Abstract
Using in vitro and in vivo assays, we define a network of Her/Hes dimers underlying transcriptional negative feedback within the zebrafish segmentation clock. Some of the dimers do not appear to be DNA-binding, whereas those dimers that do interact with DNA have distinct preferences for cis regulatory sequences. Dimerization is specific, with Hes6 serving as the hub of the network. Her1 binds DNA only as a homodimer but will also dimerize with Hes6. Her12 and Her15 bind DNA both as homodimers and as heterodimers with Hes6. Her7 dimerizes strongly with Hes6 and weakly with Her15. This network structure engenders specific network dynamics and imparts greater influence to the Her7 node. Computational analysis supports the hypothesis that Her7 disproportionately influences the availability of Hes6 to heterodimerize with other Her proteins. Genetic experiments suggest that this regulation is important for operation of the network. Her7 therefore has two functions within the zebrafish segmentation clock. Her7 acts directly within the delayed negative feedback as a DNA-binding heterodimer with Hes6. Her7 also has an emergent function, independent of DNA binding, in which it modulates network topology via sequestration of the network hub.
Keywords: Segmentation, Somite, Zebrafish, Segmentation clock, Her/Hes, Dimer network
INTRODUCTION
Somitogenesis is the process of segmenting the vertebral and myogenic progenitors during vertebrate embryogenesis. The segmental pattern is generated in the presomitic mesoderm (PSM) by an oscillating gene network called the segmentation clock while posterior to anterior gradients of Fgf and Wnt signaling maintain cells in a labile state until the segmental pattern is established (Pourquié, 2011). The segmentation clock causes cells of the PSM to undergo repeated cycles of gene expression and repression. The most conserved of these oscillating genes in zebrafish, mouse and chick are members of the hairy-enhancer of split related family (hes or her) (Krol et al., 2011). Her/Hes are basic helix-loop-helix (bHLH) proteins that dimerize, bind DNA and repress transcription. These genes were among the first oscillating genes identified in each species and are crucial for somitogenesis (Bessho et al., 2003; Bessho et al., 2001; Dunwoodie et al., 2002; Henry et al., 2002; Holley et al., 2000; Holley et al., 2002; Jouve et al., 2000; Oates and Ho, 2002; Palmeirim et al., 1997; Sawada et al., 2000). The hairy genes also oscillate in humans, and mutation of hes7 leads to a malformed vertebral column (Sparrow et al., 2008; Sparrow et al., 2010; William et al., 2007). In zebrafish, the segmentation clock is thought to center on cycles of her gene transcription and self-repression (Holley et al., 2002; Lewis, 2003; Oates and Ho, 2002). Indeed, a mutation of zebrafish hes6 lengthens the period of the zebrafish segmentation clock (Schroter and Oates, 2010). Models suggest that stable oscillations depend upon the intrinsic delay caused by transcription and translation of the her genes (Lewis, 2003; Monk, 2003). This aspect of the model was recently supported by experiments in the mouse showing that deletion of the introns of hes7 perturbs the segmentation clock (Takashima et al., 2011). In mouse, hes1, hes5, hes7 and hey1 oscillate in the PSM, but only hes7 has been shown to be necessary for somitogenesis (Bessho et al., 2001; Dunwoodie et al., 2002; Jouve et al., 2000; Krol et al., 2011). hes7 was further shown to oscillate antiphase to Hes7 protein in vivo, and the appropriate half-lives of Hes7 and Hes1 are required for oscillation in vivo and in vitro, respectively (Bessho et al., 2003; Bessho et al., 2001; Hirata et al., 2004; Hirata et al., 2002).
In zebrafish, seven her genes have been shown to oscillate – of these her1, her7, her11, her12 and her15 are the best characterized (Gajewski et al., 2006; Gajewski et al., 2003; Henry et al., 2002; Holley et al., 2000). hes6 (previously her13.2) does not oscillate but rather is expressed in a posterior to anterior gradient in the tailbud (Kawamura et al., 2005). hes6 and her1 expression are both promoted by Fgf signaling and thus may function to integrate the zebrafish oscillator with tissue maturation (Ishimatsu et al., 2010; Kawamura et al., 2005). Abrogation of her/hes gene function via morpholino knockdown or mutant analyses suggests that while the different her/hes genes may have distinct roles in somitogenesis, there is some functional redundancy (Gajewski et al., 2006; Gajewski, et al., 2003; Henry et al., 2002; Holley et al., 2002; Kawamura et al., 2005; Oates and Ho, 2002; Schroter and Oates, 2010; Shankaran et al., 2007; Sieger et al., 2006; Sieger et al., 2004).
To understand the zebrafish segmentation clock network, a better characterization of the biochemical properties of the Her proteins is necessary. These proteins have four highly conserved domains: the basic domain, the helix-loop-helix, the orange domain and the WRPW motif. The basic domain forms a continuous α-helix with the N-terminal helix of the HLH domain and contacts the major groove of the class B E-box sequence CACGYG. The HLH domain has been shown largely to mediate dimerization, whereas the orange domain can influence the specificity of dimer formation (Fischer and Gessler, 2007; Kageyama et al., 2007). Lastly, the WRPW motif at the C-terminus of the Hairys recruits the co-repressor Groucho to DNA (Paroush et al., 1994). bHLH proteins also repress transcription via competitive dimerization, in which some dimers are unable to bind DNA (Cinquin and Page, 2007; Fisher et al., 1996; Pichon et al., 2004).
Here, we explore two biochemical parameters that may underlie the differing functions of Her1, Her7, Her11, Her12, Her15 and Hes6 and use computational and genetic analyses built upon the biochemical data to understand the significance of the network topology. First, we examine dimerization among these proteins in vitro using electrophoretic mobility shift assays (EMSAs) and in vivo via bimolecular fluorescence complementation (BiFC). Second, we test the ability of each dimer to bind a number of putative E-boxes and find variation in preference for cis regulatory sequences. Our data reveal relationships between the dimer network topology and the function of these integrated components within the zebrafish segmentation clock.
MATERIALS AND METHODS
Zebrafish maintenance
Zebrafish breeding and care followed standard protocols (Nüsslein-Volhard and Dahm, 2002) as approved by Yale IACUC. The wild-type strains used were Tü and TLF.
In vitro transcription and translation
mRNA and proteins were synthesized as in Brend and Holley (Brend and Holley, 2009) with the following change: proteins were synthesized using the EasyXpress Protein Synthesis Kit (Qiagen) according to the manufacturer’s instructions. Her proteins were epitope-tagged on their amino termini. The epitope-tagged proteins were detected with the following primary antibodies via western blot: anti-Flag M2 monoclonal (Sigma), anti-c-myc 9E10 monoclonal (Covance), anti-V5 V5-10 monoclonal (Sigma), anti-His (Sigma), anti-VSVG (Sigma), anti-HA (Sigma). Blots were developed using an anti-mouse IgG peroxidase-conjugated secondary antibody (Sigma) and ECL Plus Detection Reagents (GE Healthcare). Protein levels among the lysates were not normalized (supplementary material Fig. S1A).
Electrophoretic mobility shift assay
The EMSAs were performed as previously described (Brend and Holley, 2009). Kodak film was exposed for between 1 and 72 hours. Oligonucleotide sequences are provided in supplementary material Fig. S4L. Many of the gel shifts contain multiple bands. Given the number of Her dimer combinations and probes being tested, we did not optimize each binding reaction to obtain more photogenic gels. Each interpretable gel shift is supported by supershift controls.
Bimolecular fluorescence complementation
Constructs were cloned into the pCS2 vector with the N-terminal or C-terminal half of Venus YFP on the 3′ end of each her gene (Saka et al., 2008). mRNA was synthesized using the mMESSAGE mMACHINE SP6 mRNA synthesis kit (Ambion). All combinations of her genes were injected into the one-cell stage of TLF zebrafish embryos at 200 ng/μl, 400 ng/μl or 800 ng/μl each. Western blot on lysates of injected embryos indicated that the fusion proteins were expressed at different levels (supplementary material Fig. S1B,C). Embryos developed until 20% epiboly at 31°C and were analyzed for fluorescence using a Stemi SV6 fluorescent dissecting microscope (Zeiss). Each injection was performed two to three times at the different concentrations depending on whether fluorescence was observed. Representative embryos were mounted in methylcellulose and images taken on an Axioskop 2 mot plus widefield microscope (Zeiss).
Morpholino injections and luciferase assay
Percentage knockdown was calculated using a modified luciferase assay (Kamachi et al., 2008). The predicted 5′ UTR of each her gene plus 11 N-terminal amino acids were amplified by PCR and cloned into pCS2+ in frame with firefly luciferase lacking the endogenous ATG. Zebrafish embryos at the one-cell stage were injected with mRNA encoding chimeric herUTR-firefly luciferase and unmodified renilla luciferase (80 ng/μl firefly, 1.25 ng/μl renilla RNA). Injection volume was calibrated using a micrometer, with ∼500 pl injected per embryo. Half of the embryos were also injected with morpholino specific to the 5′ UTR [(0.6 mM her13.2 MO1: 5′-TGCAGTTCAGGACGCTTGAATGGG-3′ (Kawamura et al., 2005), 0.4 mM her7-antiATG MO: 5′-CATTGCACGTGTACTCCAATAGTTG-3′ (Gajewski et al., 2003)]. Embryos were incubated at 22°C or 31°C until dome stage, then 13 embryos per sample were manually dechorionated and de-yolked at room temperature. Luminescence was determined using the Dual-Luciferase Reporter Assay (Promega) and detected with a Wallac Victor 2 1420 Multi-Label Counter (Perkin Elmer). Results were averaged among three experimental replicates and are provided with standard deviation. We expect this percentage inhibition to accurately reflect the degree of knockdown of the endogenous gene because the morpholino levels are saturating: we calculate that at an average injection concentration, after 16 cell divisions the embryos are in the segmentation stages and have ∼700,000 morpholino molecules per cell. Even factoring in morpholino degradation, the morpholino should be in vast excess of her mRNA numbers, which are estimated to range from ∼10 to ∼1000 (Cinquin, 2007; Lewis, 2003). For in situ hybridization, injected embryos were incubated at 22°C or 31°C until the 8-10 somite stage and processed using standard protocols. Phenotypes were categorized double blind by two observers in parallel with little resulting variation in phenotype distributions.
Computational methods
A system of equations describing dimerization of the Her proteins at quasi-equilibrium was solved under the assumptions of well-mixed kinetics and conserved total numbers of each protein species since synthesis and degradation are much slower than binding and unbinding of monomers. The input parameters were the total concentration of the Her1, Her7, Her12, Hes6 and Her15 protein and the dissociation constants for formation of the dimers. The output parameters were the concentrations of the free monomers, the Her1, Her12, Hes6 and Her15 homodimers, as well as the Her1/Hes6, Her7/Hes6, Her7/Her15, Her12/Hes6 and Hes6/Her15 heterodimers. We performed sensitivity analysis by calculating derivatives of the steady-state dimer concentrations with respect to the dissociation constants and total protein concentrations (Rabitz et al., 1983). Analysis was performed 1000 times and sensitivity values were averaged across all runs. Since the values of the total protein concentration and of the dissociation constants are unknown, the total concentration of protein for each species was sampled from a log-normal distribution logN(μ,σ2) with (μ,σ)=(0,1). Likewise, the dissociation constants were sampled from a log-normal distribution with (μ,σ)=(–5,2). Because the Her1/Hes6 and Her7/Her15 dimers were detected weakly via BiFC and not at all by EMSA, the Kd values for these two dimers were assumed to be, on average, tenfold weaker than the other dimers. Therefore, these values were sampled from a log normal distribution with (μ,σ)=(–4,2). A custom Mathematica script was used for all calculations (available upon request).
RESULTS
Her dimerization and DNA binding in vitro
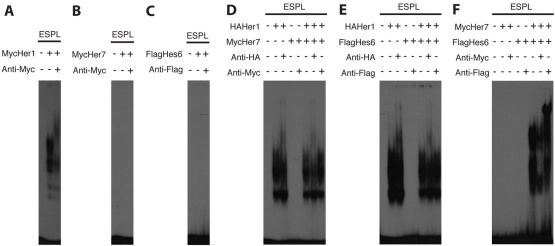
To determine which Her proteins can dimerize in vitro and bind DNA, we performed EMSAs using a probe containing a high affinity binding site for Drosophila Enhancer of split E(spl) proteins (Brend and Holley, 2009; Jennings et al., 1999). We mixed radiolabeled probe with epitope-tagged Her proteins, with or without epitope-specific antibodies. We resolved these reactions on native polyacrylamide gels and assayed for mobility shifts and super-shifts with the antibody. We examined all single extracts and pairwise combinations of Her1, Her7, Her11, Her12, Her15 and Hes6 (Fig. 1; see supplementary material Fig. S2). Only seven of the 21 possible combinations form dimers that can detectably bind DNA. These data indicate unique patterns of dimerization and DNA binding. Her1 bound only DNA as a homodimer (Fig. 1A), whereas Her7 bound DNA as a heterodimer with Hes6 only (Fig. 1F). Hes6 is more promiscuous in that it heterodimerizes and binds DNA with Her7, Her12 and Her15 (Fig. 1F; supplementary material Fig. S2). her12 and her15 were created by a relatively recent gene duplication (Shankaran et al., 2007). They bound DNA as heterodimers with Hes6 and as homodimers, but no evidence of a Her12/Her15 heterodimer was found (supplementary material Fig. S2). The Her11 gel shifts were weak and ambiguous. It may have bound DNA as a homodimer and perhaps, but not likely, as a heterodimer with Hes6. The amount of synthesized protein detected via the western blot (supplementary material Fig. S1) was not predictive of the EMSA signal. For example, Her1 was relatively poorly synthesized in vitro, but produced one of the strongest gel shifts, requiring one-sixth the exposure time of the other EMSA gels. These data suggest that there may be significant differences in the affinities between Her monomers and/or between Her dimers and DNA. By extension, there may be relatively weak interactions not detected by EMSA. In summary, we observed a range of dimer specificity, as Her1 and Her7 were highly specific and Hes6 was rather promiscuous.
Fig. 1.
Her proteins form specific DNA-binding dimers in vitro. (A-C) EMSA showing that Her1 by itself, presumably as a homodimer, binds DNA (A), whereas Her7 and Hes6 do not bind DNA by themselves (B and C, respectively). (D-F) Her1 does not detectibly heterodimerize with Her7 (D) or Hes6 (E), but Her7 and Hes6 are able to form a heterodimer (F) based on the presence and shifting of bands with each respective antibody when the two proteins are combined.
Her dimerization in vivo
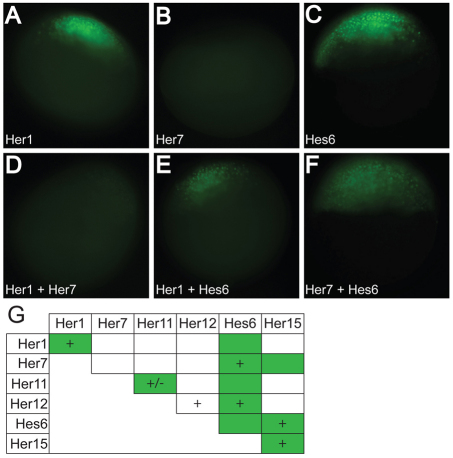
We next sought to corroborate our in vitro data by using BiFC to visualize Her dimerization in live embryos (Saka et al., 2008). In this assay, the coding sequence of the N-terminal or C-terminal halves of Venus YFP are fused to the C-terminus of each of the Her coding sequences. mRNAs encoding these constructs are co-injected into the zebrafish embryo and dimerization is revealed by reconstitution of the YFP and fluorescence (Fig. 2A-F; supplementary material Fig. S3). All combinations of possible Her dimers were tested. We also examined each Her combination with complementary swaps of the N-terminal and C-terminal halves of YFP, e.g. Her7-N-YFP + Hes6-C-YFP and Her7-C-YFP + Hes6-N-YFP, and each combination produced consistent results. Fluorescence was determined during early gastrulation, as the embryos do not gastrulate normally owing to the mRNA injection. Using BiFC, we were able to confirm six of the seven dimer species observed in vitro (Fig. 2G). The amount of protein detected via the western blot on mRNA-injected embryos was not predictive of the signal from the BiFC assay. For example, Her11 is not detectable in the western blot (supplementary material Fig. S1), but its fluorescence signal as both a homodimer and heterodimer with Hes6 was substantially stronger than the BiFC signal produced by the Her1 homodimer (supplementary material Fig. S3). Only 200 ng/μl of the Her11 constructs were injected to observe fluorescence, whereas 800 ng/μl of Her1 were injected to see a signal. These data suggest that there may be significant differences in the affinities between Her monomers. Moreover, there may be relatively weak interactions not revealed by the BiFC assay. We were unable to detect the Her12 homodimer using BiFC (Fig. 2G; supplementary material Fig. S3). While the EMSA assay may be more sensitive, it is also possible that the formation of the Her12 homodimer requires or is stabilized by DNA binding. Surprisingly, we observed several Hes6 dimers using BiFC that we did not detect via EMSA. We interpret these data as indicating that the Hes6 homodimer and Hes6 heterodimers with Her1 and Her11 are either unable to bind DNA or do so very weakly. The Her1/Hes6 heterodimer had been previously identified via immunoprecipitation (Kawamura et al., 2005). We hypothesize that the Hes6/Hes6 and Her1/Hes6 dimers act as a sink that reduces the number of dimers that can effectively bind DNA and repress transcription.
Fig. 2.
Her proteins form DNA-binding and non-DNA-binding dimers in vivo. (A-F) Representative images of Her1 (A), Her7 (B), Hes6 (C), Her1+Her7 (D), Her1+Hes6 (E) and Her7+Hes6 (F) proteins. (G) A chart summarizing the dimers that bind DNA in vitro (denoted by a +) and the dimers that form in vivo (denoted by a green box). All the dimers observed in vitro, except Her12/Her12, were observed in vivo in addition to the presence of four novel dimers. (+/–) Her11 weakly binds DNA, and although we have observed binding in multiple independent experiments, we frequently did not detect the faint gel shift.
Distinct DNA binding preferences by different Her dimers
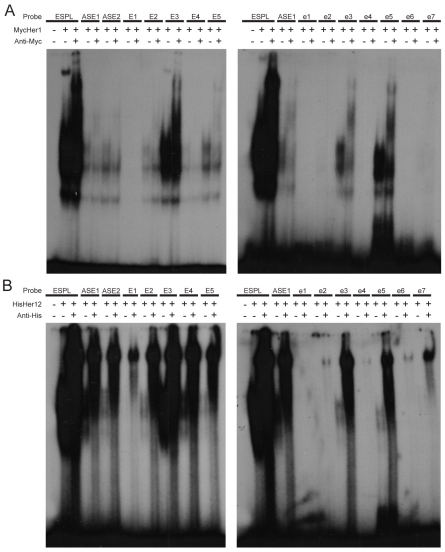
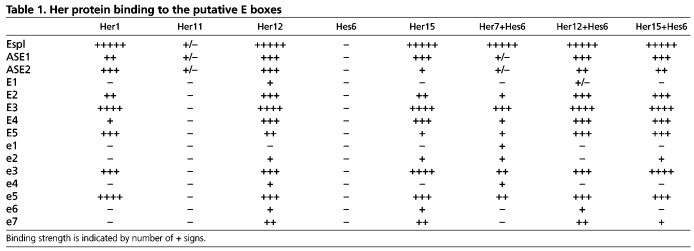
The Her proteins bind class B E-boxes but these sequences can vary from the consensus and there is evidence that the flanking sequences influence protein-DNA interaction (Grove et al., 2009). Within the 3.7 kb genomic sequence upstream of her7, we identified 11 putative Her binding sites. Five of the sites, named E1-E5, match the consensus sequence whereas seven sites, named e1-e7, deviate from the consensus (supplementary material Fig. S4). We tested each of the dimer combinations that bound the E(spl) consensus site for the ability to bind the putative E-boxes. We observed several differences between the Her dimers in sequence preference and binding strength. Her1 homodimers are the most selective, only binding to six sequences: four consensus sites and two non-consensus sites (Fig. 3A). Her12 homodimers display the broadest DNA-binding ability and interact with ten of 11 sites (Fig. 3B). Each dimer binds to a unique subset of consensus and non-consensus sites (Table 1; supplementary material Fig. S4). We examined whether Her7 or Hes6 alone could bind related N-boxes, but found no evidence of specific binding (supplementary material Fig. S4J,K). Similarly, Her7 alone was unable to bind any of the putative E-boxes upstream of her7 (supplementary material Fig. S4F). We also tested the Hes6 BiFC dimer to see if the BiFC stabilized the homodimer and allowed it to bind DNA. However, no DNA binding was observed, supporting the interpretation that the Hes6 homodimer acts as a sink (supplementary material Fig. S4E).
Fig. 3.
Different Her dimers preferentially bind specific E boxes. (A,B) EMSA of Her1 (A) and Her12 (B) homodimers binding to the five consensus (E1-E5) and seven non-consensus (e1-e7) E boxes upstream of her7. The Her1 homodimer binds to E2, E3, E4, E5, e3 and e5, whereas the Her12 homodimer binds to E1, E2, E3, E4, E5, e2, e3, e4, e5, e6 and e7.
Table 1.
Her protein binding to the putative E boxes
Although our EMSA assay was not quantitative, we were able to draw some qualitative conclusions about binding strengths by exposing the EMSA blots for a range of times. The strength of binding is compared internally to three control probes: the E(spl) probe, which bound most strongly to all dimers, and two sites that we identified upstream of her1 (ASE1 and ASE2) that bind less robustly (Brend and Holley, 2009; Gajewski et al., 2003). Of the new E-boxes, all dimers bound most strongly to E3 (Table 1; supplementary material Fig. S4), which has flanking residues that most closely match those of the E(spl) probe. The strength of Her binding to E4 and E5 varied with each dimer. For example, the Her1 homodimer bound with greater strength to the E5 probe than the E4 probe, whereas the Her15 homodimer bound more strongly to the E4 probe. The Her12 homodimer was the only dimer to bind the E1 probe (Fig. 3B; Table 1; supplementary material Fig. S4). Overall, we conclude that there are differences in binding preferences and speculate that the existence of multiple Her dimers in zebrafish contributes to robustness of the negative feedback by expanding the number of cis regulatory sites utilized by the segmentation clock.
From network topology to segmentation clock function
Counterintuitive to the dimerization data, the her7 knockdown phenotype is stronger than the hes6 knockdown (Oates and Ho, 2002; Schroter and Oates, 2010; Sieger et al., 2006). Given that Her7 binds DNA as a heterodimer with Hes6, whereas Hes6 can also bind DNA as a heterodimer with Her12 and Her15, how can the her7 knockdown phenotype be stronger than the hes6 knockdown? Her7 heterodimerizes with Hes6 and Her15, and the Her7/Her15 interaction appears to be relatively weak. Therefore, knockdown of her7 could disproportionately free the Hes6 hub to form other dimers and thus have a broad indirect effect on network topology that may be responsible for the stronger her7 loss of function phenotype. If loss of her7 leads to a disruption of the segmentation clock by freeing Hes6 to form other dimers, then a testable prediction is that knockdown of hes6 should actually reduce the severity of the her7 knockdown. This proposed rescue would obviously not be due to restoration of the Her7/Hes6 heterodimer ability to bind DNA but rather to a rebalancing of the dimer composition within the remaining network.
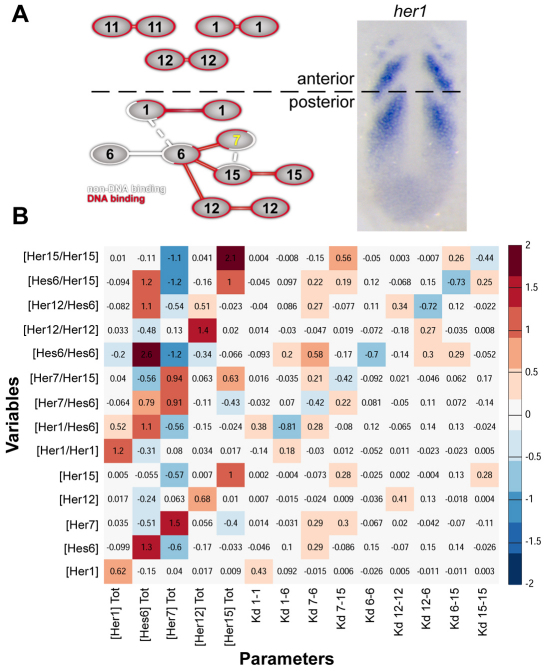
To examine the hypothesis that Her7 could regulate network topology by sequestering the Hes6 hub, we performed a computational sensitivity analysis based on our dimerization data. To simplify the network analysis, we assumed well-mixed kinetics and conserved total numbers of each protein species, as synthesis and degradation are much slower than binding and unbinding of monomers. Given that the values of the total protein concentration and of the dissociation constants are unknown, these values were randomly selected from a log-normal distribution. If a particular dimer gave a strong signal in either the EMSA or BiFC assay relative to the amount of protein present, we classified it as a ‘strong’ interaction, otherwise it was classified as ‘weak’. The two weak dimers are the Her1/Hes6 and Her7/Her15 heterodimers and they are distinguished by the dashed lines in the network diagram (Fig. 4). In the sensitivity analysis, the range of dissociation constants sampled by these two dimers was an order of magnitude weaker than for all the other dimers. We focus on the posterior PSM because the network in the anterior PSM is composed of three homodimers (Fig. 4A). We found that when dimer formation was favored, changes in protein concentrations had greater effects on dimer composition than changes in dimerization constants. In particular, we found that the total concentration of Her7 and Hes6 had the greatest impact on the dimer network (Fig. 4B). Of the concentrations affected more than 20% by a change of Her1, only one is of a monomer/dimer that does not directly contain Her1 (Hes6/Hes6). Only two of six changes in response to modification of Her15 are indirect. However, five of 11 changes due to modification of Hes6 are indirect, and seven of ten changes due to alteration of Her7 are indirect. Her7 thus has by far the largest indirect effect on dimer composition. In addition, the effects of varying total Hes6 and Her7 concentration on dimer composition are opposite for all species except the Hes6/Her7 heterodimer (Fig. 4B). As we initially hypothesized, the sensitivity analysis suggests that loss of her7 increases the number of Hes6 monomers that are free to dimerize with other Her proteins. Ergo, loss of her7 has the opposite effect on network dimer composition as loss of hes6. Further, as the different dimers have distinct preferences for DNA-binding sites, a change in dimer composition could affect which cis regulatory sequences are most utilized by the clock.
Fig. 4.
Sensitivity analysis of the dimer network. (A) Schematic of Her dimer network. DNA-binding dimers are connected in red, non-DNA-binding dimers are connected in white. Networks present in the anterior or posterior PSM are segregated and indicated on the wild-type expression of her1. Weak dimers are connected by dashed lines. (B) Summary of local sensitivity analysis of posterior PSM Her/Hes network. Input parameters (total protein concentration and Kd) are listed along the bottom and output variables (Her monomer and dimer concentrations) along the left. Sensitivity values were calculated as the fractional change in each output variable (y) for a given fractional change in the corresponding input parameter (x), to give (Δy/y)/(Δx/x), averaged across 1000 randomly sampled parameter values. Red values indicate a positive relationship, whereas blue values indicate a negative relationship between changes in parameter value and effects on Her concentration. For example, a 1% increase in total Hes6 concentration decreases Her7/Her15 by 0.56%.
We tested the hypothesis that knockdown of hes6 would rescue knockdown of her7 using antisense morpholinos. The severity of loss of hes6 phenotype, via mutation or morpholino knockdown, is temperature sensitive; thus we performed knockdowns at both permissive (31°C) and restrictive (22°C) temperatures (Schroter and Oates, 2010). We first used a luciferase assay to estimate the efficacy of the morpholino knockdown (Kamachi et al., 2008). These previously characterized her7 and hes6 morpholinos achieved 94-97% knockdown in the luciferase assay (supplementary material Fig. S5). There was no significant difference in knockdown efficiency at 22°C or 31°C, nor when morpholinos were co-injected.
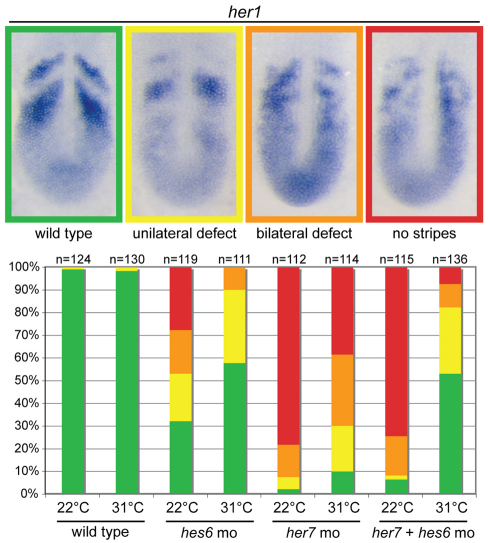
Knockdown of her7 eliminates the her1 stripes produced by the segmentation clock, whereas the hes6 knockdown produces significant alterations in the her1 stripes only at 22°C and at much lower frequency than knockdown of her7 (Fig. 5). Strikingly, and in confirmation of the hypothesis, knockdown of hes6 with her7 at 31°C rescues her1 stripe formation in over half of the embryos and attenuates the severity of the defects in the remaining embryos (Fig. 5). A similar rescue of striped deltaC expression was also observed (supplementary material Fig. S6). Thus, these genetic data suggest that loss of the Her7 regulation of the Hes6 hub, independent of loss of Her7/Hes6 DNA binding, strongly contributes to the her7 knockdown phenotype.
Fig. 5.
Knockdown of hes6 rescues segmentation clock defects due to knockdown of her7. Representative images are provided for four categories of her1 expression with the color coding corresponding to the histogram. The histogram displays the distribution of the four categories of her1 expression observed in single and double knockdown experiments performed at 22°C and 31°C. The her7 knockdown phenotype is stronger than the hes6 knockdown, and although both are temperature sensitive, the hes6 phenotype is much more affected by temperature. The only condition in which her7 knockdown results in a significant proportion of wild-type segmentation clock patterns is the her7+hes6 double knockdown at 31°C.
DISCUSSION
The Her dimer network functions within the segmentation clock to cyclically repress her transcription. Interpretation of genetic interaction data within this network is sensitive to a number of factors. Traditionally, use of null alleles in double mutant combinations allowed the construction of genetic pathways: if the double mutant phenotype is only as strong as one of the single mutants, then the genes are assigned to the same genetic pathway. Conversely, if the double mutant is stronger than the single mutants, then the two genes are assigned to parallel, redundant pathways. These experiments are in contrast to the utilization of hypomorphic alleles in genetic enhancer screens. In these experiments, second mutations are sought that can exacerbate the phenotype of the original hypomorphic allele. The expectation is that this second mutation is in the same pathway as the original mutation. Thus, depending on the types of alleles used and the context of the experiment, a double mutant combination that is more severe than either of the single mutant alleles can be interpreted as indicating that the two genes are either in a single genetic pathway or two parallel pathways. Interpretation of genetic interaction data in vertebrates is further complicated by the prevalence of genetic redundancy and the use of reverse genetic techniques that may not produce null-phenotypes. The dimerization network data serve as a guide that reduces ambiguity in interpreting the genetic interactions between double knockdown of different her genes. In addition to more clearly demonstrating that different dimers act in succession or in parallel, the integration of these data reveals a more nuanced relationship between network structure and network function.
Overall, the dimerization data indicate that Hes6 is the hub of a dimer network that contains both DNA-binding and non-DNA-binding dimers (Fig. 4A). Previous morpholino knockdown studies found that double knockdown of hes6 and her1, but not hes6 and her7, leads to a synergistic perturbation of somitogenesis (Sieger et al., 2006). Our biochemical data indicate that Her7 must dimerize with Hes6 to effectively bind DNA, so simply inhibiting hes6 indirectly eliminates the ability of her7 to repress transcription. Therefore, double knockdown of hes6 and her7 has no increase in the segmentation defect. By contrast, her1 represses transcription in parallel to the hes6 hub. Consequently, simultaneous knockdown of her1 and hes6 causes a greater disruption of somitogenesis. A second example is the strong morphological phenotype observed after concomitant knockdown of both her11 and her7 (Sieger et al., 2004). Her11 is only expressed in the anterior PSM, whereas Her7 heterodimerizes with Hes6 and Her15, both of which are expressed exclusively in the posterior PSM. Thus, her7 and her11 function at distinct times, and the morphological defect is likely to be caused by the combined perturbation of subdomains of the network that are active at successive time points (Fig. 4A). In these two instances of genetic interactions, as well as the lack of synergy in the her7 + hes6 double knockdown, the morpholino knockdowns appear to behave as null alleles, and the synergies result from perturbation of distinct pathways.
her1 was the first oscillating hairy gene identified in zebrafish and, along with her7, has received disproportionate attention by researchers in the field. This focus is partly due to her1 and her7 being superior in situ hybridization markers for the zebrafish segmentation clock. However, a better understanding of the Her network somewhat justifies the emphasis on her1 and her7. Our data indicate that the non-oscillating gene Hes6 is the hub of the network, but that Her7 has a unique and important role in both binding DNA as a heterodimer with Hes6 and regulating network topology. Her1 is distinguished in that it appears to primarily bind DNA as a homodimer. Its function as a transcriptional repressor is thus in parallel with the Hes6 core of the clock network. Her12 and Her15 have similar characteristics, perhaps suggesting greater redundancy among these two genes, and exhibit potent DNA-binding activity. However, there is little indication that they have a particularly unique function within the clock network. Her11 expression is confined to the anterior half of the PSM, meaning that it is not involved in the earlier oscillatory cycles thought to be most important for establishing segmental periodicity.
A network of Her dimers functioning within a negative feedback loop is thought to underpin the zebrafish segmentation clock. The in vivo and in vitro protein interaction data presented here resolve connectivity within this network. Her1 binds DNA strongly as a homodimer but will also dimerize with Hes6. Her12 and Her15 bind DNA both as homodimers and as heterodimers with Hes6. Her7 forms a DNA-binding dimer with Hes6 and a weak heterodimer with Her15. Using a simple sensitivity analysis based on the dimerization data, we are able to substantiate a model that explains the her7 and hes6 single- and double-loss-of-function phenotypes. These analyses suggest that the structure of the network begets a network dynamic, where Hes6 is the hub of the dimer network and the Her7 node disproportionately regulates Hes6 availability and by doing so indirectly adjusts the balance of dimers in the entire network. This modulation of the network topology is an emergent property of the system and is necessary for the function of the zebrafish segmentation clock.
Supplementary Material
Acknowledgments
We thank Jim Smith for the BiFC constructs and Craig Crews for use of the Wallac Multi-Label Counter.
Footnotes
Funding
Research support was provided by a National Institutes of Health predoctoral genetics training grant [T32 GM007499 to A.M.T. and J.S.S.], by an National Science Foundation Graduate Research Fellowship (J.S.S.) and by the National Institute of Child Health and Human Development [RO1 HD045738 to S.A.H.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.073544/-/DC1
References
- Bessho Y., Sakata R., Komatsu S., Shiota K., Yamada S., Kageyama R. (2001). Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 15, 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y., Hirata H., Masamizu Y., Kageyama R. (2003). Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 17, 1451–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brend T., Holley S. A. (2009). Expression of the oscillating gene her1 is directly regulated by Hairy/Enhancer of Split, T-box and Suppressor of Hairless proteins in the zebrafish segmentation clock. Dev. Dyn. 238, 2745–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinquin O. (2007). Repressor dimerization in the zebrafish somitogenesis clock. PLoS Comput. Biol. 3, e32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinquin O., Page K. M. (2007). Generalized, switch-like competitive heterodimerization networks. Bull. Math. Biol. 69, 483–494 [DOI] [PubMed] [Google Scholar]
- Dunwoodie S. L., Clements M., Sparrow D. B., Sa X., Conlon R. A., Beddington R. S. (2002). Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 129, 1795–1806 [DOI] [PubMed] [Google Scholar]
- Fischer A., Gessler M. (2007). Delta-Notch-and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 35, 4583–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A. L., Ohsako S., Caudy M. (1996). The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4 amino acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 16, 2670–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski M., Sieger D., Alt B., Leve C., Hans S., Wolff C., Rohr K. B., Tautz D. (2003). Anterior and posterior waves of cyclic her1 gene expression are differentially regulated in the presomitic mesoderm of zebrafish. Development 130, 4269–4278 [DOI] [PubMed] [Google Scholar]
- Gajewski M., Elmasri H., Girschick M., Sieger D., Winkler C. (2006). Comparative analysis of her genes during fish somitogenesis suggests a mouse/chick-like mode of oscillation in medaka. Dev. Genes Evol. 216, 315–332 [DOI] [PubMed] [Google Scholar]
- Grove C. A., De Masi F., Barrasa M. I., Newburger D. E., Alkema M. J., Bulyk M. L., Walhout A. J. (2009). A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell 138, 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C. A., Urban M. K., Dill K. K., Merlie J. P., Page M. F., Kimmel C. B., Amacher S. L. (2002). Two linked hairy/Enhancer of split-related zebrafish genes, her1 and her7, function together to refine alternating somite boundaries. Development 129, 3693–3704 [DOI] [PubMed] [Google Scholar]
- Hirata H., Yoshiura S., Ohtsuka T., Bessho Y., Harada T., Yoshikawa K., Kageyama R. (2002). Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840–843 [DOI] [PubMed] [Google Scholar]
- Hirata H., Bessho Y., Kokubu H., Masamizu Y., Yamada S., Lewis J., Kageyama R. (2004). Instability of Hes7 protein is crucial for the somite segmentation clock. Nat. Genet. 36, 750–754 [DOI] [PubMed] [Google Scholar]
- Holley S. A., Geisler R., Nüsslein-Volhard C. (2000). Control of her1 expression during zebrafish somitogenesis by a Delta-dependent oscillator and an independent wave-front activity. Genes Dev. 14, 1678–1690 [PMC free article] [PubMed] [Google Scholar]
- Holley S. A., Jülich D., Rauch G. J., Geisler R., Nüsslein-Volhard C. (2002). her1 and the notch pathway function within the oscillator mechanism that regulates zebrafish somitogenesis. Development 129, 1175–1183 [DOI] [PubMed] [Google Scholar]
- Ishimatsu K., Takamatsu A., Takeda H. (2010). Emergence of traveling waves in the zebrafish segmentation clock. Development 137, 1595–1599 [DOI] [PubMed] [Google Scholar]
- Jennings B. H., Tyler D. M., Bray S. J. (1999). Target specificities of Drosophila enhancer of split basic helix-loop-helix proteins. Mol. Cell. Biol. 19, 4600–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouve C., Palmeirim I., Henrique D., Beckers J., Gossler A., Ish Horowicz D., Pourquié O. (2000). Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development 127, 1421–1429 [DOI] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Kobayashi T. (2007). The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251 [DOI] [PubMed] [Google Scholar]
- Kamachi Y., Okuda Y., Kondoh H. (2008). Quantitative assessment of the knockdown efficiency of morpholino antisense oligonucleotides in zebrafish embryos using a luciferase assay. Genesis 46, 1–7 [DOI] [PubMed] [Google Scholar]
- Kawamura A., Koshida S., Hijikata H., Sakaguchi T., Kondoh H., Takada S. (2005). Zebrafish hairy/enhancer of split protein links FGF signaling to cyclic gene expression in the periodic segmentation of somites. Genes Dev. 19, 1156–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol A. J., Roellig D., Dequeant M. L., Tassy O., Glynn E., Hattem G., Mushegian A., Oates A. C., Pourquie O. (2011). Evolutionary plasticity of segmentation clock networks. Development 138, 2783–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. (2003). Autoinhibition with transcriptional delay: a simple mechanism for the zebrafish somitogenesis oscillator. Curr. Biol. 13, 1398–1408 [DOI] [PubMed] [Google Scholar]
- Monk N. A. (2003). Oscillatory expression of Hes1, p53, and NF-kappaB driven by transcriptional time delays. Curr. Biol. 13, 1409–1413 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Dahm R. (eds) (2002). Zebrafish. Oxford, UK: Oxford University Press; [Google Scholar]
- Oates A. C., Ho R. K. (2002). Hairy/E(spl)-related (Her) genes are central components of the segmentation oscillator and display redundancy with the Delta/Notch signaling pathway in the formatoin of anterior segmental boundaries in the zebrafish. Development 129, 2929–2946 [DOI] [PubMed] [Google Scholar]
- Palmeirim I., Henrique D., Ish-Horowicz D., Pourquié O. (1997). Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91, 639–648 [DOI] [PubMed] [Google Scholar]
- Paroush Z., Finley R. L., Kidd T., Wainwright S. M., Ingham P. L., Brent R., Ish-Horowicz D. (1994). Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with Hairy-related bHLH proteins. Cell 79, 805–815 [DOI] [PubMed] [Google Scholar]
- Pichon B., Taelman V., Bellefroid E. J., Christophe D. (2004). Transcriptional repression by the bHLH-Orange factor XHRT1 does not involve the C-terminal YRPW motif. Biochim. Biophys. Acta 1680, 46–52 [DOI] [PubMed] [Google Scholar]
- Pourquié O. (2011). Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell 145, 650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitz H., Kramer M., Dacol D. (1983). Sensitivity analysis in chemical kinetics. Annu. Rev. Phys. Chem. 34, 419–461 [Google Scholar]
- Saka Y., Hagemann A. I., Smith J. C. (2008). Visualizing protein interactions by bimolecular fluorescence complementation in Xenopus. Methods 45, 192–195 [DOI] [PubMed] [Google Scholar]
- Sawada A., Fritz A., Jiang Y.-J., Yamamoto A., Yamasu K., Kuroiwa A., Saga Y., Takeda H. (2000). Zebrafish Mesp family genes, mesp a and mesp b are segmentally expressed in the presomitic mesoderm, Mesp b confers the anterior identity to the developing somites. Development 127, 1691–1702 [DOI] [PubMed] [Google Scholar]
- Schroter C., Oates A. C. (2010). Segment number and axial identity in a segmentation clock period mutant. Curr. Biol. 20, 1254–1258 [DOI] [PubMed] [Google Scholar]
- Shankaran S. S., Sieger D., Schroter C., Czepe C., Pauly M. C., Laplante M. A., Becker T. S., Oates A. C., Gajewski M. (2007). Completing the set of h/E(spl) cyclic genes in zebrafish: her12 and her15 reveal novel modes of expression and contribute to the segmentation clock. Dev. Biol. 304, 615–632 [DOI] [PubMed] [Google Scholar]
- Sieger D., Tautz D., Gajewski M. (2004). her11 is involved in the somitogenesis clock in zebrafish. Dev. Genes Evol. 214, 393–406 [DOI] [PubMed] [Google Scholar]
- Sieger D., Ackermann B., Winkler C., Tautz D., Gajewski M. (2006). her1 and her13.2 are jointly required for somitic border specification along the entire axis of the fish embryo. Dev. Biol. 293, 242–251 [DOI] [PubMed] [Google Scholar]
- Sparrow D. B., Guillen-Navarro E., Fatkin D., Dunwoodie S. L. (2008). Mutation of HAIRY-AND-ENHANCER-OF-SPLIT-7 in humans causes spondylocostal dysostosis. Hum. Mol. Genet. 17, 3761–3766 [DOI] [PubMed] [Google Scholar]
- Sparrow D. B., Sillence D., Wouters M. A., Turnpenny P. D., Dunwoodie S. L. (2010). Two novel missense mutations in HAIRY-AND-ENHANCER-OF-SPLIT-7 in a family with spondylocostal dysostosis. Eur. J. Hum. Genet. 18, 674–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y., Ohtsuka T., Gonzalez A., Miyachi H., Kageyama R. (2011). Intronic delay is essential for oscillatory expression in the segmentation clock. Proc. Natl. Acad. Sci. USA 108, 3300–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- William D. A., Saitta B., Gibson J. D., Traas J., Markov V., Gonzalez D. M., Sewell W., Anderson D. M., Pratt S. C., Rappaport E. F., et al. (2007). Identification of oscillatory genes in somitogenesis from functional genomic analysis of a human mesenchymal stem cell model. Dev. Biol. 305, 172–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.