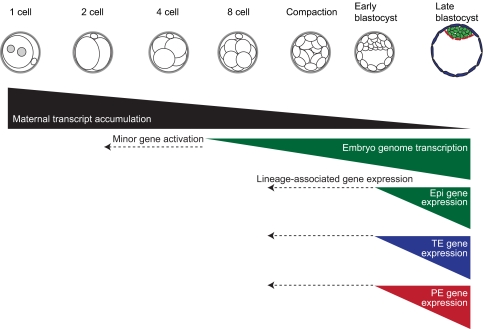
Fig. 4.
Genetic networks of human pre-implantation development. Maternal transcripts inherited from the oocyte are degraded through subsequent rounds of cell division. Human genome activation principally occurs between the 4- and 8-cell stages, and perhaps as early as the 2-cell stage. It is unclear when genes associated with the restriction of the TE or ICM cell lineage are expressed in human embryos, but data suggest that these lineage-associated genes are expressed in human embryos later than in mice at around the early blastocyst stage. Human embryos can be cultured in vitro for 7-8 days post-fertilization and, in vivo, human embryos implant around day 7. The derivation of epiblast stem-like cells from human pre-implantation embryos suggests that human embryos might be capable of reaching a more mature stage in vitro than mouse pre-implantation embryos, which can be cultured in vitro up to 4 days post-fertilization. The dashed arrows indicate the possibility of earlier minor gene activation and lineage-associated gene expression. Epi, epiblast; TE, trophectoderm; PE, primitive endoderm.
Recent studies by Vassena and colleagues have addressed the growing need for increasingly more comprehensive analyses of human pre-implantation development by performing RNA sequencing analysis on individual embryos and generating a searchable database called the Human Embryo Resource (HumER) (Vassenna et al., 2011). It remains unclear whether dynamic changes in gene expression before the 4- to 8-cell stage are the result of maternal transcript degradation, activation per se, are linked to embryo arrest or other aspects of embryogenesis that affect the quality of the embryos used for analysis. Finally, results differ when one examines gene expression in single embryos versus single blastomeres (Wong et al., 2010). Thus, it is likely that the translation of human embryo analyses to applications in ART or stem cell biology and regenerative medicine will require further comprehensive transcriptional analyses, including confirmation of whether human embryos undergo successive waves of genome activation, transcriptional analysis of single cells including those dissected from ICM and TE, and comparisons between human and mouse pre-implantation embryos on the same analytic platform.
Aneuploidy in human pre-implantation embryos
Aneuploidy (see Glossary, Box 1) is common in humans and is the leading cause of all human birth defects as well as miscarriage; errors can arise in meiosis during generation of the oocyte and sperm and in the mitotic divisions of the nascent embryo. Estimates of meiotic error rates in humans are high (5-20% of oocytes) compared with other species (1/10,000 meiotic yeast cells, 1/2000-1/6000 Drosophila germ cells and 1/100-1/200 murine germ cells). Previous studies have documented that high rates of meiotic errors may occur not only in meiosis II (MII) but also in meiosis I (MI) (Hassold and Hunt, 2001; Hunt and Hassold, 2002). A recent study that examined recombination in human oocytes demonstrated that the number and distribution of the mismatch repair protein MLH1 marked regions of recombination; moreover, these authors noted the presence of ‘vulnerable’ crossover configurations in human fetal oocytes that may be associated with an increased vulnerability of oocytes to nondisjunction and ultimately to aneuploidy (Cheng et al., 2009).
In addition to meiotic errors, mitotic errors in the first few cleavage divisions appear to be even more frequent. Initial data regarding aneuploidy in cleavage stage human embryos were met with skepticism related primarily to the methods of analysis, which depended on intricate fluorescence in situ hybridization protocols or PCR-based methods that relied upon degenerate oligonucleotide priming (DOP), which is prone to the preferential amplification of DNA from single cells and allele dropout (ADO) (Munné et al., 1993; Wilton et al., 2002). Subsequent methods, however, have largely overcome these initial limitations via the analysis of large regions of adjacent DNA to permit the detection and resolution of both preferential amplification and ADO. This has allowed groups such the European Society for Human Reproduction and Embryology (ESHRE) to validate recent methods by including multiple control samples, TE and cleavage stage biopsy, and reconciliation with outcomes (Harton et al., 2011).
A recent study used array-based technology to examine the genome-wide copy number of distinct loci in cleavage stage human embryos (Vanneste et al., 2009). This study identified several types of chromosomal abnormalities that occurred in human embryos, and observed that mosaicism for whole chromosomes (aneuploidies) in one or more blastomeres occurred in more than 80% of embryos. In addition, Vanneste et al. (Vanneste et al., 2009) identified frequent chromosome segmental deletions, duplications and amplifications that were reciprocal between presumptive sister blastomeres, suggesting that there is frequent chromosome breakage and fusion during early human development, especially in the cleavage divisions. Results suggest that embryonic blastomeres differ substantially from somatic cells; apoptosis in response to errors in chromosome segregation is common in somatic cells but has not been widely observed in human embryos prior to day 5 by time-lapse imaging or immunofluorescence microscopy in spite of occasional reports that apoptosis may occur in a subset of blastomeres or embryo fragments (Boumela et al., 2011; Koo et al., 2011). Following formation of the blastocyst, however, apoptosis is a common feature of the ICM (Brison, 2000).
Altogether, these results raise fundamental questions (see Box 5): why is aneuploidy so common in humans, why does it increase with maternal age, and how might chromosome instability be dealt with during ART? Addressing these questions is a major focus of human embryology.
Box 5. Major questions regarding human aneuploidy
Why is aneuploidy so common in humans?
Potential explanations include: (1) ovarian stimulation conditions (or induced ovulation) may recruit oocytes that are of suboptimal quality and thus prone to aneuploidy; (2) culture conditions may induce chromosome missegregation; (3) men and women who undergo ART may have sperm/eggs carrying chromosome abnormalities; and (4) aneuploidy in vitro may reflect the incidence of aneuploidy in vivo. The current view is that chromosome instability observed in vitro reflects that observed in vivo, with estimates that, at most, 30% of human fertilized eggs result in a live birth and that the majority of spontaneous abortions or miscarriages have chromosomal abnormalities (Fritz et al., 2001; Macklon et al., 2002; Benkhalifa et al., 2005).
Why does aneuploidy increase with maternal age?
Although well documented, the increase in aneuploidy with age is of unknown origin. Potential causes include intrinsic degradation of key spindle components, accumulated damage due to environmental exposure, triggers of hormonal dysfunction leading to missegregation of chromosomes, and intrinsic differences in the recruitment of superior oocytes over less-optimal oocytes (Hassold et al., 1996; Robinson et al., 2000; Hunt and Hassold, 2008; Hassold and Hunt, 2009; Hunt and Hassold, 2010; Chiang et al., 2011; Ghosh et al., 2011; Selesniemi et al., 2011).
How might aneuploidy be dealt with during ART?
Attempts have been made to correlate embryo morphology with aneuploidy; however, aneuploid embryos often appear normal and suitable for transfer based on traditional IVF assessment techniques (Hardarson et al., 2003; Baltaci et al., 2006; Munné et al., 2009; Alfarawati et al., 2011). Thus, efforts have been directed at developing non-invasive approaches for detecting variations in chromosome copy number in human embryos. Currently, the most frequently used method for diagnosing aneuploidy is pre-implantation genetic screening (PGS) of blastomeres biopsied from day-3 embryos. However, PGS is invasive to the embryo, is influenced by mosaicism, which is common between blastomeres within an embryo, and is consequently used only occasionally (Baart et al., 2006; Kuo et al., 1998). Alternative approaches, such as extended culture to the blastocyst stage and analysis of chromosomal status via TE biopsy, have also been used (Schoolcraft et al., 2011). These methods have met with some success but there are concerns over epigenetic changes, embryo arrest and other factors associated with prolonged embryo culture that disrupt embryo integrity. Moreover, TE biopsy/analysis requires that embryo transfer be delayed until chromosomal testing is completed (Khosla et al., 2001; Fernández-González et al., 2009; Katari et al., 2009; Lim et al., 2009). Thus, screening of embryos for aneuploidies is still not routine in the clinic.
Human pre-implantation embryos: a source of embryonic stem cells
Deriving and culturing hESCs from pre-implantation embryos
Human pre-implantation embryos also provide a source for the derivation of embryonic stem cells (Thomson et al., 1998). Efficient methods to derive and grow hESCs remain challenging even as methods are optimized for their derivation from embryos of variable quality (Ström et al., 2007). Early steps in the derivation process are crucial, as the majority of embryos either fail to generate a viable ICM or the isolated ICM cells differentiate upon initial passaging. Although hESCs have been established from plating whole blastocysts, the derivation of hESCs more typically involves immunosurgery, microdissection or laser-mediated separation and destruction of the TE prior to culturing the ICM in vitro (Thomson et al., 1998; Ström et al., 2007; Chen et al., 2009). The benefit of TE elimination during hESC derivation is incompletely understood and might be due to the removal of negative, possibly paracrine, signals originating from the TE that cause the differentiation of ICM-derived cells in vitro. Mitotically inactivated mouse embryonic fibroblasts (MEFs) or human fibroblasts are thought to provide adherence support for hESCs and to provide factors that further enable hESCs to proliferate (see Fig. 5). The requirement for these supportive cells can be replaced by facilitating hESC attachment with extracellular matrix components such as collagen, laminin and fibronectin. In addition, the medium can be pre-conditioned by MEFs to acquire secreted growth factors that help maintain hESCs. However, the presence of non-human factors in the culture may limit the use of hESCs for transplantation therapies. More recently, chemically defined culture conditions have been developed for hESCs (Vallier et al., 2005), addressing the challenge of delivering the clinical promise of hESCs and ultimately leading to Good Manufacturing Practice (GMP)-compatible approaches with the aim of protecting patients from the risk of infection and the presence of undefined products (Vallier, 2011).
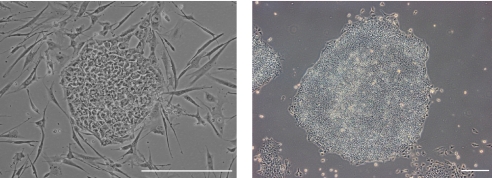
Fig. 5.
Derivation of human embryonic stem cells (hESCs). (Left) hESCs cultured on feeders (e.g. fibroblasts cells) and (right) in ‘feeder-free’ conditions (supportive fibroblast cells have been substituted with extracellular matrix components). Scale bars: left, 50 μm; right, 100 μm.
Despite advances since their initial derivation, the molecular mechanisms involved in the establishment and maintenance of hESCs are not completely understood. Moreover, hESCs have historically been difficult to culture due to dissociation-induced apoptosis (DIA), which is common when hESCs are reduced to single cells. However, recent efforts have elucidated mechanisms to inhibit DIA via the use of ROCK (Rho kinase) inhibitors (Amit et al., 2000; Hasegawa et al., 2006; Watanabe et al., 2007; Ohgushi et al., 2010). ROCK inhibitors are particularly valuable for promoting growth of human pluripotent stem cells, including hESCs, during stress-inducing processes of single-cell cloning, suspension culture and cryopreservation. Mechanistic studies suggest that ROCK inhibitors suppress DIA by alleviating cytoskeletal hyperactivation of actomyosin that results in extensive blebbing of the hESCs. Further elucidation of the intrinsic and extrinsic factors that influence hESC derivation and of the unique biology associated with culture will be fundamentally important for the utilization of these cells for regenerative medicine and as models of early human development. Conversely, understanding early human pre-implantation development, particularly the timing and molecular basis of lineage restriction and the enrichment of growth factor receptors on ICM cells, will undoubtedly lead to improvements in hESC culture that will be beneficial for the use of hESCs to generate lineage-committed cells of therapeutic relevance.
Differences between human and mouse ESCs
Remarkably, the culture conditions used to propagate hESCs, namely fibroblast growth factor (FGF2) and activin (Vallier et al., 2005), can be used to establish pluripotent stem cell lines from post-implantation mouse and rat epiblast cells (EpiSCs), just before gastrulation (Brons et al., 2007; Tesar et al., 2007). By contrast, mouse ESCs (mESCs) can be derived from the pre-implantation ICM in the presence of either leukemia inhibitory factor (LIF) and bone morphogenetic protein (BMP) (Ying et al., 2003; Smith et al., 1988; Williams et al., 1988) or in defined culture conditions in the presence of inhibitors of differentiation-inducing signals, namely FGF/Erk and glycogen synthase kinase 3 (GSK3) inhibitors (Ying et al., 2008). This suggests that species-specific mechanisms might be necessary to immortalize stem cell lines from embryos derived at distinct stages of development. What remains poorly understood is whether the different signaling requirements for mESCs versus EpiSCs or hESCs (Greber et al., 2010) have consequences for their efficiency in directed differentiation into organ-contributing cell types and whether the mechanisms of action of pluripotency factors are overlapping or distinct. Importantly, comparisons between the genome-wide transcription profile of pre-implantation human ICM versus hESCs indicate significant differences in the expression of a large set of genes, including components of important developmental signaling pathways such as the transforming growth factor beta (TGFβ), insulin growth factor (IGF) and MAPK pathways, which are uniquely expressed in the ICM but not in hESCs. This suggests that the growth factor requirements for hESC maintenance might not reflect in vivo pluripotency mechanisms, at least not those of the ICM (Reijo Pera et al., 2009). Further elucidation of key molecular, cellular and biological properties of human pre-implantation embryos and comparisons with mouse pre- and post-implantation development will be important for establishing how hESCs acquire the unique properties of post-implantation mouse epiblast cells.
The developmental potential of hESCs
Whereas mESCs can contribute to chimeras, including the germline of such chimeras, when reintroduced into a host embryo, EpiSCs exhibit little contribution to chimeras (Brons et al., 2007; Tesar et al., 2007). A possible explanation for this discrepancy is that EpiSCs and, by extension, hESCs have limited pluripotency, but this seems unlikely as hESCs and EpiSCs have been demonstrated to differentiate into all three definitive germ cell lineages both in vitro, by embryoid body (EB) assays and directed differentiation, and in vivo in teratoma assays (Thomson et al., 1998; Brons et al., 2007; Tesar et al., 2007). Another possible explanation for the limited chimerism is that EpiSCs and hESCs are developmentally asynchronous with pre-implantation embryos and are thus unable to contribute to aggregation or blastocyst injection chimeras (Brons et al., 2007). To address this, hESCs or EpiSCs could be injected into the epiblast or primitive streak of post-implantation host embryos to look for a contribution to chimeras. Alternatively, if hESCs were established that more closely resemble mESCs with regard to their gene expression and growth factor requirements, their contribution to pre-implantation chimeras would resolve the question of whether they are equivalent in pluripotency to mESCs. Such experiments evoke ethical consideration, as they involve the formation of animal-human chimeras (James et al., 2006). Another possibility is that hESCs and EpiSCs undergo fundamental alterations during derivation and prolonged culture and therefore are not biologically equivalent either to embryos or to mESCs.
An active area of human stem cell research is the development of methods to derive stem cell lines from human embryos that more closely resemble mESCs. This would be advantageous as established mESC-based directed differentiated protocols could be rapidly translated for use in mESC-like human stem cells and these cells might provide a more efficient starting population for driving differentiation. Moreover, the establishment of human stem cells that could be easily genetically manipulated, like mESCs, would be a tremendous advancement, as conventional hESCs are less efficient at homologous recombination using established techniques. However, it remains unclear whether protracted human development prior to implantation, as compared with the mouse, reflects human embryo maturation and whether this has consequences for the type of stem cell that can be derived. Understanding when and how the human ICM acquires characteristics of the mouse post-implantation epiblast will be important for researchers wishing to derive stem cell lines and to use hESCs for disease modeling and for therapeutic aims.
hESCs as a model of early human development
hESCs have been used to generate diverse lineage-committed cells of therapeutic relevance, including motoneurons to model amyotrophic lateral sclerosis (Di Giorgio et al., 2008), hematopoietic tissue for the treatment of blood disorders (Vijayaragavan et al., 2009), and cardiogenic precursors for the treatment of heart and vascular disease (Kattman et al., 2011). Despite these significant advances in stem cell research, there are still challenges to the therapeutic application of stem cells for disease modeling and cell replacement therapies. Our limited understanding of the key signals that regulate cell fate choices during early human development, which might be distinct from cues in other organisms, is a major challenge to the development of efficient directed differentiation protocols. Another challenge is that hESCs are currently maintained as heterogeneous populations in vitro and different cells might respond uniquely to extrinsic signals. Yet another limitation is that in vitro hESCs might be biased in terms of differentiation potential. This is supported by the observation that different hESC lines have distinct differences in their efficiency to generate cardiogenic precursors or pancreatic cells (Osafune et al., 2008). Recently, Bernardo and colleagues have suggested that hESCs, like EpiSCs, respond equivalently to BMP signaling and differentiate into cells that resemble extra-embryonic mesoderm, supporting the hypothesis that hESCs cannot reverse their developmental commitment and are refractory to trophoblast differentiation (Bernardo et al., 2011). This suggests that either at the embryo stage or during the derivation process, human ICM cells undergo a lineage restriction, as they resemble mouse post-implantation epiblast cells both in their molecular characteristics and response to differentiation-promoting signals.
Conclusions
In summary, the current understanding from human embryo studies is that human pre-implantation development is characterized by reprogramming and programming that encompasses fusion of the egg and sperm pronuclei, epigenetic reprogramming and modification, an extensive wave of degradation of maternal transcripts, and activation of the nascent human embryonic genome. Parameters of the first three mitotic divisions can predict the success or failure of developing to the blastocyst stage; this suggests that the success or failure to develop might be a predetermined property of early human embryonic cells. Human embryos develop cell-autonomously by the 8-cell stage but show no evidence of commitment to the TE or ICM at this stage. Human embryos are remarkably prone to errors in genetic and epigenetic programs with more than 80% of embryos carrying numerical and/or structural chromosomal abnormalities and at least 30% are characterized by errors in global epigenetic modification and abnormal gene expression.
Given the importance of embryo development in human biology and pathology, an understanding of the cellular dynamics, molecular programs and genetic and epigenetic correlates of normal and abnormal development is clearly needed. The translation of basic studies of human embryo development also promises to improve ART by enabling the identification of viable embryos to allow their transfer earlier and in fewer numbers, thereby reducing adverse outcomes such as high-risk multiple births. Studies of human embryo development will also inform directed differentiation methods using pluripotent hESCs derived from the human embryo and will be useful for establishing optimized culture conditions for the derivation of stem cells from embryos and induced pluripotent stem cells (iPSCs).
Note added in proof
Two recent papers bring new insight into the roles of FGF and MAP kinase signaling in the specification of the epiblast and primitive endoderm (hypoblast) cell lineages in human embryos. Cell sorting within the ICM of mouse embryos is thought to require FGF/MAP kinase signaling (Lanner and Rossant 2010); however, it is unclear whether this is a conserved mechanism for the specification of epiblast and primitive endoderm lineages in other species. Kuijk et al. demonstrate that pharmacological inhibition of MAP kinase signaling in bovine embryos blocks GATA6-expressing primitive endoderm cells, similar to the mouse; although, intriguingly, in human embryos MAP kinase inhibition has no effect (Kuijk et al., 2012). Roode et al. also demonstrate that the development of the primitive endoderm in human embryos is unaffected by the inhibition of FGF and MAP kinase signaling (Roode et al., 2012). These results suggest that there are species-specific mechanisms in the specification of epiblast and primitive endoderm lineages and that further insight is needed into the molecular basis of cell sorting within the human ICM.
Acknowledgments
We thank Thomas Moreau for the image of feeder-free hESCs and members of the R.A.P. and R.A.R.P. laboratories for helpful discussions
Footnotes
Funding
Work in the authors’ laboratories is funded by the California Institute for Regenerative Medicine (R.A.R.P.); the National Institutes of Health (as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research to R.A.R.P.); the UK Medical Research Council (R.A.P.); the March of Dimes Foundation (R.A.P., K.K.N. and R.A.R.P.); and the Cambridge Centre for Trophoblast Research (K.K.N.). Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
References
- Alfarawati S., Fragouli E., Colls P., Stevens J., Gutiérrez-Mateo C., Schoolcraft W., Katz-Jaffe M., Wells D. (2011). The relationship between blastocyst morphology, chromosomal abnormality, and embryo gender. Fertil. Steril. 95, 520–524 [DOI] [PubMed] [Google Scholar]
- Amit M., Carpenter M., Inokuma M., Chiu C., Harris C., Waknitz M., Itskovitz-Eldor J., Thomson J. (2000). Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev. Biol. 227, 271–278 [DOI] [PubMed] [Google Scholar]
- Baart E., Martini E., van den Berg I., Macklon N., Galjaard R., Fauser B., Opstal D. V. (2006). Preimplantation genetic screening reveals a high incidence of aneuploidy and mosaicism in embryos from young women undergoing IVF. Hum. Reprod. 21, 223–233 [DOI] [PubMed] [Google Scholar]
- Baltaci V., Satiroglu H., Kabukçu C., Unsal E., Aydinuraz B., Uner O., Aktas Y., Cetinkaya E., Turhan F., Aktan A. (2006). Relationship between embryo quality and aneuploidies. Reprod. Biomed. Online 12, 77–82 [DOI] [PubMed] [Google Scholar]
- Beaujean N., Taylor J., Gardner J., Wilmut I., Meehan R., Young L. (2004). Effect of limited DNA methylation reprogramming in the normal sheep embryo on somatic cell nuclear transfer. Biol. Reprod. 71, 185–193 [DOI] [PubMed] [Google Scholar]
- Bell C. E., Calder M. D., Watson A. J. (2008). Genomic RNA profiling and the programme controlling preimplantation mammalian development. Mol. Hum. Reprod. 14, 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkhalifa M., Kasakyan S., Clement P., Baldi M., Tachdjian G., Demirol A., Gurgan T., Fiorentino F., Mohammed M., Qumsiyeh M. (2005). Array comparative genomic hybridization profiling of first-trimester spontaneous abortions that fail to grow in vitro. Prenat. Diagn. 25, 894–900 [DOI] [PubMed] [Google Scholar]
- Berg D. K., Smith C. S., Pearton D. J., Wells D. N., Broadhurst R., Donnison M., Pfeffer P. L. (2011). Trophectoderm lineage determination in cattle. Dev. Cell 20, 244–255 [DOI] [PubMed] [Google Scholar]
- Bernardo A. S., Faial T., Gardner L., Niakan K. K., Ortmann D., Senner C. E., Callery E. M., Trotter M. W., Hemberger M., Smith J. C., et al. (2011). BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9, 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggers J., Summers M. (2008). Choosing a culture medium, making informed choices. Fertil. Steril. 90, 473–483 [DOI] [PubMed] [Google Scholar]
- Bongso A., Tan S. (2005). Human blastocyst culture and derivation of embryonic stem cell lines. Stem Cell Rev. 1, 87–98 [DOI] [PubMed] [Google Scholar]
- Boumela I., Assou S., Aouacheria A., Haouzi D., Dechaud H., Vos J. D., Handyside A., Hamamah S. (2011). Involvement of BCL2 family members in the regulation of human oocyte and early embryo survival and death, gene expression and beyond. Reproduction 141, 549–561 [DOI] [PubMed] [Google Scholar]
- Braude P., Bolton V., Moore S. (1988). Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332, 459–461 [DOI] [PubMed] [Google Scholar]
- Brison D. (2000). Apoptosis in mammalian preimplantation embryos: regulation by survival factors. Hum. Fertil. 3, 36–47 [DOI] [PubMed] [Google Scholar]
- Brons I., Smithers L., Trotter M., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S. M., Howlett S., Clarkson A., Ahrlund-Richter L., Pedersen R., et al. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 [DOI] [PubMed] [Google Scholar]
- Cedars M. (2005). Introduction to infertility. In Infertility (ed. Cedars M.). New York, NY: McGraw-Hill; [Google Scholar]
- Chen A. E., Egli D., Niakan K., Deng J., Akutsu H., Yamaki M., Cowan C., Fitz-Gerald C., Zhang K., Melton D. A., et al. (2009). Optimal timing of inner cell mass isolation increases the efficiency of human embryonic stem cell derivation and allows generation of sibling cell lines. Cell Stem Cell 4, 103–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng E. Y., Hunt P. A., Naluai-Cecchini T. A., Fligner C. L., Fujimoto V. Y., Pasternack T. L., Schwartz J. M., Steinauer J. E., Woodruff T. J., Cherry S. M., et al. (2009). Meiotic recombination in human oocytes. PLoS Genet. 5, e1000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang T., Schultz R., Lampson M. A. (2011). Age-dependent susceptibility of chromosome cohesion to premature separase activation in mouse oocytes. Biol. Reprod. 85, 1279–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke G. (2006). A.R.T. and history, 1678-1978. Hum. Reprod. 21, 1645–1650 [DOI] [PubMed] [Google Scholar]
- Di Giorgio F. P., Boulting G. L., Bobrowicz S., Eggan K. C. (2008). Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell 3, 637–648 [DOI] [PubMed] [Google Scholar]
- Dietrich J. E., Hiiragi T. (2007). Stochastic patterning in the mouse pre-implantation embryo. Development 134, 4219–4231 [DOI] [PubMed] [Google Scholar]
- Dobson A. T., Raja R., Abeyta M. J., Taylor T., Shen S., Haqq C., Reijo Pera R. A. (2004). The unique transcriptome through day 3 of human preimplantation development. Hum. Mol. Genet. 13, 1461–1470 [DOI] [PubMed] [Google Scholar]
- Duranthon V., Watson A. J., Lonergan P. (2008). Preimplantation embryo programming: transcription, epigenetics, and culture environment. Reproduction 135, 141–150 [DOI] [PubMed] [Google Scholar]
- Edwards R., Bavister B., Steptoe P. (1969). Early stages of fertilization in vitro of human eggs matured in vitro. Nature 221, 632–635 [DOI] [PubMed] [Google Scholar]
- Edwards R., Steptoe P., Purdy J. (1970). Fertilisation and cleavage in vitro of preovulatory human oocytes. Nature 227, 1307–1309 [DOI] [PubMed] [Google Scholar]
- Edwards R., Steptoe P., Purdy J. (1980). Establishing full-term human pregnancies using cleaving embryos grown in vitro. Br. J. Obstet. Gynaecol. 87, 737–756 [DOI] [PubMed] [Google Scholar]
- Edwards R. G., Hansis C. (2005). Initial differentiation of blastomeres in 4-cell human embryos and its significance for early embryogenesis and implantation. Reprod. Biomed. Online 11, 206–218 [DOI] [PubMed] [Google Scholar]
- Enders A. C., Lopata A. (1999). Implantation in the marmoset monkey: expansion of the early implantation site. Anat. Rec. 256, 279–299 [DOI] [PubMed] [Google Scholar]
- Evers J. L. (2002). Female subfertility. Lancet 360, 151–159 [DOI] [PubMed] [Google Scholar]
- Fernández-González R., de Dios Hourcade J., López-Vidriero I., Benguría A., Fonseca F. D., Gutiérrez-Adán A. (2009). Analysis of gene transcription alterations at the blastocyst stage related to the long-term consequences of in vitro culture in mice. Reproduction 137, 271–283 [DOI] [PubMed] [Google Scholar]
- Flach G., Johnson M. H., Braude P. R., Taylor R. A., Bolton V. N. (1982). The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J. 1, 681–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French D. B., Sabanegh E. S., Jr, Goldfarb J., Desai N. (2010). Does severe teratozoospermia affect blastocyst formation, live birth rate, and other clinical outcome parameters in ICSI cycles? Fertil. Steril. 93, 1097–1103 [DOI] [PubMed] [Google Scholar]
- Fritz B., Hallermann C., Olert J., Fuchs B., Bruns M., Aslan M., Schmidt S., Coerdt W., Müntefering H., Rehder H. (2001). Cytogenetic analyses of culture failures by comparative genomic hybridisation (CGH)-Re-evaluation of chromosome aberration rates in early spontaneous abortions. Eur. J. Hum. Genet. 9, 539–547 [DOI] [PubMed] [Google Scholar]
- Fujioka T., Yasuchika K., Nakamura Y., Nakatsuji N., Suemori H. (2004). A simple and efficient cryopreservation method for primate embryonic stem cells. Int. J. Dev. Biol. 48, 1149–1154 [DOI] [PubMed] [Google Scholar]
- Fulka H., Mrazek M., Tepla O., Fulka J., Jr (2004). DNA methylation pattern in human zygotes and developing embryos. Reproduction 128, 703–708 [DOI] [PubMed] [Google Scholar]
- Galan A., Montaner D., Poo M., Valbuena D., Ruiz V., Aguilar C., Dopazo J., Simon C. (2010). Functional genomics of 5- to 8-cell stage human embryos by blastomere single-cell cDNA analysis. PLoS ONE 5, e13615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner D., Lane M., Schoolcraft W. (2000). Culture and transfer of viable blastocysts, a feasible proposition for human IVF. Hum. Reprod. 6 Suppl., 9–23 [PubMed] [Google Scholar]
- Gardner R. (2001). Specification of embryonic axes begins before cleavage in normal mouse development. Development 128, 839–847 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Hong C., Feingold E., Ghosh P., Bhaumik P., Dey S. (2011). Epidemiology of down syndrome, new insight into the multidimensional interactions among genetic and environmental risk factors in the oocyte. Am. J. Epidemiol. 174, 1009–1016 [DOI] [PubMed] [Google Scholar]
- Greber B., Wu G., Bernemann C., Joo J. Y., Han D. W., Ko K., Tapia N., Sabour D., Sterneckert J., Tesar P., Schöler H. R. (2010). Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell 6, 215–226 [DOI] [PubMed] [Google Scholar]
- Hamatani T., Daikoku T., Wang H., Matsumoto H., Carter M. G., Ko M. S., Dey S. K. (2004). Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation. Proc. Natl. Acad. Sci. USA 101, 10326–10331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T., Ko M., Yamada M., Kuji N., Mizusawa Y., Shoji M., Hada T., Asada H., Maruyama T., Yoshimura Y. (2006). Global gene expression profiling of preimplantation embryos. Hum. Cell 19, 98–117 [DOI] [PubMed] [Google Scholar]
- Hansis C., Grifo J. A., Tang Y., Krey L. C. (2002). Assessment of beta-HCG, beta-LH mRNA and ploidy in individual human blastomeres. Reprod. Biomed. Online 5, 156–161 [DOI] [PubMed] [Google Scholar]
- Hardarson T., Caisander G., Sjögren A., Hanson C., Hamberger L., Lundin K. (2003). A morphological and chromosomal study of blastocysts developing from morphologically suboptimal human pre-embryos compared with control blastocysts. Hum. Reprod. 18, 399–407 [DOI] [PubMed] [Google Scholar]
- Hardy K., Wright C. S., Franks S., Winston R. M. (2000). In vitro maturation of oocytes. Br. Med. Bull. 56, 588–602 [DOI] [PubMed] [Google Scholar]
- Harton G., Rycke M. D., Fiorentino F., Moutou C., SenGupta S., Traeger-Synodinos J., Harper J. C. and the European Society for Human Reproduction and Embryology (ESHRE) PGD Consortium (2011). ESHRE PGD consortium best practice guidelines for amplification-based PGD. Hum. Reprod. 26, 33–40 [DOI] [PubMed] [Google Scholar]
- Hasegawa K., Fujioka T., Nakamura Y., Nakatsuji N., Suemori H. (2006). A method for the selection of human embryonic stem cell sublines with high replating efficiency after single-cell dissociation. Stem Cells 24, 2649–2660 [DOI] [PubMed] [Google Scholar]
- Hassold T., Hunt P. (2001). To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291 [DOI] [PubMed] [Google Scholar]
- Hassold T., Hunt P. (2009). Maternal age and chromosomally abnormal pregnancies: what we know and what we wish we knew. Curr. Opin. Pediatr. 21, 703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T., Abruzzo M., Adkins K., Griffin D., Merrill M., Millie E., Saker D., Shen J., Zaragoza M. (1996). Human aneuploidy: incidence, origin, and etiology. Environ. Mol. Mutagen. 28, 167–175 [DOI] [PubMed] [Google Scholar]
- Hertig A., Rock J., Adams E., Mulligan W. (1954). On the preimplantation stages of the human ovum: a description of four normal and four abnormal specimens ranging from the second to the fifth day of development. Contrib. Embryol. Carnegie Instn. 35, 119–220 [Google Scholar]
- Hertig A. T., Rock J., Adams E. C. (1956). A description of 34 human ova within the first 17 days of development. Am. J. Anat. 98, 435–493 [DOI] [PubMed] [Google Scholar]
- Hoffman D., Zellman G., Fair C., Mayer J., Zeitz J., Gibbons W., Turner T. G., Jr (2003). Cryopreserved embryos in the United States and their availability for research. Fertil. Steril. 79, 1063–1069 [DOI] [PubMed] [Google Scholar]
- Hunt C. J., Timmons P. M. (2007). Cryopreservation of human embryonic stem cell lines. Methods Mol. Biol. 368, 261–270 [DOI] [PubMed] [Google Scholar]
- Hunt P. A., Hassold T. J. (2002). Sex matters in meiosis. Science 296, 2181–2183 [DOI] [PubMed] [Google Scholar]
- Hunt P., Hassold T. (2008). Human female meiosis: what makes a good egg go bad? Trends Genet. 24, 86–93 [DOI] [PubMed] [Google Scholar]
- Hunt P., Hassold T. (2010). Female meiosis: coming unglued with age. Curr. Biol. 20, R699–R702 [DOI] [PubMed] [Google Scholar]
- Ilic D., Genbacev O., Krtolica A. (2007). Derivation of hESC from intact blastocysts. Curr. Protoc. Stem Cell Biol. 1:1A.2.1–1A.2.18 [DOI] [PubMed] [Google Scholar]
- Inge G., Brinsden P., Elder K. (2005). Oocyte number per live birth in IVF, were Steptoe and Edwards less wasteful? Hum. Reprod. 20, 588–592 [DOI] [PubMed] [Google Scholar]
- James D., Noggle S. A., Swigut T., Brivanlou A. H. (2006). Contribution of human embryonic stem cells to mouse blastocysts. Dev. Biol. 295, 90–102 [DOI] [PubMed] [Google Scholar]
- Jedrusik A., Parfitt D. E., Guo G., Skamagki M., Grabarek J. B., Johnson M. H., Robson P., Zernicka-Goetz M. (2008). Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. 22, 2692–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H. (2003). IVF, past and future. Reprod. Biomed. Online 6, 375–381 [DOI] [PubMed] [Google Scholar]
- Kalista T., Freeman H. A., Behr B., Reijo Pera R. A., Scott C. T. (2011). Donation of embryos for human development and stem cell research. Cell Stem Cell 8, 360–362 [DOI] [PubMed] [Google Scholar]
- Katari S., Turan N., Bibikova M., Erinle O., Chalian R., Foster M., Gaughan J., Coutifaris C., Sapienza C. (2009). DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum. Mol. Genet. 18, 3769–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S. J., Witty A. D., Gagliardi M., Dubois N. C., Niapour M., Hotta A., Ellis J., Keller G. (2011). Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8, 228–240 [DOI] [PubMed] [Google Scholar]
- Khosla S., Dean W., Reik W., Feil R. (2001). Preimplantation embryos and its long-term effects on gene expression and phenotype. Hum. Reprod. Update 7, 419–427 [DOI] [PubMed] [Google Scholar]
- Koo J.-J., Choi S.-Y., Jeong H.-J., Roh S.-I. (2011). Fragmentation of embryos is associated with both necrosis and apoptosis. Fertil. Steril. 96, 187–192 [DOI] [PubMed] [Google Scholar]
- Kuijk E. W., van Tol L. T. A., Van de Velde H., Wubbolts R., Welling M., Geijsen N., Roelen B. A. J. (2012). The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development 139, 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H., Ogilvie C., Handyside A. (1998). Chromosomal mosaicism in cleavage-stage human embryos and the accuracy of single-cell genetic analysis. J. Assist. Reprod. Genet. 15, 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanner F., Rossant J. (2010). The role of FGF/Erk signaling in pluripotent cells. Development 137, 3351–3360 [DOI] [PubMed] [Google Scholar]
- Lazzari G., Wrenzycki C., Herrmann D., Duchi R., Kruip T., Niemann H., Galli C. (2002). Cellular and molecular deviations in bovine in vitro-produced embryos are related to the large offspring syndrome. Biol. Reprod. 67, 767–775 [DOI] [PubMed] [Google Scholar]
- Leese H. (2002). Quiet please, do not disturb: a hypothesis of embryo metabolism and viability. BioEssays 24, 845–849 [DOI] [PubMed] [Google Scholar]
- Lim D., Bowdin S., Tee L., Kirby G., Blair E., Fryer A., Lam W., Oley C., Cole T., Brueton L., et al. (2009). Clinical and molecular genetic features of Beckwith-Wiedemann syndrome associated with assisted reproductive technologies. Hum. Reprod. 24, 741–747 [DOI] [PubMed] [Google Scholar]
- Lin T., Yen J., Gong K., Hsu T., Chen L. (2003). IGF-1/IGFBP-1 increases blastocyst formation and total blastocyst cell number in mouse embryo culture and facilitates the establishment of a stem-cell line. BMC Cell Biol. 4, 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklon N., Geraedts J., Fauser B. (2002). Conception to ongoing pregnancy: the ‘black box’ of early pregnancy loss. Hum. Reprod. Update 8, 333–343 [DOI] [PubMed] [Google Scholar]
- Memili E., First N. L. (2000). Zygotic and embryonic expression in cow: a review of timing and mechanisms of early gene expression as compared with other species. Zygote 8, 87–96 [DOI] [PubMed] [Google Scholar]
- Mercader A., Valbuena D., Simón C. (2006). Human embryo culture. Methods Enzymol. 420, 3–18 [DOI] [PubMed] [Google Scholar]
- Mitalipov S., Kuo H., Hennebold J., Wolf D. (2003). Oct-4 expression in pluripotent cells of the rhesus monkey. Biol. Reprod. 69, 1785–1792 [DOI] [PubMed] [Google Scholar]
- Moffett A., Loke C. (2006). Immunology of placentation in eutherian mammals. Nat. Rev. Immunol. 6, 584–594 [DOI] [PubMed] [Google Scholar]
- Morris S. A., Teo R. T., Li H., Robson P., Glover D. M., Zernicka-Goetz M. (2010). Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc. Natl. Acad. Sci. USA 107, 6364–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motosugi N., Bauer T., Polanski Z., Solter D., Hiiragi T. (2005). Polarity of the mouse embryo is established at blastocyst and is not prepatterned. Genes Dev. 19, 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munné S., Lee A., Rosenwaks Z., Grifo J., Cohen J. (1993). Diagnosis of major chromosome aneuploidies in human preimplantation embryos. Hum. Reprod. 8, 2185–2191 [DOI] [PubMed] [Google Scholar]
- Munné S., Tomkin G., Cohen J. (2009). Selection of embryos by morphology is less effective than by a combination of aneuploidy testing and morphology observations. Fertil. Steril. 91, 943–945 [DOI] [PubMed] [Google Scholar]
- Niakan K. K., Ji H., Maehr R., Vokes S. A., Rodolfa K. T., Sherwood R. I., Yamaki M., Dimos J. T., Chen A. E., Melton D. A., et al. (2010). Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 24, 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norwitz E. R., Schust D. J., Fisher S. J. (2001). Implantation and the survival of early pregnancy. N. Engl. J. Med. 345, 1400–1408 [DOI] [PubMed] [Google Scholar]
- Ohgushi M., Matsumura M., Eiraku M., Murakami K., Aramaki T., Nishiyama A., Muguruma K., Nakano T., Suga H., Ueno M., et al. (2010). Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell 7, 225–239 [DOI] [PubMed] [Google Scholar]
- Osafune K., Caron L., Borowiak M., Martinez R. J., Fitz-Gerald C. S., Sato Y., Cowan C. A., Chien K. R., Melton D. A. (2008). Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 26, 313–315 [DOI] [PubMed] [Google Scholar]
- Palermo G., Joris H., Devroey P., Van Steirteghem A. C. (1992). Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte. Lancet 340, 17–18 [DOI] [PubMed] [Google Scholar]
- Piotrowska K., Wianny F., Pedersen R., Zernicka-Goetz M. (2001). Blastomeres arising from the first cleavage division have distinguishable fates in normal mouse development. Development 128, 3739–3748 [DOI] [PubMed] [Google Scholar]
- Plachta N., Bollenbach T., Pease S., Fraser S., Pantazis P. (2011). Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat. Cell Biol. 13, 117–123 [DOI] [PubMed] [Google Scholar]
- Plusa B., Piliszek A., Frankenberg S., Artus J., Hadjantonakis A. K. (2008). Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development 135, 3081–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racowsky C. (2002). High rates of embryonic loss, yet high incidence of multiple births in human ART: Is this paradoxical? Theriogenol. 57, 87–96 [DOI] [PubMed] [Google Scholar]
- Ralston A., Rossant J. (2008). Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev. Biol. 313, 614–629 [DOI] [PubMed] [Google Scholar]
- Reijo Pera R. A., DeJonge C., Bossert N., Yao M., Hwa Yang J. Y., Asadi N. B., Wong W., Wong C., Firpo M. T. (2009). Gene expression profiles of human inner cell mass cells and embryonic stem cells. Differentiation 78, 18–23 [DOI] [PubMed] [Google Scholar]
- Reubinoff B. E., Pera M. F., Vajta G., Trounson A. O. (2001). Effective cryopreservation of human embryonic stem cells by the open pulled straw vitrification method. Hum. Reprod. 16, 2187–2194 [DOI] [PubMed] [Google Scholar]
- Robertson S., Sjöblom C., Jasper M., Norman R., Seamark R. (2001). Granulocyte-macrophage colony-stimulating factor promotes glucose transport and blastomere viability in murine preimplantation embryos. Biol. Reprod. 64, 1206–1215 [DOI] [PubMed] [Google Scholar]
- Robinson W., Christian S., Kuchinka B., Peñaherrera M., Das S., Schuffenhauer S., Malcolm S., Schinzel A., Hassold T., Ledbetter D. (2000). Somatic segregation errors predominantly contribute to the gain or loss of a paternal chromosome leading to uniparental disomy for chromosome 15. Clin. Genet. 57, 349–358 [DOI] [PubMed] [Google Scholar]
- Roode M., Blair K., Snell P., Elder K., Marchant S., Smith A., Nichols J. (2012). Human hypoblast formation is not dependent on FGF signalling. Dev. Biol. 361, 358–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother F., Shmidt T., Popova E., Krivokharchenko A., Hügel S., Vilianovich L., Ridders M., Tenner K., Alenina N., Köhler M., et al. (2011). Importin α7 is essential for zygotic genome activation and early mouse development. PLoS ONE 6, e18310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathananthan A., Osianlis T. (2010). Human embryo culture and assessment for the derivation of embryonic stem cells (ESC). Methods Mol. Biol. 584, 1–20 [DOI] [PubMed] [Google Scholar]
- Schoolcraft W. B., Treff N. R., Stevens J. M., Ferry K., Katz-Jaffe M., Scott R. T., Jr (2011). Live birth outcome with trophectoderm biopsy, blastocyst vitrification, and single-nucleotide polymorphism microarray-based comprehensive chromosome screening in infertile patients. Fertil. Steril. 96, 638–640 [DOI] [PubMed] [Google Scholar]
- Selesniemi K., Lee H., Muhlhauser A., Tilly J. (2011). Prevention of maternal aging-associated oocyte aneuploidy and meiotic spindle defects in mice by dietary and genetic strategies. Proc. Natl. Acad. Sci. USA 108, 12319–12324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons D., Fortier A., Cross J. (2007). Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev. Biol. 304, 567–578 [DOI] [PubMed] [Google Scholar]
- Sjöblom C., Wikland M., Robertson S. (1999). Granulocyte-macrophage colony-stimulating factor promotes human blastocyst development in vitro. Hum. Reprod. 14, 3069–3076 [DOI] [PubMed] [Google Scholar]
- Sjöblom C., Wikland M., Robertson S. (2002). Granulocyte-macrophage colony-stimulating factor (GM-CSF). acts independently of the beta common subunit of the GM-CSF receptor to prevent inner cell mass apoptosis in human embryos. Biol. Reprod. 67, 1817–1823 [DOI] [PubMed] [Google Scholar]
- Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M., Rogers D. (1988). Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336, 688–690 [DOI] [PubMed] [Google Scholar]
- Spallanzani L. (1785). Esperimenti che servono nella storio della genera-zione di animali e piante. Geneva: Barthelmi Ciro; [Google Scholar]
- Spanos S., Becker D., Winston R., Hardy K. (2000). Anti-apoptotic action of insulin-like growth factor-I during human preimplantation embryo development. Biol. Reprod. 63, 1413–1420 [DOI] [PubMed] [Google Scholar]
- Steptoe P. C., Edwards R. G. (1978). Birth after the reimplantation of a human embryo. Lancet 2, 366 [DOI] [PubMed] [Google Scholar]
- Ström S., Inzunza J., Grinnemo K., Holmberg K., Matilainen E., Strömberg A. M., Blennow E., Hovatta O. (2007). Mechanical isolation of the inner cell mass is effective in derivation of new human embryonic stem cell lines. Hum. Reprod. 22, 3051–3058 [DOI] [PubMed] [Google Scholar]
- Tarkowski A. K., Suwinska A., Czolowska R., Ozdzenski W. (2010). Individual blastomeres of 16- and 32-cell mouse embryos are able to develop into foetuses and mice. Dev. Biol. 348, 190–198 [DOI] [PubMed] [Google Scholar]
- Taylor D. M., Ray P. F., Ao A., Winston R. M., Handyside A. H. (1997). Paternal transcripts for glucose-6-phosphate dehydrogenase and adenosine deaminase are first detectable in the human preimplantation embryo at the three-to-four-cell stage. Mol. Reprod. Dev. 48, 442–448 [DOI] [PubMed] [Google Scholar]
- Tesar P., Chenoweth J., Brook F., Davies T., Evans E., Mack D., Gardner R., McKay R. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 [DOI] [PubMed] [Google Scholar]
- Tesarík J., Kopecný V., Plachot M., Mandelbaum J. (1987). High-resolution autoradiographic localization of DNA-containing sites and RNA synthesis in developing nucleoli of human preimplantation embryos: a new concept of embryonic nucleologenesis. Development 101, 777–791 [DOI] [PubMed] [Google Scholar]
- Thomson J., Itskovitz-Eldor J., Shapiro S., Waknitz M., Swiergiel J., Marshall V., Jones J. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- Torres-Padilla M., Parfitt D., Kouzarides T., Zernicka-Goetz M. (2007). Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445, 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso C., Bosch E., Rubio C., Remohí J., Simón C., Pellicer A. (2003). The origin of biochemical pregnancies: lessons learned from preimplantation genetic diagnosis. Fertil. Steril. 79, 449–450 [DOI] [PubMed] [Google Scholar]
- Vallier L. (2011). Serum-free and feeder-free culture conditions for human embryonic stem cells. Methods Mol. Biol. 690, 57–66 [DOI] [PubMed] [Google Scholar]
- Vallier L., Alexander M., Pedersen R. A. (2005). Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 118, 4495–4509 [DOI] [PubMed] [Google Scholar]
- Vanneste E., Voet T., Caignec C. L., Ampe M., Konings P., Melotte C., Debrock S., Amyere M., Vikkula M., Schuit F., et al. (2009). Chromosome instability is common in human cleavage-stage embryos. Nat. Med. 15, 577–583 [DOI] [PubMed] [Google Scholar]
- Vanneste E., Melotte C., Voet T., Robberecht C., Debrock S., Pexsters A., Staessen C., Tomassetti C., Legius E., D’Hooghe T., et al. (2011). PGD for a complex chromosomal rearrangement by array comparative genomic hybridization. Hum. Reprod. 26, 941–949 [DOI] [PubMed] [Google Scholar]
- Vassena R., Boué S., González-Roca E., Aran B., Auer H., Veiga A., Belmonte J. (2011). Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development 138, 3699–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaragavan K., Szabo E., Bosse M., Ramos-Mejia V., Moon R. T., Bhatia M. (2009). Noncanonical Wnt signaling orchestrates early developmental events toward hematopoietic cell fate from human embryonic stem cells. Cell Stem Cell 4, 248–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Piotrowska K., Ciemerych M. A., Milenkovic L., Scott M. P., Davis R. W., Zernicka-Goetz M. (2004). A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell 6, 133–144 [DOI] [PubMed] [Google Scholar]
- Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J., Nishikawa S., Nishikawa S., Muguruma K., et al. (2007). A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25, 681–686 [DOI] [PubMed] [Google Scholar]
- Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., Wagner E. F., Metcalf D., Nicola N. A., Gough N. M. (1988). Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336, 684–687 [DOI] [PubMed] [Google Scholar]
- Wilton L., Williamson R., McBain J., Edgar D., Voullaire L. (2002). Birth of a healthy infant after preimplantation confirmation of euploidy by comparative genomic hybridization. N. Engl. J. Med. 345, 1537–1541 [DOI] [PubMed] [Google Scholar]
- Wong C., Loewke K., Bossert N., Behr B., DeJonge C., Baer T., Reijo Pera R. R. (2010). Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat. Biotechnol. 28, 1115–1121 [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Nichols J., Chambers I., Smith A. (2003). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292 [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F., Schultz R. (2005). RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev. Biol. 283, 40–57 [DOI] [PubMed] [Google Scholar]
- Zhang P., Zucchelli M., Bruce S., Hambiliki F., Stavreus-Evers A., Levkov L., Skottman H., Kerkela E., Kere J., Hovatta O. (2009). Transcriptome profiling of human preimplantation development. PLoS ONE 4, e7844 [DOI] [PMC free article] [PubMed] [Google Scholar]