Abstract
The SIX family of homeodomain-containing DNA-binding proteins play crucial roles in both Drosophila and vertebrate retinal specification. In flies, three such family members exist, but only two, Sine oculis (So) and Optix, are expressed and function within the eye. In vertebrates, the homologs of Optix (Six3 and Six6) and probably So (Six1 and Six2) are also required for proper eye formation. Depending upon the individual SIX protein and the specific developmental context, transcription of target genes can either be activated or repressed. These activities are thought to occur through physical interactions with the Eyes absent (Eya) co-activator and the Groucho (Gro) co-repressor, but the relative contribution that each complex makes to overall eye development is not well understood. Here, we attempt to address this issue by investigating the role that each complex plays in the induction of ectopic eyes in Drosophila. We fused the VP16 activation and Engrailed repressor domains to both So and Optix, and attempted to generate ectopic eyes with these chimeric proteins. Surprisingly, we find that So and Optix must initially function as transcriptional repressors to trigger the formation of ectopic eyes. Both factors appear to be required to repress the expression of non-retinal selector genes. We propose that during early phases of eye development, SIX proteins function, in part, to repress the transcription of non-retinal selector genes, thereby allowing induction of the retina to proceed. This model of repression-mediated induction of developmental programs could have implications beyond the eye and might be applicable to other systems.
Keywords: Sine oculis, Optix, Retinal determination, Drosophila
INTRODUCTION
Drosophila eye development is initiated during embryogenesis when groups of cells expressing the Pax genes eyeless (ey), twin of eyeless (toy), eyegone (eyg) and twin of eyegone (toe) are set aside from the rest of the embryo (Aldaz et al., 2003; Czerny et al., 1999; Jang et al., 2003; Quiring et al., 1994). Concomitant with the onset of these gene expression profiles, the eye anlage are organized into monolayer epithelia called the eye-antennal imaginal discs (Cohen, 1993; Held, 2002). During the second larval instar, the Toy and Ey proteins cooperate to activate directly the expression of sine oculis (so), the founding member of the Sine oculis homeobox (SIX) family of DNA-binding proteins, and its paralog Optix (Michaut et al., 2003; Niimi et al., 1999; Punzo et al., 2002; Ostrin et al., 2006). Loss of so within the eye primordium results in the complete elimination of the compound eyes (Milani, 1941; Milani, 1951; Cheyette et al., 1994; Serikaku and O’Tousa, 1994). Unfortunately, retinal phenotypes of Optix loss-of-function mutants have not been reported; thus, its role in eye development is unclear and a direct comparison with so is not yet possible. It has been shown, however, that both genes bind to identical DNA sequences and are capable of inducing ectopic eye formation when forcibly expressed within non-retinal tissues (Seimiya and Gehring, 2000; Weasner et al., 2007; Berger et al., 2008; Noyes et al., 2008).
Ey also activates the expression of eyes absent (eya), which encodes a transcriptional co-activator and protein tyrosine phosphatase (Halder et al., 1998; Li et al., 2003; Michaut et al., 2003; Rayapueddi et al., 2003; Tootle et al., 2003; Ostrin et al., 2006; Jemc and Rebay, 2007a). Removal of eya from the eye primordium also leads to a complete block in retinal development (Bonini et al., 1993). So and Eya form a biochemical complex that serves to activate a number of targets that are important for retinal development (Giot et al., 2003; Jemc and Rebay, 2007b; Pappu et al., 2005; Pauli et al., 2005; Pignoni et al., 1997; Tanaka-Matakatsu and Du, 2008; Zhang et al., 2006). Similar biochemical interactions have been reported for murine homologs of So (Six1 and Six2) and Eya proteins (Heanue et al., 1999; Ikeda et al., 2002; Ohto et al., 1999).
So and its vertebrate orthologs do not serve as dedicated transcriptional activators. Expression of a Six1-En fusion protein, which is predicted to function as a transcriptional repressor, mimics a subset of the phenotypes that are associated with expression of wild-type Six1 (Brugmann et al., 2004). This So/Six1 mediated repression is thought to be dependent, in part, upon interactions with Groucho (Gro), a member of the TLE family of transcriptional co-repressors. The data supporting a So-Gro complex rely heavily on yeast two-hybrid assays with some studies using truncated proteins (Kenyon et al., 2005a; Brugmann et al., 2004; Kobayashi et al., 2001). However, studies using insect cell lines indicate that Gro is capable of binding to So through the eh-1 motif within the SIX protein-protein interaction domain (Zhu et al., 2002; Silver et al., 2003). In the Drosophila retina, the So-Gro complex has been postulated to repress transcription of dachshund (dac), which encodes a DNA-binding protein that shares homology with members of the Ski/Sno family of transcriptional co-repressors (Salzer and Kumar, 2009). Gro has also been reported to interact synergistically with Six1 to repress the expression of genes that are important for the formation of the epidermis and neural crest (Brugmann et al., 2004). There is evidence, however, to indicate that So/Six1-mediated repression may also involve additional mechanisms and interactions with other transcriptional co-repressors. For example, in murine cell lines Six1 can repress transcription of reporter constructs either on its own or through interactions with Dach1, the vertebrate homolog of Drosophila dac (Li et al., 2003). The biochemical and genetic data suggest that So and its orthologs Six1/2 can either activate or repress target genes.
The most extensive evidence for SIX proteins functioning as transcriptional repressors within the retina comes from studies of Optix and its mammalian orthologs Six3 and Six6 (Gallardo et al., 1999; Jean et al., 1999; Oliver et al., 1995; Seimiya and Gehring, 2000; Seo et al., 1999; Toy et al., 1998). Six3 and Six6 appear to bind physically to Gro/TLE family members, and these interactions are crucial for promoting retinal growth and differentiation (Kobayashi et al., 2001; Lopez-Rios et al., 2003; Zhu et al., 2002). Six6 also interacts with Dach1 to repress transcription of the cyclin-dependent protein kinase inhibitor p27kip1 (Cdkn1b – Mouse Genome Informatics), thereby promoting proliferation of retinal precursor cells (Li et al., 2002). However, like Six1 and Six2, Six6 is also capable of activating transcription via interactions with Eya family members (Li et al., 2003). Similarly, Six3 is required to activate Pax6 expression during lens induction and specification (Liu et al., 2006). These data further the contention that, in general, interactions with specific co-factors allow SIX proteins to either activate or repress transcription of downstream target genes. The evidence surrounding Drosophila Optix presents a slightly different picture. In directed yeast two-hybrid assays, a truncated version of Optix is capable of binding to Gro but not to Eya (Kenyon et al., 2005a; Kenyon et al., 2005b). Optix and Eya also do not functionally interact during the induction of ectopic eye development (Seimiya and Gehring, 2000; Salzer and Kumar, 2010). These results imply that Optix may influence gene expression differently than other SIX proteins by functioning as a dedicated transcriptional repressor.
In this report, we attempt to determine the relative contributions that activating and repressing SIX protein complexes make during eye development. We used the ability of So and Optix to initiate ectopic eye formation as an assay for determining whether SIX proteins initiate eye development using either activation or repression mechanisms. Although expression of some individual retinal determination network members, such as Ey, can promote ectopic eye formation in a broad range of tissues and at very high frequencies, So and Optix are capable of transforming non-retinal tissues only in limited circumstances and at relatively low frequencies (Halder et al., 1995; Seimiya and Gehring, 2000; Weasner et al., 2007). It was thought that this deficiency could be compensated for (at least in the case of So) by the co-expression of Eya because a synergistic increase in the range and frequency of ectopic eye formation was reported when both genes were simultaneously expressed (Pignoni et al., 1997; Seimiya and Gehring, 2000). However, we have failed to detect this strong synergistic effect (Salzer and Kumar, 2010). The conflicting data from these papers highlights the uncertainty that surrounds the mechanism by which So initiates ectopic eye formation. By contrast, the ability of Optix to induce ectopic eyes is presumed to be through its function as a transcriptional repressor (Kenyon et al., 2005a; Seimiya and Gehring, 2000). It seems unlikely that these two proteins could bind to identical target sequences but then induce ectopic eyes using diametrically opposing mechanisms. In this paper, we attempt to resolve this conflict.
We fused the VP16 activation and Engrailed (En) repression domains to So and Optix in order to create strong transcriptional activators and repressors. Quite surprisingly, expression of So-En and Optix-En (but not So-VP16 nor Optix-VP16) is sufficient to generate ectopic eyes within the developing antenna and head capsule. We demonstrate that several activities of So-En and Optix-En mimic that of the wild-type proteins, suggesting that both So and Optix function as transcriptional repressors to initiate ectopic eye development. A potential target for both So and Optix appears to be the antennal/head capsule selector gene cut (ct), as its expression is downregulated by both So-En and Optix-En. As a consequence, ey expression is de-repressed within the antennal disc, and eye formation is initiated through the activation of the RD network. We propose a model in which the formation of ectopic eyes occurs in two steps. So and Optix first function as transcriptional repressors to shut down non-retinal developmental programs. Once the eye program is initiated, So and Optix then switch their activities and promote eye development as transcriptional activators. Although the results in this manuscript describe SIX protein involvement in ectopic retinal formation, our model also supports the use of this mechanism during normal eye development.
MATERIALS AND METHODS
Fly stocks
The following 16 fly stocks were used in this study: (1) UAS-SoFL, (2) UAS-So-VP16NT, (3) UAS-So-VP16CT, (4) UAS-So-ENNT, (5) UAS-OptixFL, (6) UAS-Optix-VP16NT, (7) UAS-Optix-ENNT, (8) UAS-GFP, (9) UAS-Ey, (10) UAS-Cut, (11) cb49-GAL4, (12) ey-GAL4, (13) GMR-GAL4, (14) dpp-GAL4, (15) Act5C-GAL4 and (16) so[1]. All UAS-So and UAS-Optix variant fly strains were generated via standard methods with the exception of UAS-So-VP16CT, which was generated using the phiC31 integration system (Bischof and Basler, 2008; Bischof et al., 2007; Venken et al., 2006).
Constructs
The So full-length and deletion constructs are described elsewhere (Weasner et al., 2007). So-VP16 proteins were generated by fusing the VP16 activation domain to either the N-terminal (So-VP16NT) or C-terminal (So-VP16CT) of full-length So. The So-EN protein was generated by fusing the Engrailed repressor domain to the N-terminal (So-ENNT) of full-length So. Fusing either VP16 or EN to the N-terminal of full-length Optix generated the Optix-VP16NT and Optix-ENNT fusion proteins.
Immunostaining, antibodies and microscopy
Imaginal discs were dissected from developing larvae and fixed in 2% paraformaldehyde for 45 minutes at room temperature. Tissues were blocked in a solution containing 10% goat serum and 0.1% Triton, incubated with primary antibodies at room temperature overnight, washed in PBS + 0.1% Triton, incubated with secondary antibodies for 2-4 hours at room temp, washed in PBS + 0.1% Triton and mounted in Vectashield. Images were taken with a Zeiss Axioplan II fluorescent microscope. Antibodies used were mouse anti-Ey, mouse anti-Ct and rat anti-Elav. F-actin was visualized with phalloidin. For scanning electron microscopy, adult flies were serially passaged through 25% ethanol, 50% ethanol, 75% ethanol, 100% ethanol, 50% ethanol:50% hexamethyldisilazane (HMDS) and 100% HMDS, coated with gold-palladium and viewed with a JEOL 5800LV SEM. For light microscope imaging of adult flies, individuals were immobilized by chilling at –80°C and then viewed on a Zeiss Discovery Microscope.
Immunoprecipitations
Kc167 cells were transfected with 0.4 μg of each of the following plasmids by using Qiagen Effectene Transfection Reagent and induced after 24 hours with 1 mM CuSO4: (1) mt-GAL4, (2) UAS-SoFL-Myc, (3) UAS-So ΔSD-Myc, (4) UAS-DSix4FL-Myc, (5) UAS-OptixFL-Myc, (6) UAS-Optix ΔSD-Myc, (7) UAS-Eya-HA and (8) UAS-Gro-HA. For immunoprecipitation studies, nuclear proteins were extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents. After conducting a pre-clear step with protein G agarose beads, the supernatant was incubated with 1 μg rabbit anti-HA antibody at 4°C overnight followed by an additional 3 hour incubation at 4°C with protein G agarose beads. The samples were centrifuged and washed three times in RIPA buffer + 25% acetonitrile, boiled in SDS loading buffer + β mercaptoethanol and resolved on 10% SDS-PAGE gels. All proteins were visualized on immunoblots using mouse anti-HA or mouse anti-cMyc primary antibodies, goat anti-mouse HRP secondary antibodies and the SuperSignal West Pico Chemiluminescent Substrate.
Confirmation of protein production in cell lines and embryos
DNA constructs containing So-VP16NT, So-VP16CT and Optix-VP16NT were cloned into a modified Gateway expression vector. S2 cells were transfected with 0.4 μg of plasmids containing mt-GAL4 and each of the following plasmids: (1) UAS-So-VP16NT, (2) UAS-So-VP16CT, (3) UAS-Optix-VP16NT and induced after 24 hours with 1 mM CuSO4. Protein lysates were also prepared from 100 μl of 0-12 hour embryos of the genotypes Act5C-GAL4/UAS-So-VP16NT and Act5C-GAL4/UAS-Optix-VP16NT. All samples were treated as described above for Kc167 cells. Proteins were detected using mouse anti-VP16 and mouse anti-Actin antibodies.
Transcriptional activation assay
Full-length and deletion constructs were assayed in yeast for the presence of intrinsic transactivation activity. Each construct was cloned into the pDEST32 bait vector, which contains the GAL4 DNA-binding domain and the ADH1 promoter, in order to allow constitutive expression of the cDNA of interest. These ARS/CEN-based vectors are low copy number expression vectors, which result in the bait proteins being expressed at relatively low levels. The bait plasmids were transformed into MaV203 yeast cells. All transformations were plated on media deficient for the amino acid leucine to ensure incorporation of the bait plasmid (which carries the Leu2 gene as a selectable marker) into the yeast cells. UAS-lacZ and UAS-HIS3 reporters are contained within the yeast genome. The presence of transcriptional activation was measured by the presence or absence of β-galactosidase activity. Activation strength was measured by the ability of transformed cells to grow on increasing levels of 3-amino-1,2,4 triazol (3AT), which inhibits histidine biosynthesis. Each assay was replicated five times. Each well and yeast colony in Fig. 1 is a representative example of the five replicates.
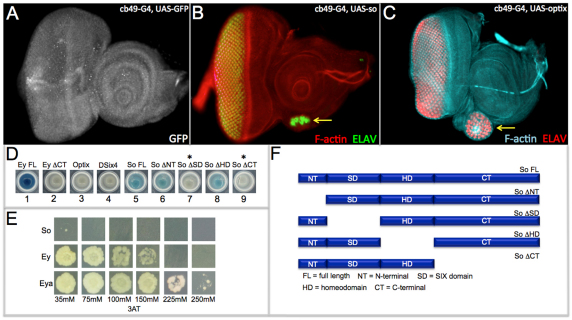
Fig. 1.
Sine oculis and Optix have limited intrinsic ability to activate transcription. (A-C) Confocal images of third instar larval eye-antennal discs. cb49-GAL4 drives expression of the GFP reporter at low levels in all cells of the eye-antennal disc. Arrows in B and C mark the location of ectopic eyes. (D) Transcriptional activation of UAS-lacZ in yeast by Optix-FL, DSix4-FL, So-FL, So-ΔNT, So-ΔSD, So-ΔHD and So-ΔCT. Asterisks indicate that So-ΔSD and So-ΔCT failed to activate transcription of the UAS-lacZ reporter. (E) Transcriptional activation of UAS-HIS3 by So, Ey and Eya in yeast. Assay is carried out in the presence of increasing amounts of the 3AT histidine biosynthesis inhibitor to determine the relative strength of activation potential. (F) Schematic of So full-length and deletion constructs.
RESULTS
Forcible expression of so and Optix with the cb49-GAL4 driver is sufficient to induce ectopic eye formation within the developing antenna (Fig. 1A-C, arrows) (Weasner et al., 2007). The ability of either SIX protein to induce eye development is not dependent upon the presence of the Eya transcriptional co-activator, as it is not expressed within the antenna and is not supplied in this experiment. This suggests that So may either interact with other transcriptional co-activators within the antennal disc or may itself be capable of activating transcription of downstream target genes. We conducted yeast two-hybrid screens in which the full-length wild-type versions of So and Optix were used as bait to screen a third larval instar library enriched with cDNAs from the eye-antennal disc. We recovered several potential new interacting proteins, but none of these factors is predicted to be transcriptional co-activators (data not shown). This result cannot rule out the possibility that So and Optix proteins interact with other transcriptional co-activators in vivo. We then used a yeast transcriptional assay to determine whether either SIX protein harbors intrinsic transcriptional activation potential. Plasmids containing full-length SoFL, OptixFL and DSix4FL sequences fused to the GAL4 DNA-binding domain were transformed into yeast and each chimeric protein was tested for its ability to activate a UAS-lacZ reporter. Full-length Ey protein (EyFL) is a strong transcriptional activator and served as a positive control in this assay (Fig. 1D, well 1). Deletion of the CT segment of Ey removes the transcriptional activation domain (Ey ΔCT). This protein variant fails to activate the UAS-lacZ reporter and serves as a negative control (Fig. 1D, well 2). Both Optix and DSix4 are incapable of activating the UAS-lacZ reporter, suggesting that neither protein functions on its own to activate transcription (Fig. 1D, wells 3 and 4). SoFL activates transcription of the reporter weakly (Fig. 1D, well 5). The differences amongst the SIX family members is consistent with DSix4 and Optix being more closely related to each other when compared with So (Datta et al., 2011). Using a series of deletion constructs (Fig. 1F), we mapped the location of the potential sites for transcriptional activation activity to the SIX protein-protein interaction domain and the CT segment. Deletion of either region (So-ΔSD, So-ΔCT) eliminated the ability of the protein to activate the reporter (Fig. 1D, wells 7 and 9), while removal of the other protein domains had no effect on the transactivation potential of So (Fig. 1D, wells 6 and 8).
The frequency at which ectopic eyes are induced by So is significantly lower than that of other retinal determination proteins (Halder et al., 1995; Weasner et al., 2007; Weasner et al., 2009). We set out to determine the relative strength of the activation potential of So in relationship to Ey and Eya. MaV203 cells containing a stably integrated UAS-HIS3 reporter were transformed with plasmids containing sequences for the GAL4 DNA-binding domain fused to either so, ey or eya (So-GAL4DB, Eya-GAL4DB, Ey-GAL4DB). The transformed cells were plated on media lacking the amino acid histidine and were grown in the presence of increasing amounts of the 3-amino-1,2,4 triazol (3AT) histidine biosynthesis inhibitor. Growth on plates containing increasing amounts of inhibitor allows for a comparison between different transcriptional activators and a determination of relative strengths. As previously described, the Ey-GAL4DB fusion protein will grow in media containing up to 150 mM 3AT (Fig. 1E, middle row) (Weasner et al., 2009). By contrast, cells containing the So-GAL4DB fusion protein grew weakly on plates containing 35 mM 3AT and failed to grow on plates containing higher concentrations of 3AT (Fig. 1E, top row) suggesting that So, on its own, is a relatively weak transcriptional activator. We determined that cells containing the Eya-GAL4DB fusion protein grew on media containing 250 mM 3AT (Fig. 1E, bottom row). Despite the higher activation potential, the So-Eya complex does not induce ectopic eyes at an equal or higher frequency nor in as wide a range of tissues as Ey (Halder et al., 1995; Weasner et al., 2007; Salzer and Kumar, 2010).
Even though expression of either So or Optix is sufficient to induce ectopic eyes in the antennal disc (Fig. 1B,C) (Seimiya and Gehring, 2000; Weasner et al., 2007), the results from our transcriptional strength assays suggest that neither SIX protein is capable of strongly activating the expression of downstream target genes (Fig. 1D, wells 3 and 5). As we failed to identify candidate genes encoding additional transcriptional co-activators that could interact with either So or Optix within the antenna, we fused the activation domain of VP16 to both proteins (So-VP16, Optix-VP16) thereby generating strong transcriptional activators that bypass the requirement for interactions with other co-factors (Fig. 2A,E). We created two versions of the So-VP16 activator: one in which VP16 was fused to the N terminus (So-VP16NT) and another in which VP16 was added to the C terminus (So-VP16CT). A single Optix-VP16 activator was generated: in this case VP16 was fused to the N terminus (Optix-VP16NT). We also created strong transcriptional repressors by fusing the repression domain of Engrailed to both SIX proteins (So-ENNT, Optix-ENNT) to serve as negative controls (Fig. 2A,E).
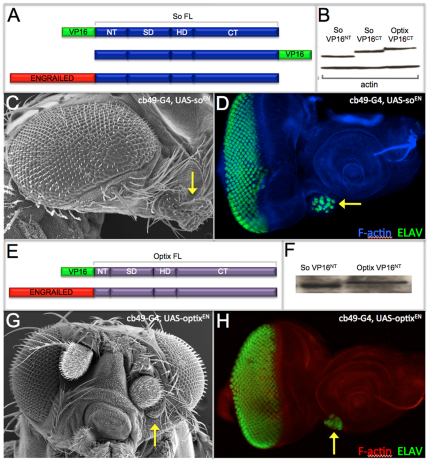
Fig. 2.
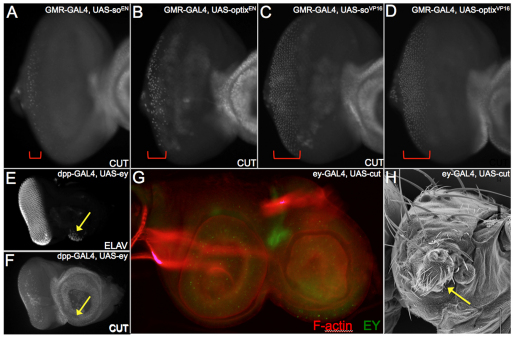
Transcriptional repressor forms of So and Optix initiate ectopic eye formation. (A,E) Schematic of So-VP16, So-EN, Optix-VP16 and Optix-EN constructs used in this study. (B,F) Immunoblot (B) and western blot (F) demonstrating that So-VP16 and Optix-VP16 proteins are made in S2 cells and Drosophila embryos. (C,D,G,H) Scanning electron microscopy and confocal images of ectopic eyes (arrows) formed by expression of So-EN and Optix-EN.
We expected that expression of So-VP16 and Optix-VP16 would be sufficient to induce ectopic eyes, while So-EN and Optix-EN would fail to promote eye development. To our surprise, expression of neither So-VP16 nor Optix-VP16 was sufficient to induce ectopic eye formation (data not shown). The fact that these strong transcriptional activators could not induce ectopic eyes is consistent with a lack of a strong synergistic interaction between Eya and both SIX proteins during forced expression assays (Seimiya and Gehring, 2000; Salzer and Kumar, 2010). We attempted to use our yeast transcriptional activation assay to assess the relative strength of So-VP16 to that of So-Eya, but both So-VP16 constructs are lethal to yeast cells. We did confirm, however, that So-VP16 and Optix-VP16 fusion proteins are produced and stable in both S2 cells and in embryonic lysates (Fig. 2B,F), and that the proteins are functional in other genetic assays (Fig. 3B,C,E).
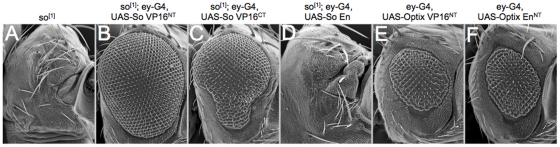
Fig. 3.
An activator form of So rescues all retinal defects associated with the so[1] loss-of-function mutant allele. (A-F) Scanning electron microscopy images of adult Drosophila compound eyes and heads.
We did observe the formation of ectopic eyes when So-ENNT and Optix-ENNT were expressed within the antenna (Fig. 2C,D,G,H). These results are supported by data from other studies indicating that the Optix C-terminal segment, which contains a putative repression domain, is absolutely required for the formation of ectopic eyes (Weasner and Kumar, 2009). We also demonstrated that, although a So protein variant consisting of just the SIX and homeodomain (SoSD-HD) cannot induce ectopic eyes, the addition of the C-terminal of Optix to this truncated So protein (So-OptixCT) restores its ability to induce ectopic retinal formation (Weasner et al., 2007). Based on the results presented in this report, as well as those from our earlier studies, we conclude that So and Optix function in part as transcriptional repressors during the induction of ectopic eyes.
We set out to integrate our results with the known biochemical and genetic interactions that have been reported for the fly SIX proteins. In order to address this issue, we placed both So-VP16 and So-EN under the control of the ey-GAL4 driver and attempted to restore eye development to the so1 loss-of-function mutant. so1 flies harbor a deletion within an eye-specific enhancer and are characterized by the complete absence of compound eyes (Fig. 3A) (Milani, 1941; Cheyette et al., 1994; Niimi et al., 1999). Expression of both So-VP16NT and So-VP16CT transcriptional activators restores eye development to nearly wild-type levels (Fig. 3B,C), whereas expression of the So-ENNT transcriptional repressor failed to do so (Fig. 3D). A clean stand-alone Optix null allele has not been reported, so we were unable to conduct similar rescue experiments with the Optix-VP16 and Optix-EN constructs. However, we did express both constructs ahead of the furrow in wild-type retinas using ey-GAL4. Expression of either construct disrupted the structure of the compound eye. The resulting rough eyes were nearly indistinguishable from each other (Fig. 3E,F), which could indicate that Optix, in addition to functioning as a repressor could also function as an activator through contact with other binding partners. From these results, we conclude that both So-VP16 and Optix-VP16 are functional and therefore the absence of ectopic eyes in assays using these constructs indicates that the initial stages of eye specification require a repression step.
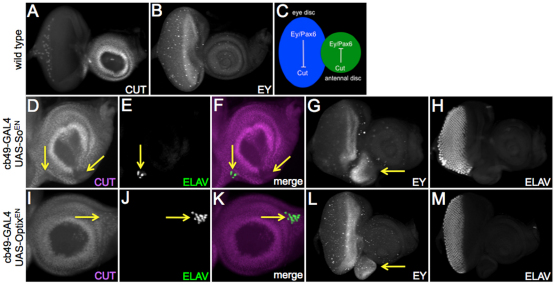
What role does transcriptional repression via retinal determination genes play in the induction of ectopic eye development? It has been suggested that the division of the eye-antennal primordium into distinct regions requires segregation of selector proteins into region specific compartments. One such selector protein is Ct, a homeodomain-containing transcription factor that is expressed within and required for the specification of the antenna (Bodmer et al., 1987; Blochinger et al., 1993). The onset of its expression within the antenna coincides with the restriction of Ey to just the eye field (Kenyon et al., 2003). For the remainder of eye development, these two proteins are distributed in mutually exclusive patterns (Fig. 4A,B), thereby raising the possibility that Ey (or a downstream RD gene) normally inhibits the transcription of Ct, and vice versa (Fig. 4C). We find that in regions where we have ectopic eye formation, we see a corresponding downregulation of ct (Fig. 4D-F,I-K, arrows). Moreover, ectopic ey is also observed with overexpression of So or Optix (Fig. 4G,L) supporting the idea that one function of Ct might be to repress ey within the antennal field. It should be pointed out that activation of ey does not always result in ectopic eye formation (Fig. 4H,M). This is also true of most retinal determination genes: in many cases, the retinal determination network will be activated but ectopic eyes will be seen only in a fraction of animals (Salzer and Kumar, 2010).
Fig. 4.
The antennal selector gene cut is a potential target of both Sine oculis and Optix. (A,B,D-M) Confocal images of third larval instar eye-antennal discs. Arrows in D,I indicate repression of ct expression; in G,L, they indicate de-repression of ey expression; in E,J,F,K, they indicate the formation of photoreceptors. (C) Model in which Ey and Cut mutually repress each other’s transcription.
The ability to repress ct is not limited to the antenna as expression of So-EN and Optix-EN within the developing cone cells (using GMR-GAL4) dramatically reduces ct expression in the eye disc (Fig. 5A,B, bracket). By contrast, ct expression is not altered when the So-VP16 activator is expressed in the eye field with GMR-GAL4 (Fig. 5C,D, bracket). This could suggest that activation of ct by So may require additional co-factors or DNA-binding proteins that were not supplied in this experiment. Kenyon and colleagues had suggested that Ey and Ct may mutually repress each other’s transcription (Kenyon et al., 2003). We placed ey under the control of the dpp-GAL4 driver and induced expression within the antennal disc. Although we see ectopic eye formation, there are negligible effects on Ct protein levels (Fig. 5E,F arrow). We conclude that the retinal determination network can repress ct expression at the level of So and Optix. This contention is supported by the observation that ct transcription is de-repressed when so and/or eya are removed in retinal loss-of-function clones (Salzer and Kumar, 2009). By contrast, expression of ct within the retinal field completely eliminated ey expression and converted the eye into a duplicate antenna (Fig. 5G,H). Moreover, co-expression of cut with so using the cb49-GAL4 driver prevents ectopic eye formation (supplementary material Fig. S1).
Fig. 5.
Regulatory relationship between Eyeless, Sine oculis and Cut in the eye-antennal disc. (A-G) Confocal images of third instar eye-antennal discs. Brackets in A-D mark the zones of ct-positive cone cells. (H) Scanning electron microscopy of an adult head. Arrows indicate a partial antennal segment in the place of the eye.
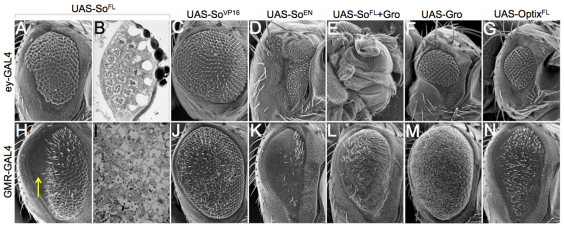
The collective evidence from this and other studies indicates that So is capable of serving as both a transcriptional activator and a repressor (this report) (Pignoni et al., 1997; Silver et al., 2003; Kenyon et al., 2005a; Salzer and Kumar, 2009). We expressed both So-VP16 and So-EN ahead of the furrow with ey-GAL4 and behind the furrow with GMR-GAL4, and compared the structure of the retina with animals in which full-length So had been overexpressed (Fig. 6). The logic behind this experiment is that any similarities between So FL and either So-VP16 or So-EN-induced phenotypes might shed light onto how So is functioning ahead of and behind the furrow. Expression of SoFL ahead of the furrow yields a small eye with most of the remaining ommatidia having relatively normal numbers of photoreceptors and positioning (Fig. 6A,B). Similarly, expression of So-VP16 yields a size reduction and mild to moderate roughening of the retinal surface (Fig. 6C). By contrast, expression of So-En completely eliminates all retinal tissue, which is nearly identical to situations in which So and Gro are co-expressed and qualitatively similar to overexpression of Gro alone (Fig. 6D-F). Expression of Optix ahead of the furrow gives a small rough eye that is equivalent in size to that of Gro (Fig. 6G). As expression of So-VP16 results in a phenotype that is more similar to that of full-length wild-type So, we suggest that So is probably functioning as a transcriptional activator anterior to the furrow. This is consistent with the rescue of so[1] mutants by So-VP16 (Fig. 3A-C). Moreover, co-expression of cut with so using the cb49-GAL4 driver prevents ectopic eye formation (supplementary material Fig. S1).
Fig. 6.
Expression of activator (So-VP16) and repressor (So-EN) forms of Sine oculis differentially affect the developing eye. (A,C-H,J-N) Scanning electron microscopy images of adult compound eyes. Arrow in H indicates glazed portion of eye. (B,I) Light microscope images of adult compound eye retinal sections.
Expression of SoFL behind the furrow leads to a severe roughening of the anterior half of the eye and a near complete elimination of retinal tissue in the posterior half (Fig. 6H, arrow). Sections of the adult retina reveal complete degeneration of all photoreceptors, as witnessed by the missing rhabdomeres and absent corneal lens at the posterior edge of the eye (Fig. 6I). Expression of So-VP16 in developing photoreceptors leads to a roughening of the eye that is qualitatively distinct from SoFL (Fig. 6J). Expression of So-EN leads to a ‘glazed’ eye in which the eye field, while present, is completely devoid of ommatidia (Fig. 6K). Animals expressing both So and Gro behind the furrow exhibit a glazed eye appearance, as well and are replete with tiny bristles (Fig. 6L). Expression of Gro by itself behind the furrow gives a slightly glazed eye that also contains many tiny bristles (Fig. 6M). Although the phenotypes of SoFL may be phenotypically similar to that of So-EN and So+Gro, we cannot definitively say that So functions as either a repressor or an activator behind the furrow. It is possible that both activating and repressing complexes may function in differentiating photoreceptor cells. Expression of full-length Optix (OptixFL) gives a phenotype that is similar to that of So-EN, suggesting that Optix may function as a repressor when mis-expressed behind the furrow (Fig. 6N).
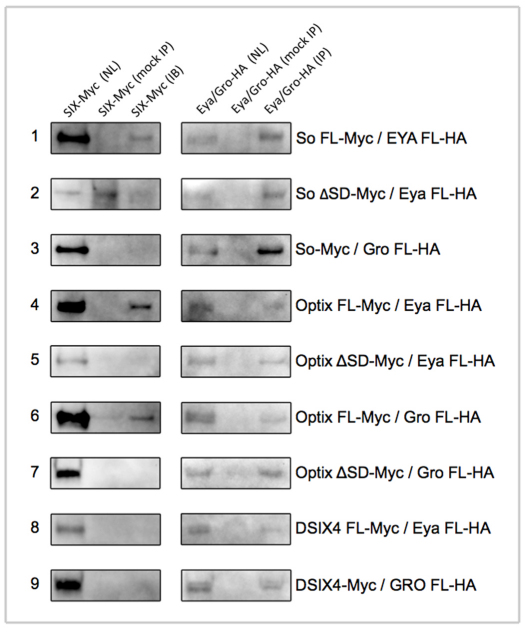
Finally, we set out to determine the complete set of physical interactions between all three DSIX proteins and both Eya and Gro. We transfected Kc167 cells with constructs encoding individual SIX proteins and either Eya or Gro. We find that both So and Optix (but not DSix4) can physically interact with Eya (Fig. 7, rows 1, 4 and 8) and that these interactions occur through the SIX protein-protein interaction domain (Fig. 7, rows 2 and 5). Our results showing that So binds to Eya is supported by data from yeast 2-hybrid and in vitro GST pull-down assays (Pignoni et al., 1997; Kenyon et al., 2005a; Kenyon et al., 2005b). However, our data demonstrating that Optix can interact with Eya stands in contrast to a report that failed to observe a genetic interaction between the two genes (Seimiya and Gehring, 2000) and a yeast 2-hybrid assay that used a truncated Optix protein (Kenyon et al., 2005a). Our ability to detect binding between Optix and Eya is probably due to the use of a full-length protein and the use of an in vivo Drosophila cell culture system. The formation of an Optix-Eya complex lends support to the possibility that Optix is not a dedicated repressor and, like its vertebrate orthologs Six3/6, has dual transcriptional activities. As expected we can also detect physical interactions between Gro and the SIX domain of Optix (Fig. 7, rows 6 and 7). However, in our Kc167 cell assay, we failed to observe the formation of So-Gro or DSix4-Gro complexes (Fig. 7, rows 3 and 9). As the data presented in this report support a role for So as a transcriptional repressor, it is possible that, although So does not bind to Gro in Kc167 cells, it may still do so within imaginal discs. Alternatively, So may repress transcription of target genes by either binding to other transcriptional co-repressors such as C-terminal Binding Protein (CtBP) and dNAB, or by interacting directly with the general transcription factor machinery.
Fig. 7.
Formation of SIX-Eya and SIX-Gro transcriptional complexes. Immunoblots of biochemical interactions from Kc167 cells between the Drosophila SIX proteins, Eya and Gro. Production of individual proteins is shown in the nuclear lysate (NL) lanes. Successful isolation of individual proteins is shown in the immunoprecipitation (IP) lanes. Specificity of the pull-downs is shown in the mock IP lanes. Protein interactions are shown in the immunoblot (IB) lanes.
DISCUSSION
SIX proteins are thought to act as both transcriptional activators and repressors during eye development. This conclusion is based on the observed effects that these proteins have on individual target genes, as well as on biochemical and genetic interactions with Eya and Gro family members. However, the overall contribution that these activation and repression complexes make to eye development is poorly understood. In Drosophila, the SIX proteins So and Optix are important for compound eye development and are capable of inducing ectopic eyes when mis-expressed in non-retinal tissues. This feature is not unique to the SIX proteins, as all members of the retinal determination network are capable of coaxing non-retinal cells into adopting an eye fate. The prevailing view is that ectopic eyes result from simply activating the retinal determination network at higher levels than endogenous gene regulatory networks such as those that control specification of the antennae, wings, legs, halteres and genitals. In support of this model is the observation that the frequency at which ectopic eyes are induced by So is synergistically increased by co-expression with Eya, a strong transcriptional co-activator and binding partner (Pignoni et al., 1997; Seimiya and Gehring, 2000).
There is some evidence to suggest that this transcriptional activation model may not fully describe the mechanism by which genes of the retinal determination network, particularly So and Optix, induce ectopic eye formation. First, So and Optix can initiate ectopic eye development on their own and without the co-expression of Eya (Seimiya and Gehring, 2000; Weasner et al., 2007). Second, in a large-scale study we have failed to observe strong synergistic interactions between So and Eya (Salzer and Kumar, 2010). And third, Optix which binds to an identical DNA sequence does not appear to synergize functionally nor form a biochemical complex with Eya (Seimiya and Gehring, 2000; Kenyon et al., 2005a). In fact the evidence surrounding Optix has suggested that it may function as a dedicated transcriptional repressor. Here, we have attempted to determine the relative roles that SIX-dependent activation and repression complexes play in the induction of ectopic eyes.
Using a yeast transcriptional activation assay, we first determined that, on its own, Optix is incapable of activating transcription of a reporter construct (Fig. 1D, well 3), whereas So can only activate transcription of the reporter weakly (Fig. 1D, well 5). We conclude from these assays that neither So nor Optix possesses sufficient intrinsic activation potential to induce ectopic eyes. To determine whether either SIX protein requires interactions with a robust transcriptional co-activator, we fused the VP16 activation domain to both So and Optix, thereby creating strong transcriptional activators and bypassing the requirement to interact with other co-factors. As a control, we fused the Engrailed (EN) domain to both proteins, creating chimeras that are predicted to be strong transcriptional repressors (Fig. 2A,E). These constructs were assayed for their ability to induce ectopic eye formation within the antennal disc. We find that, in both cases, only the repressor version of each SIX protein (So-EN and Optix-EN) can promote ectopic eye development (Fig. 2C,D,G,H). We conclude from these results that at least one step in the induction of ectopic eyes by So and Optix requires a transcriptional repression complex. As both proteins recognize nearly identical consensus target sequences (Berger et al., 2008; Noyes et al., 2008), our results are consistent with there being a common mechanism for both proteins to promote eye development.
What is the nature of the repressive complexes that are formed by So and Optix? Our biochemical assays suggest that Optix may repress transcription of target genes through interactions with Gro. We were unable to detect similar interactions between So and Gro, thereby raising the possibility that So represses transcription by forming complexes with other co-repressors and histone deacetylases, or through direct interactions with the general transcription factor machinery components. Binding between Optix and Gro appears to occur through the SIX protein-protein interaction domain. We have previously shown that the C-terminal region of Optix (downstream of the homeodomain) also contains a repression domain (Weasner and Kumar, 2009). We do not know how these two repression domains are related. It is possible that, in addition to the Gro interaction, other co-repressors could also bind to the C-terminal tail. There could also be physical interactions between these two regions of Optix. Further experimentation, including structural analysis, will be required to distinguish these possibilities.
Do So and Optix have a common transcriptional target(s) within the antenna, and why is the repression of this gene(s) necessary for the induction of ectopic eyes? One hint comes from earlier observations suggesting that the eye selector protein Ey and the antennal selector protein Ct mutually repress one another’s transcription. This results in the exclusive distribution of Ey and Ct proteins within the eye and antennal fields, respectively. This reciprocal expression pattern is important for maintaining distinct eye and antennal fates (Kenyon et al., 2003). However, we find that ct is more likely to be a target of So and Optix than Ey (Fig. 4D,E,J,K, Fig. 5A,B,E,F). As a result of ct being inhibited by So and Optix, ey expression is de-repressed within the antenna which in turn leads to the induction of ectopic eyes (Fig. 4F-H,L-N).
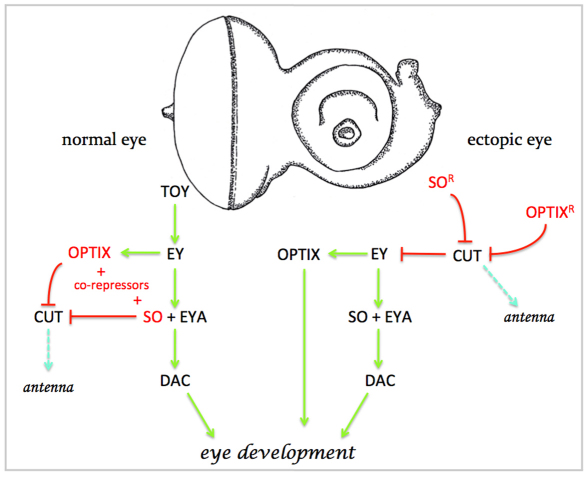
We therefore propose a model in which the first step in the formation of ectopic eyes is the shutting off of the non-retinal endogenous developmental program: in the case of this study it is the antennal program that must be disabled (Fig. 8). In our model, SoFL and OptixFL induce ectopic eyes by functioning as transcriptional repressors (by binding with co-repressors that are endogenously expressed within the antennal disc) to downregulate non-retinal developmental programs, thereby relieving repression of ey (Fig. 8). Our data support this model because: (1) the So-En and Optix-En repressors are the only versions that will induce ectopic eyes (Fig. 2C,D,G,H); (2) expression of these molecules is sufficient to inhibit ct expression (Fig. 4D,E,J,K); and (3) ey is ectopically activated within the antenna (Fig. 4F,L). In our model, Ey then activates the entire retinal determination network by directly activating so, Optix, eya and dac transcription (Halder et al., 1998; Michaut et al., 2003; Niimi et al., 1999; Ostrin et al., 2006; Pappu et al., 2005; Punzo et al., 2002). According to our model, the So-Eya complex then functions as a transcriptional activator that, in turn, triggers the expression of several downstream targets and results in the formation of an ectopic eye (Jemc and Rebay, 2007b; Pauli et al., 2005; Tanaka-Matakatsu and Du, 2008; Zhang et al., 2006). Similarly, Optix can switch its activity and also function as a transcriptional activator (possibly through interactions with Eya) and promote eye formation.
Fig. 8.
Model comparing normal and ectopic eye development. In this model, the first step in ectopic eye development is the repression of the antennal/head capsule specifying gene ct. In normal eye development, this is achieved through interactions between the SIX proteins So and Optix with co-repressors. In the second step, Ey activates the entire retinal determination network through So-Eya and Optix-Eya complexes. These two steps then lead to the promotion of eye development.
How do the initial steps of ectopic eye formation differ from normal eye development? As Ey and its paralog Toy are present in the eye disc from mid-embryogenesis (Czerny et al., 1999; Quiring et al., 1994), the eye anlagen are funneled to an eye fate from very early on in development, while the antennal fate program is repressed. This can be seen in the repression of ct expression within the eye and its restriction to the antennal segment (Kenyon et al., 2003). We propose a two-step model in which the unifying first step for both normal and ectopic eye development is the repression of non-retinal tissue fates. This is followed by the activation of the eye-specifying developmental program by Ey/Pax6. If our model is correct, it would then imply that tissues develop by not only activating the appropriate developmental programs but also repressing inappropriate ones. The study of ectopic eye induction provides a simple yet elegant way to define these repressive mechanisms.
Supplementary Material
Acknowledgments
We thank Janice Fischer, Craig Micchelli, Chrysoula Pitsouli, Larry Zipursky and the Bloomington Drosophila Stock Center for fly strains; the Developmental Studies Hybridoma bank for antibodies; and Adi Salzberg for the DNA constructs containing the VP16 activation and En repressor domains.
Footnotes
Funding
This work was supported by a grant from the National Eye Institute [R01 EY014863 to J.P.K.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.077255/-/DC1
References
- Aldaz S., Morata G., Azpiazu N. (2003). The Pax-homeobox gene eyegone is involved in the subdivision of the thorax of Drosophila. Development 130, 4473–4482 [DOI] [PubMed] [Google Scholar]
- Altmann C. R., Chow R. L., Lang R. A., Hemmati-Brivanlou A. (1997). Lens induction by Pax-6 in Xenopus laevis. Dev. Biol. 185, 119–123 [DOI] [PubMed] [Google Scholar]
- Berger M. F., Badis G., Gehrke A. R., Talukder S., Philippakis A. A., Pena-Castillo L., Alleyne T. M., Mnaimneh S., Botvinnik O. B., Chan E. T., et al. (2008). Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133, 1266–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Basler K. (2008). Recombinases and their use in gene activation, gene inactivation, and transgenesis. Methods Mol. Biol. 420, 175–195 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blochlinger K., Jan L. Y., Jan Y. N. (1993). Postembryonic patterns of expression of cut, a locus regulating sensory organ identity in Drosophila. Development 117, 441–450 [DOI] [PubMed] [Google Scholar]
- Bodmer R., Barbel S., Sheperd S., Jack J. W., Jan L. Y., Jan Y. N. (1987). Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell 51, 293–307 [DOI] [PubMed] [Google Scholar]
- Bonini N. M., Leiserson W. M., Benzer S. (1993). The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72, 379–395 [DOI] [PubMed] [Google Scholar]
- Brugmann S. A., Pandur P. D., Kenyon K. L., Pignoni F., Moody S. A. (2004). Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development 131, 5871–5881 [DOI] [PubMed] [Google Scholar]
- Cheyette B. N., Green P. J., Martin K., Garren H., Hartenstein V., Zipursky S. L. (1994). The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12, 977–996 [DOI] [PubMed] [Google Scholar]
- Cohen S. M. (1993). Imaginal disc development. In The Development of Drosophila melanogaster (ed. Bate M., Arias A. Martinez.), pp. 747–841 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Czerny T., Halder G., Kloter U., Souabni A., Gehring W. J., Busslinger M. (1999). twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell 3, 297–307 [DOI] [PubMed] [Google Scholar]
- Datta R. R., Cruickshank T., Kumar J. P. (2011). Differential selection within the Drosophila retinal determination network and evidence for functional divergence between paralog pairs. Evol. Dev. 13, 58–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo M. E., Lopez-Rios J., Fernaud-Espinosa I., Granadino B., Sanz R., Ramos C., Ayuso C., Seller M. J., Brunner H. G., Bovolenta P., et al. (1999). Genomic cloning and characterization of the human homeobox gene SIX6 reveals a cluster of SIX genes in chromosome 14 and associates SIX6 hemizygosity with bilateral anophthalmia and pituitary anomalies. Genomics 61, 82–91 [DOI] [PubMed] [Google Scholar]
- Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., et al. (2003). A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 [DOI] [PubMed] [Google Scholar]
- Halder G., Callaerts P., Gehring W. J. (1995). Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267, 1788–1792 [DOI] [PubMed] [Google Scholar]
- Halder G., Callaerts P., Flister S., Walldorf U., Kloter U., Gegring W. J. (1998). Eyeless initiates the expression of both sine oculis and eyes absent during drosophila compound eye development. Development 125, 2181–2191 [DOI] [PubMed] [Google Scholar]
- Heanue T. A., Reshef R., Davis R. J., Mardon G., Oliver G., Tomarev S., Lassar A. B., Tabin C. J. (1999). Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 13, 3231–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held L. I. (2002). Imaginal Discs: The Genetic and Cellular Logic of Pattern Formation (Developmental and Cell Biology Series), Vol. 39 Cambridge, UK: Cambridge University Press; [Google Scholar]
- Ikeda K., Watanabe Y., Ohto H., Kawakami K. (2002). Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol. Cell. Biol. 22, 6759–6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C. C., Chao J. L., Jones N., Yao L. C., Bessarab D. A., Kuo Y. M., Jun S., Desplan C., Beckendorf S. K., Sun Y. H. (2003). Two Pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development 130, 2939–2951 [DOI] [PubMed] [Google Scholar]
- Jean D., Bernier G., Gruss P. (1999). Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech. Dev. 84, 31–40 [DOI] [PubMed] [Google Scholar]
- Jemc J., Rebay I. (2007a). The eyes absent family of phosphotyrosine phosphatases: properties and roles in developmental regulation of transcription. Annu. Rev. Biochem. 76, 513–538 [DOI] [PubMed] [Google Scholar]
- Jemc J., Rebay I. (2007b). Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev. Biol. 310, 416–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon K. L., Ranade S. S., Curtiss J., Mlodzik M., Pignoni F. (2003). Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev. Cell 5, 403–414 [DOI] [PubMed] [Google Scholar]
- Kenyon K. L., Li D. J., Clouser C., Tran S., Pignoni F. (2005a). Fly SIX-type homeodomain proteins Sine oculis and Optix partner with different cofactors during eye development. Dev. Dyn. 234, 497–504 [DOI] [PubMed] [Google Scholar]
- Kenyon K. L., Yang-Zhou D., Cai C. Q., Tran S., Clouser C., Decene G., Ranade S., Pignoni F. (2005b). Partner specificity is essential for proper function of the SIX-type homeodomain proteins Sine oculis and Optix during fly eye development. Dev. Biol. 286, 158–168 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Nishikawa K., Suzuki T., Yamamoto M. (2001). The homeobox protein Six3 interacts with the Groucho corepressor and acts as a transcriptional repressor in eye and forebrain formation. Dev. Biol. 232, 315–326 [DOI] [PubMed] [Google Scholar]
- Li X., Perissi V., Liu F., Rose D. W., Rosenfeld M. G. (2002). Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science 297, 1180–1183 [DOI] [PubMed] [Google Scholar]
- Li X., Oghi K. A., Zhang J., Krones A., Bush K. T., Glass C. K., Nigam S. K., Aggarwal A. K., Maas R., Rose D. W., Rosenfeld M. G. (2003). Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 426, 247–254 [DOI] [PubMed] [Google Scholar]
- Liu W., Lagutin O. V., Mende M., Streit A., Oliver G. (2006). Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 25, 5383–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rios J., Tessmar K., Loosli F., Wittbrodt J., Bovolenta P. (2003). Six3 and Six6 activity is modulated by members of the groucho family. Development 130, 185–195 [DOI] [PubMed] [Google Scholar]
- Michaut L., Flister S., Neeb M., White K. P., Certa U., Gehring W. J. (2003). Analysis of the eye developmental pathway in Drosophila using DNA microarrays. Proc. Natl. Acad. Sci. USA 100, 4024–4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani R. (1941). Two new eye-shape mutant alleles in Drosophila melanogaster. Dros. Inf. Serv. 14, 52 [Google Scholar]
- Milani R. (1951). Linkage data: the locus so of Drosophila melanogaster. Dros. Inf. Serv. 25, 79 [Google Scholar]
- Niimi T., Seimiya M., Kloter U., Flister S., Gehring W. J. (1999). Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development 126, 2253–2260 [DOI] [PubMed] [Google Scholar]
- Noyes M. B., Christensen R. G., Wakabayashi A., Stormo G. D., Brodsky M. H., Wolfe S. A. (2008). Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133, 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto H., Kamada S., Tago K., Tominaga S. I., Ozaki H., Sato S., Kawakami K. (1999). Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol. Cell. Biol. 19, 6815–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver G., Mailhos A., Wehr R., Copeland N. G., Jenkins N. A., Gruss P. (1995). Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development 121, 4045–4055 [DOI] [PubMed] [Google Scholar]
- Ostrin E. J., Li Y., Hoffman K., Liu J., Wang K., Zhang L., Mardon G., Chen R. (2006). Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 16, 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu K. S., Ostrin E. J., Middlebrooks B. W., Sili B. T., Chen R., Atkins M. R., Gibbs R., Mardon G. (2005). Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development 132, 2895–2905 [DOI] [PubMed] [Google Scholar]
- Pauli T., Seimiya M., Blanco J., Gehring W. J. (2005). Identification of functional sine oculis motifs in the autoregulatory element of its own gene, in the eyeless enhancer and in the signalling gene hedgehog. Development 132, 2771–2782 [DOI] [PubMed] [Google Scholar]
- Pignoni F., Hu B., Zavitz K. H., Xiao J., Garrity P. A., Zipursky S. L. (1997). The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91, 881–891 [DOI] [PubMed] [Google Scholar]
- Punzo C., Seimiya M., Flister S., Gehring W. J., Plaza S. (2002). Differential interactions of eyeless and twin of eyeless with the sine oculis enhancer. Development 129, 625–634 [DOI] [PubMed] [Google Scholar]
- Quiring R., Walldorf U., Kloter U., Gehring W. J. (1994). Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science 265, 785–789 [DOI] [PubMed] [Google Scholar]
- Rayapureddi J. P., Kattamuri C., Steinmetz B. D., Frankfort B. J., Ostrin E. J., Mardon G., Hegde R. S. (2003). Eyes absent represents a class of protein tyrosine phosphatases. Nature 426, 295–298 [DOI] [PubMed] [Google Scholar]
- Salzer C. L., Kumar J. P. (2009). Position dependent responses to discontinuities in the retinal determination network. Dev. Biol. 326, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer C. L., Kumar J. P. (2010). Identification of retinal transformation hot spots in developing Drosophila epithelia. PLoS ONE 5, e8510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimiya M., Gehring W. J. (2000). The Drosophila homeobox gene optix is capable of inducing ectopic eyes by an eyeless-independent mechanism. Development 127, 1879–1886 [DOI] [PubMed] [Google Scholar]
- Seo H. C., Curtiss J., Mlodzik M., Fjose A. (1999). Six class homeobox genes in drosophila belong to three distinct families and are involved in head development. Mech. Dev. 83, 127–139 [DOI] [PubMed] [Google Scholar]
- Serikaku M. A., O’Tousa J. E. (1994). sine oculis is a homeobox gene required for Drosophila visual system development. Genetics 138, 1137–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S. J., Davies E. L., Doyon L., Rebay I. (2003). Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol. Cell. Biol. 23, 5989–5999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M., Du W. (2008). Direct control of the proneural gene atonal by retinal determination factors during Drosophila eye development. Dev. Biol. 313, 787–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootle T. L., Silver S. J., Davies E. L., Newman V., Latek R. R., Mills I. A., Selengut J. D., Parlikar B. E. W., Rebay I. (2003). The transcription factor eyes absent is a protein tyrosine phosphatase. Nature 426, 299–302 [DOI] [PubMed] [Google Scholar]
- Toy J., Yang J. M., Leppert G. S., Sundin O. H. (1998). The optx2 homeobox gene is expressed in early precursors of the eye and activates retina-specific genes. Proc. Natl. Acad. Sci. USA 95, 10643–10648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J., He Y., Hoskins R. A., Bellen H. J. (2006). P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747–1751 [DOI] [PubMed] [Google Scholar]
- Weasner B., Salzer C., Kumar J. P. (2007). Sine oculis, a member of the SIX family of transcription factors, directs eye formation. Dev. Biol. 303, 756–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weasner B. M., Weasner B. P., DeYoung S. M., Michaels S. D., Kumar J. P. (2009). Transcriptional activities of the Pax6 gene eyeless regulate tissue specificity of ectopic eye formation in Drosophila. Dev. Biol. 334, 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weasner B. P., Kumar J. P. (2009). The non-conserved C-terminal segments of Sine Oculis Homeobox (SIX) proteins confer functional specificity. Genesis 47, 514–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Ranade S., Cai C. Q., Clouser C., Pignoni F. (2006). Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development 133, 4881–4889 [DOI] [PubMed] [Google Scholar]
- Zhu C. C., Dyer M. A., Uchikawa M., Kondoh H., Lagutin O. V., Oliver G. (2002). Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development 129, 2835–2849 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.