Abstract
achaete-scute homologs (ash) regulate neural development in all bilaterian model animals indicating that they represent a component of the ancestral neurogenic pathway. We test this by investigating four ash genes during development of a basal metazoan, the cnidarian sea anemone Nematostella vectensis. Spatiotemporal expression of ash genes in the early embryo and larval stages suggests that they regulate neurogenesis. More specifically, NvashA is co-expressed with neural genes in the embryonic ectoderm. Knockdown of NvashA results in decreased expression of eight neural markers, including the six novel neural targets identified here. Conversely, overexpression of NvashA induces increased expression of all eight genes, but only within their normal axial domains. Overexpression of NvashB-D differentially increases expression of NvashA targets. The expression patterns and differential ability of ash genes to regulate neural gene expression reveals surprising molecular complexity in these ‘simple’ animals. These data suggest that achaete-scute homologs functioned in the ancestral metazoan neurogenic pathway and provide a foundation to investigate further the evolution of neurogenesis and the origin of complex central nervous systems.
Keywords: NvashA, Achaete-scute, Neurogenesis, Nematostella, Cnidarian, Proneural
INTRODUCTION
The evolution of centralized nervous systems in bilaterians, a diverse group of animals that includes insects and man, was a key innovation that led to overwhelming success and diversity. Gene expression patterns and limited functional studies suggest that the metazoan (all multicellular animals) nervous system arose early in animal evolution. However, basal metazoans such as ctenophores (comb jellies) and cnidarians (sea anemones, coral, jellyfish and hydras) do not possess highly organized central nervous systems, but rather have nerve cell populations that are distributed throughout their bodies forming a ‘nerve net’ (Galliot et al., 2009; Jager et al., 2010). Thus, comparing neurogenic mechanisms deployed in basal metazoans to the later branching bilaterian taxa will provide insight into conserved ancestral mechanisms of neural development and improve our understanding of the origins and evolution of central nervous systems.
We focus on the cnidarian sea anemone Nematostella vectensis. Cnidarians are the sister taxa to the bilaterians (Dunn et al., 2008; Hejnol et al., 2009) and, thus, are the metazoan group most closely related to bilaterians. Within the cnidarians Nematostella belongs to the anthozoan class (anemones and corals), a group that radiated early in the cnidarian lineage (Collins et al., 2006; Putnam et al., 2007). Studying the anthozoans in addition to the hydrozoans (e.g. Hydra) provides a better data set to reconstruct the ancestral cnidarian characteristics, which is essential for comparative analysis to understand bilaterian evolution and development. Thus, anthozoans are an informative group to investigate with regard to understanding ancestral metazoan developmental mechanisms related to the evolution of the bilaterian animals.
The nervous system of cnidarians comprises endodermal and ectodermal nerve nets, a unique metazoan feature in that most animals possess only ectodermal neurogenic programs. The neural cell types found in cnidarians include sensory cells, ganglion cells and cnidocytes (a cnidarian-specific mechanosensory cell) (Galliot et al., 2009; Kass-Simon and Scappaticci, 2002; Marlow et al., 2009; Watanabe et al., 2009). Nematostella neurogenesis begins during early embryonic stages and can be first detected by ectodermal expression of a neural marker Nvelav followed by the neurotransmitter NvanthoRFamide at mid to late gastrula stages (Marlow et al., 2009) (Fig. 2). During the swimming planula larval stage, ectodermal neural markers expand, endodermal expression of Nvelav initiates and cross-reactive antibodies against 5-HT, FMRFamide and GABA can be detected in both the endoderm and ectoderm (Marlow et al., 2009). By polyp stages, both endodermal and ectodermal nerve nets are well developed. However, functional analyses of the mechanisms that generate elements of either endodermal or ectodermal nervous system are currently lacking in Nematostella. Of particular interest is early embryonic ectodermal neural development because they are the first neural cells that appear during Nematostella development and nearly all bilaterian neurons derive from embryonic ectodermal tissue.
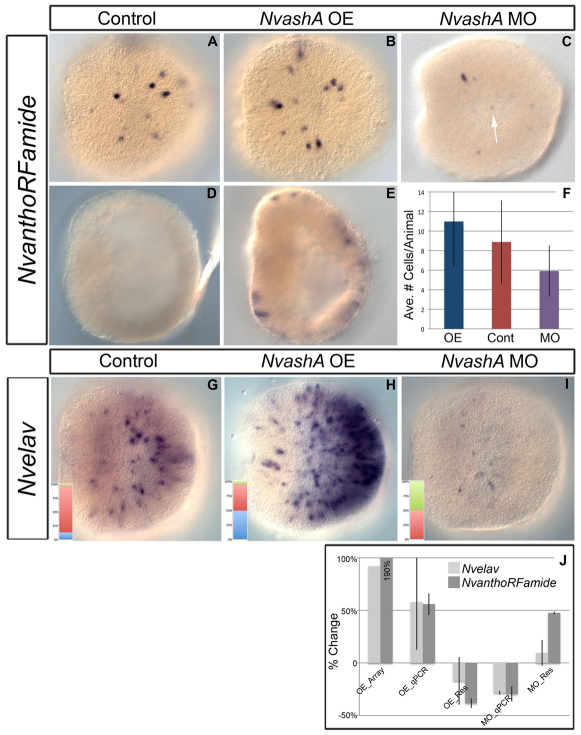
Fig. 2.
NvashA regulates NvanthoRFamide and Nvelav expression in 24 hpf embryos. All images are of late gastrula at 24 hpf. Shown are lateral views with oral to the left. (A-E) NvanthoRFamide expression (A-E) in control (A,D), NvashA:venus misexpressing embryos (B,E), and NvashA morphant animals (C). Premature expression of NvanthoRFamide was detected in early gastrula of NvashA:venus-injected animals (E), whereas no expression was detected in wild-type embryos of the same age (D). (F) Quantification of the average number of NvanthoRFamide positive cells/animal for animals represented in A-C. Overexpression (OE) is in blue, control (Cont) in red and morpholino (MO) in purple. n=43, 42, 47 for OE, Cont, MO, respectively. t-test values for OE and MO compared with control were P=0.03 and P=0.0002, respectively. Error bars represent s.e.m. (G-I) Nvelav expression in control (G), NvashA:venus-injected (H) and NvashA MO-injected (I) animals. The insets are quantification of the percentage of animals displaying wild-type (red), overexpression (blue) and reduced (green) expression levels. n=71, 75, 72 for control, NvashA:venus and NvashA morphants, respectively. (J) Changes in gene expression for Nvelav (light gray) and NvanthoRFamide (dark gray) in NvashA:venus overexpressing animals shown by microarray (OE_Array) and qPCR (OE_qPCR) and in NvashA MO-injected animals (MO_qPCR). Average of replicate experiments is shown for qPCR experiments. Error bars represent s.e.m. The NvashA:venus overexpression phenotype could be reversed by co-expression of NvashA translation-blocking MO, which recognized the endogenous and in vitro transcribed mRNAs (OE_Res). The MO phenotype could be reversed by co-injecting a truncated NtruncNvashA:venus mRNA (MO_Res), which is not recognized by the NvashA MO.
Proneural basic helix-loop-helix (bHLH) class transcription factors are likely to be conserved regulators of metazoan neural development. Bilaterian proneural genes are grouped into two main families: the atonal family and the achaete-scute family (for a review, see Bertrand et al., 2002). The proneural genes were so named for early expression in neural tissue and because they are necessary and sufficient to promote neural development in all bilaterian animals investigated thus far (Bertrand et al., 2002). The neurogenic role of proneural genes probably pre-dated the emergence of bilaterians. In the cnidarians, Hydra vulgaris (Grens et al., 1995; Hayakawa et al., 2004; Lindgens et al., 2004) and Podocoryne carnea (Seipel et al., 2004) achaete-scute homologs are expressed in differentiating neural cell types, including cnidocytes, in adult polyps. Also, an ancestral atonal family gene identified in sponges (which do not possess neurons) (Richards et al., 2008) and a cnidarian achaete-scute homolog (Grens et al., 1995) both induce neural gene expression in xenotopic experiments conducted in insects and vertebrates.
Nematostella possesses four definitive achaete-scute family genes (Simionato et al., 2007) and at least nine genes that group within the atonal super-family. However, the orthology of the atonal-like genes remains unresolved. Here, we focus on the achaete-scute genes because of the unambiguous assignment of the four distinct Nematostella achaete-scute homologs (Simionato et al., 2007).
In this study, we focus primarily on embryonic ectodermal neural development, because it is the first observable neurogenesis in Nematostella and it begins before gastrulation is complete (Marlow et al., 2009). Thus, the origin tissue of neural cells can be unambiguously assigned as ectodermal. We demonstrate that the spatiotemporal expression patterns of NvashA-D are all consistent with being regulators of cnidarian neural development, but NvashA is uniquely co-expressed with neural markers in the early embryonic ectoderm. Knockdown and overexpression of NvashA shows that NvashA is both necessary and sufficient for proper expression of neural marker genes, demonstrating that it is a bona fide proneural transcription factor in Nematostella. We also observe increases in neural gene expression in response to ectopic overexpression of NvashA. However, the increased marker expression is restricted to their normal axial domains. Our work in the cnidarian Nematostella vectensis strongly suggests that the proneural activity of ash genes pre-dates the emergence of the bilaterian central nervous systems and represents an ancient conserved component of metazoan neurogenesis.
MATERIALS AND METHODS
Nvash genes used in this study
Previous work identified four achaete-scute homologs in Nematostella vectensis (nem 13, 17, 23 and 27) (Simionato et al., 2007). The Nematostella genome v1.0 gene ID number is listed for each one in parenthesis. We have renamed them as follows: nem17=NvashA (247879), nem23=NvashB (207481), nem27=NvashC (220832) and nem13=NvashD (210540). NvashB also has a Genbank ID number (BAJ13484). Previously identified genes are also used NvanthoRFamide (200150) and Nvelav (214798) (Marlow et al., 2009). The remaining genes are identified by gene number within this paper.
Embryo manipulations and in situ analysis
All embryos were grown to 24 hours post-fertilization (hpf). They were initially maintained at ∼20°C for 4 hours, then moved to 25°C for 20 hours prior to fixation. Fixation, in situ probe synthesis, immunohistochemical detection, and in situ hybridization were carried out as described previously (Layden et al., 2010). Double fluorescent in situ hybridization was carried out as described previously (Rentzsch et al., 2008) with the following modifications: after washing, embryos were incubated overnight at 4°C with sheep anti-fluorescein-POD (1:500; Roche) and mouse anti-DIG (1:500; Jackson ImmunoResearch Laboratories, 200-002-156). The following day, embryos were incubated with anti-sheep-POD (Jackson ImmunoResearch Laboratories, 313-036-003) at 1:500 for 2 hours at room temperature followed by development of the fluorescein-labeled probe. Embryos were then washed as described previously (Rentzsch et al., 2008) and incubated in anti-mouse-POD (Jackson ImmunoResearch Laboratories, 315-036-045) (1:500) overnight at 4°C. The following day, DIG-labeled probes were detected as described previously (Rentzsch et al., 2008). Immunohistochemical images were acquired on a Zeiss Axioskop 2 in conjunction with Axiocam HRc and Axio vision 4.7 software (Carl Zeiss LLC, Thornwood, NY). Fluorescent images were acquired on a Zeiss 710 confocal microscope in conjunction with Zen software (Carl Zeiss LLC).
The NvashA:venus fusion construct was generated by cloning NvashA in frame with the venus coding sequence using the Gateway cloning vector system (Invitrogen). mRNA was in vitro transcribed using the Ambion mMessage Machine T3 transcription kit (Ambion, AM1348) and purified using Megaclear columns (Ambion, AM1908) followed by phenol:chloroform clean up and isopropanol precipitation. NvashA:venus mRNA was injected into zygotes at 200 ng/μl. Embryos were sorted at 20 hpf and only animals demonstrating ubiquitous NvashA:venus expression were kept for analysis (supplementary material Fig. S1). Fluorescein-labeled NvashA translation blocking morpholino (MO; 5′-AGCTCATGTCTTCTGTTTCACTCAT-3′; Gene Tools, LLC, Philomath, OR, USA) was injected at 1 mM. An amount of liquid approximately equal to 5% egg volume was injected for all injections. Controls for each injection were either uninjected control or dextran-injected controls. A control MO (5′-AGAGGAAGAATAACATACCCTGTCC-3′) was also injected at a concentration of 1 mM and gene expression was compared with uninjected control animals.
To demonstrate that MO phenotypes were due to specific knockdown of NvashA and not an off-target effect, we co-injected a truncated in vitro synthesized NvashA mRNA (NtruncNvashA:venus), which is missing the first 24 nucleotides and thus lacks the binding sequence for the NvashA MO.
To demonstrate that NvashA overexpression phenotypes are due specifically to the overexpression of NvashA, we co-injected the NvashA:venus mRNA with 1 mM of the NvashA translation-blocking MO, which recognizes both the in vitro transcribed and endogenous mRNA.
Counting of NvanthoRFamide cells and scoring of morphant and overexpression phenotypic classes were conducted on a Zeiss AxioSkop 2. Statistical analysis was carried out by t-tests comparing NvanthoRFamide counts between treated and control animals.
Quantitative PCR (qPCR) and microarray analysis
RNA for qPCR and microarray experiments was isolated with TriPure (F. Hoffeman-La Roche, 11667157001). Genomic DNA contamination was removed using Qiagen RNAse free DNAse (Qiagen, Valencia, CA, USA, 79254) for 15 minutes at 37°C. cDNA for qPCR was generated from 1 μg total RNA with the Advantage RT-PCR kit (Clontech, 639506). qPCR analysis was carried out using a Roche LightCycler 480 and LightCycler 480 SYBR Green 1 Master mix (Roche, 04887352001). Changes in gene expression between the treated and control animals were calculated by comparing the relative fold change (FC) ratios. Actin and/or GADPH housekeeping genes were used to normalize fold change. Relative fold change was converted to percentage change by [–1+(FC ratio)]. qPCR primers were ordered for each gene and efficiencies determined using tenfold serial dilution series. All primers used for qPCR analysis in this study ranged from 85% to 115% efficiency values. Sequences of qPCR primers used can be found in supplementary material Table S1. Each qPCR analysis was repeated in biological replicates injected in independent sessions. To conduct the microarrays, 10 μg of total RNA was used to generate cDNA with the Invitrogen superscript III double-stranded cDNA synthesis kit (11917-010). One microgram of cDNA was used for generating labeled cDNA with the Nimblegen one color DNA labeling kit (05 223 555 01). Labeled cDNA was hybridized to a whole-genome array generated for our laboratory by Roche Nimblegen (Iceland). The array was scanned with a GenePix Personal 4100a scanner in conjunction with GenePix Pro 6.0 software (Molecular Devices, Sunnyvale, CA, USA) and analyzed using Nimblescan software (Roche Nimblegen). Gene expression levels were normalized in the Nimblescan software according to the methods of Bolstad et al. and Irizarry et al. (Bolstad et al., 2003; Irizarry et al., 2003). Fold change and percentage change were calculated as described above using expression values from normalized array data.
RESULTS
Spatiotemporal expression of NvashA suggests that it regulates Nematostella vectensis embryonic ectodermal neurogenesis
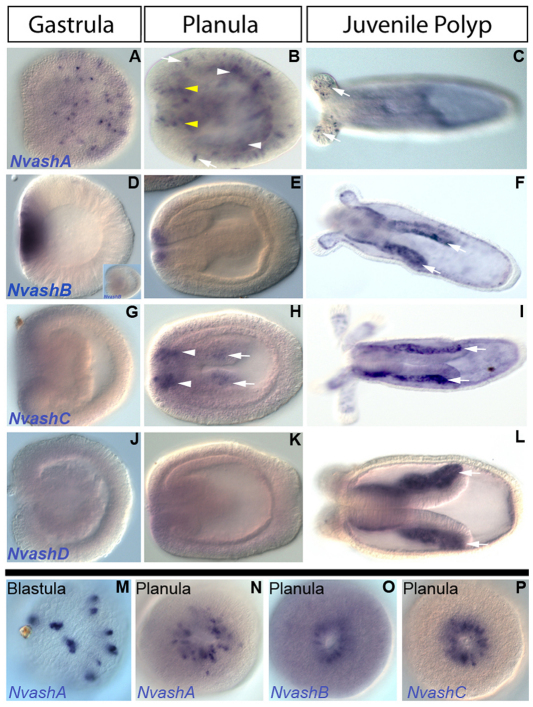
To determine whether cnidarian achaete-scute homolog(s) regulate embryonic ectodermal neural development, we screened for Nvash expression in embryonic ectoderm prior to or coincident with neural marker expression. NvashA displays this appropriate spatiotemporal pattern. NvashA expression was first detected in blastula stages in paired cells distributed in a ‘salt and pepper’ pattern (Fig. 1M). By mid-gastrula, the paired expression was not apparent and single NvashA-positive cells were distributed throughout the ectoderm (Fig. 1A). Ectodermal ‘salt and pepper’ expression was maintained throughout planula larval stages (Fig. 1B, arrows), but was only detected in the proximal tentacles of the juvenile polyp (Fig. 1C, arrows). NvashA tentacular expression is reminiscent of the achaete-scute homolog Cnash expression reported in sensory neurons at the base of the tentacle in the adult Hydra (Hayakawa et al., 2004). NvashA endodermal ‘salt and pepper’ expression was also observed in the early planula and was widespread throughout the presumptive body column endoderm by 48 hpf (Fig. 1B, white arrowheads). Additionally, a domain enriched with NvashA was detected in the oral ectoderm during planula larval stages (Fig. 1N), which is reminiscent of NvanthoRFamide expression in the oral ectoderm of planula and polyp stage animals (Marlow et al., 2009) and might correlate with the developing oral nerve ring. The oral domain appears to expand aborally, possibly in endodermal cells adjacent to the developing pharyngeal ectoderm (Fig. 1E, yellow arrowhead). It is not clear whether expansion represents nascent expression of NvashA transcripts in underlying endodermal cells or intrusion of NvashA-expressing ectodermal cells into the endoderm. The spatiotemporal distribution of NvashA expression in a salt and pepper pattern in both endodermal and ectodermal domains is consistent with the notion that NvashA regulates ectodermal and endodermal neural development.
Fig. 1.
Expression patterns of Nematostella achaete-scute homologs during development. (A-P) Lateral views with oral to the left (A-L), oral views (N-P), and view of blastula (M). NvashA expression (A-C,M,N) is detected in paired cells distributed in a salt and pepper pattern in the blastula (M, arrows), in an ectodermal salt and pepper pattern in the gastrula (A) and planula (B, arrows) and in individual cells at the base of the tentacular ectoderm (C, arrows). Salt and pepper endodermal staining is also detected in the planula endoderm (B, white arrowheads) and in cells of the planula oral ectoderm (B, yellow arrowheads). NvashB expression (D-F,O) is in the presumptive endoderm in gastrula stages (D), but not by late gastrula (D, inset), in the oral ectoderm of the planula larva (E,O) and in the lateral portion of the directive mesenteries (F, arrows). NvashC is detected initially in the oral ectoderm of the late gastrula stage and extends aborally with the outgrowth of the directive mesenteries (H, arrows). Expression is maintained in the lateral portion of the directive mesenteries (I, arrows). NvashD expression is first detected in the lateral portion of the directive mesenteries in the juvenile polyp (L, arrows).
NvashB-D expression patterns suggest roles in planula and polyp neurogenesis
NvashB expression was first detected in late blastula stages after NvashA in the presumptive endoderm and was maintained through the initiation of gastrulation (Fig. 1D). By mid gastrula, NvashB was no longer detected by in situ hybridization (Fig. 1D, inset). In the planula larval stages, NvashB was expressed in an oral ectodermal ring (Fig. 1E,O) in a pattern similar to that observed for NvashA. NvashB expression was also detected in the lateral domain of the directive mesenteries (Fig. 1F, arrows), which is likely to be derived from endoderm, at polyp stages. The mesenteries are sites from which cnidocyte-containing nematosomes originate (Williams, 1975; Williams, 1979). Expression in this domain of the mesentery would be consistent with the observed Cnash expression in developing cnidocytes in adult Hydra vulgaris (Grens et al., 1995). Lateral mesentery expression was observed in variable proportions over a number of experiments. We suspect that this variability reflects transient expression and might be associated with a wave(s) of cnidocyte/nematosome development.
NvashC expression was first detected in the early planula in the oral ectoderm (Fig. 1P). This expression then appears, like that of NvashA, to extend into the endoderm surrounding the developing pharynx (Fig. 1H, arrowheads) and expand down the presumptive mesenteries (Fig. 1H, arrows). By polyp stages,, the only detectable expression for NvashC was in the lateral domains of the developing mesenteries (Fig. 1I, arrows).
NvashD expression appears to be weakly expressed in an orally enriched domain in late gastrula and planula stages (Fig. 1K) and in a lateral mesentery domain (Fig. 1F, arrows) in juvenile polyps. The NvashB (Fig. 1F, arrows) and NvashD (Fig. 1I, arrows) expression in the lateral mesentery was probably due to nascent expression because, unlike NvashC expression, which was detectable in the presumptive and newly formed mesenteries (Fig. 1H,I, arrows), NvashB and NvashD expression were not detectable until the mesenteries had already formed. The lateral mesentery expression for all three NvashB-D genes is consistent with these genes regulating cnidocyte development and, thus, aspects of Nematostella neural development.
NvashA regulates NvanthoRFamide+ and Nvelav+ neurons
We examined the role of NvashA on the regulation of the two earliest known neural genes characterized in Nematostella, NvanthoRFamide and Nvelav (Marlow et al., 2009). A translation-blocking morpholino (MO) and in vitro synthesized NvashA:venus mRNA were injected individually into Nematostella zygotes to determine whether reduced and increased levels of NvashA affected the expression of these markers. Gene expression was scored using qPCR, whole-genome microarray and mRNA in situ hybridization at 24 hpf. Both Nvelav and NvanthoRFamide levels were decreased following MO injection (Fig. 2C,F,I,J), but not after injection of a control MO (supplementary material Fig. S2). There was a 30% reduction in NvanthoRFamide transcript levels when assayed by qPCR (Fig. 2J; MO_qPCR dark gray). The average number of cells expressing NvanthoRFamide was likewise reduced by ∼30% (Fig. 2F; 5.9 cells compared with 8.8 cells in wild type; purple and red bars, respectively) and, though slight, this difference is highly statistically significant (P=0.0002). Additionally, many of the remaining cells had a decrease in expression levels of NvanthoRFamide (Fig. 2C, arrow, for example) compared with control animals developed in parallel (Fig. 2A).
Injection of NvashA MO also reduced Nvelav levels by 28% (Fig. 2J; MO_qPCR, light gray). We were unable to accurately count the number of Nvelav cells because of cell numbers in wild-type animals. However, following in situ hybridization against Nvelav transcripts, phenotypic classes could be assigned as severely upregulated, wild type, or severely downregulated. Though crude, most animals fall unambiguously into these classes, thereby allowing us to confidently assess Nvelav phenotypes in mRNA in situ hybridization experiments. In NvashA morphant animals, approximately half of the animals displayed a severely downregulated Nvelav phenotype compared with wild-type animals (compare Fig. 2G and 2I; quantifications shown in insets n=71 wild type and n=72 MO). It is clear that Nvelav and NvanthoRFamide are not completely knocked down in NvashA morphant animals. However, morphant phenotypes for both genes can be reversed by co-injection of a truncated NtruncNvashA:venus in vitro synthesized mRNA (Fig. 2J), which is not recognized by the NvashA translation-blocking MO. Currently, we cannot resolve whether residual expression in morphants is due to the presence of maternal NvAshA protein, inefficient MO knockdown or parallel pathways regulating NvanthoRFamide and Nvelav expression.
Overexpression of NvashA by injection of the NvashA:venus mRNA resulted in increased NvanthoRFamide expression levels determined by microarray (∼200% increase; Fig. 2J, OE_Array, dark gray) and by qPCR (∼56% increase; Fig. 2J, OE_qPCR, dark gray). In situ analysis revealed a slight increase in the average number of NvanthoRFamide-positive cells in NvashA:Venus-overexpressing animals (Fig. 2F; 10.9 cells/animal compared with 8.8 cells in wild type; blue and red bars, respectively; P=0.03). Because the number of cells in NvashA:venus-injected animals was not dramatically increased, we tested whether or not temporal expression of NvanthoRFamide could be altered by misexpression of NvashA:venus mRNA. Precocious expression of NvanthoRFamide was detected in all the embryos in the early gastrula of microinjected animals and was never observed at this early stage in control animals (Fig. 2E and 2D, respectively).
Expression levels for Nvelav were also upregulated in NvashA:venus-injected animals (Fig. 2H,J). Microarray analysis revealed ∼90% increase in Nvelav transcript levels (Fig. 2J; OE_Array, light gray), whereas qPCR experiments showed an average increase of ∼60% (Fig. 2J; OE_qPCR, light gray). Approximately half of the NvashA:venus-injected animals displayed the severely upregulated Nvelav phenotype (Fig. 2H) and the remaining half appeared similar to wild type by in situ hybridization. Both the increase of NvanthoRFamide and Nvelav expression observed in NvashA:venus-overexpressing embryos could be reversed by co-injection with the translation-blocking NvashA MO, which recognizes both the endogenous transcript and the in vitro synthesized NvashA:venus mRNA (Fig. 2J, OE_Res). Thus, we conclude that NvashA is required for proper expression of both NvanthoRFamide and Nvelav and is sufficient to expand the number of Nvelav- and NvanthoRFamide-positive cells.
Identification and characterization of additional embryonic target genes regulated by NvashA
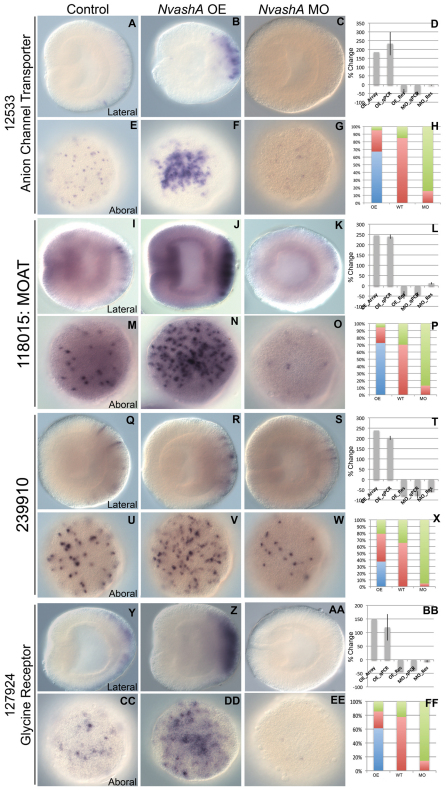
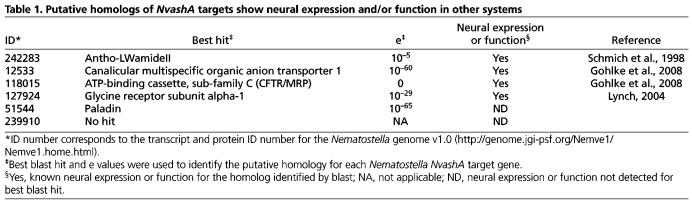
We performed a single preliminary whole-genome microarray on NvashA overexpression animals to generate a list of possible targets positively regulated by NvashA (Fig. 3D,L,T,BB, OE_Array; data not shown). Because the array data was preliminary, we only chose the top 20 upregulated genes and assayed for salt and pepper expression in 24 hpf wild-type embryos. Six genes displayed this specific expression pattern (Table 1) (Figs 3, 4). To determine the likely functional identity of each putative target, the NCBI protein database was blasted using the predicted Nematostella protein sequence. Two of the identified targets, the multiple organic anion transporter (MOAT) (ID:118015) (Fig. 3A-H) and a multispecific anion channel transporter (ID:12533) (Fig. 3I-L) are both part of the ATP-binding cassette family of membrane transporters and are targets of the mouse achaete-scute homolog Mash1 (Ascl1 – Mouse Genome Informatics) during telencephalon development (Gohlke et al., 2008). Additionally, we identified the post-synaptic glycine neurotransmitter receptor (ID:127924) (Fig. 3Y-FF), an LWamide-like neuropeptide (ID:242283) (Fig. 4I-P), Paladin-like gene (ID:51544) (Fig. 4A-H) and one unique Nematostella gene exhibiting no known homology or conserved domains (ID:239910) (Fig. 3Q-X). All of the previously identified genes (118015, 12533, 127924, 242283, 51544) except the paladin-like 51544 gene have neural expression or function described in other systems (Table 1, references). However, Paladin genes are implicated in neural crest cell migration (Roffers-Agarwal et al., 2009). Based on predicted gene function, the six identified genes are likely to function in neuronal cell types.
Fig. 3.
Aboral ectodermal targets of NvashA identified in embryonic ectoderm. (A-FF) Expression of 12533 (A-C,E-G), 118015 (I-K,M-O), 239910 (Q-S,U-W), and 127924 (Y-AA,CC-EE) in wild-type (A,E,I,M,Q,U,Y,CC), NvashA:venus overexpressing (OE) (B,F,J,N,R,V,Z,DD) and NvashA morphant (MO) (C,G,K,O,S,W,AA,EE) animals. The phenotypes for in situ hybridization experiments were quantified by distribution of increased (blue), wild-type (red) or decreased (green) levels of expression and cell number for NvashA:venus (OE), wild-type (WT) and NvashA morphant (MO) animals (H,P,X,FF). [For H, n=61 (OE), n=33 (WT) n=45 (MO); for P, n=55 (OE), n=37 (WT), n=31 (MO); for X, n=57 (OE), n=75 (WT), n=130 (MO); for FF, n=62 (OE), n=49 (WT), n=42 (MO).] The percent change of expression levels was also quantified (D,L,T,BB). Shown is the percent change observed by microarray for OE animals (OE_Array) and by qPCR (OE_qPCR, MO_qPCR) compared with wild type. The reversal of the NvashA:venus phenotype by co-injecting NvashA translation-blocking MO (OE_Res) and the reversal of NvashA morphant phenotype by co-injection of MO and NtruncNvashA:venus (MO_Res) are also shown. With the exception of the OE_Array, all qPCR percent changes shown are the average of two biological replicates. Error bars represent s.e.m. Oral is to the left in A-C,I-K,Q-S,Y-AA, and aboral views are shown in E-G,M-O,U-W,CC-EE.
Table 1.
Putative homologs of NvashA targets show neural expression and/or function in other systems
Fig. 4.
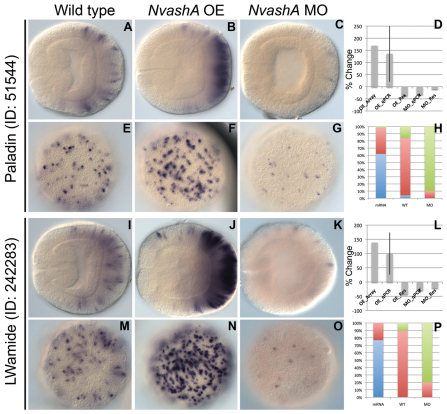
Additional targets of NvashA identified in embryonic ectoderm. (A-P) Expression of 51544 (A-C,E-G) and 242283 (I-K,M-O) in wild-type (A,E,I,M), NvashA:venus overexpressing (OE) (B,F,J,N), and NvashA morphant (MO) (C,G,K,O) animals. The phenotypes for in situ hybridization experiments were quantified by distribution of increased (blue), wild-type (red) or decreased (green) levels of expression and cell number for NvashA:venus (OE), wild-type (WT) and NvashA morphant (MO) animals (H,P). [For H, n=42 (OE), n=50 (WT), n=50 (MO; for P, n=52 (OE), n=37 (WT), n=42 (MO).] The percent change of expression levels was also quantified (D,L). Shown are the percent change observed by microarray for OE animals (OE_Array) and by qPCR (OE_qPCR, MO_qPCR) compared with wild-type controls. The reversal of the NvashA:venus phenotype by co-injecting NvashA translation-blocking MO (OE_Res) and the reversal of NvashA morphant phenotype by co-injection of MO and NtruncNvashA:venus (MO_Res) are also shown. With the exception of the OE_Array, all qPCR percent changes shown are the average of two biological replicates. Error bars represent s.e.m. Oral is to the left in A-C,I-K, and aboral views are shown E-G,M-O.
The expression pattern for four of the six (ID: 12533, 118015, 239910, 127924) genes is restricted to a salt and pepper pattern only in the aboral region corresponding to the presumptive apical tuft sensory organ domain (Fig. 3A,E,I,M,Q,U,Y,CC) (Rentzsch et al., 2008). The remaining two predicted NvashA target genes (ID: 242283 and 51544) have a slightly different expression pattern (Fig. 4). The 242283 and 51544 genes show aborally enriched salt and pepper expression like the genes described above; however, the expression domain extends more orally, similar to the broad expression observed in the Nvelav pattern (Fig. 2G). Thus, the six putative NvashA targets fall into one of two expression patterns. They are either expressed in a restricted domain at the aboral pole (12533, 118015, 239910, 127924) (Fig. 3A,I,Q,Y) or expressed throughout the entire aboral half of the embryo (242283, 51544) (Fig. 4A,I).
To confirm that each of the putative target genes was in fact downstream of NvashA, we performed qPCR and in situ hybridization analysis in both NvashA morphant and NvashA:venus-microinjected animals (Figs 3, 4). Knockdown of NvashA resulted in a statistically significant (P<0.05) decrease in expression in biological replicates for each gene (12533: Fig. 3C,D,G,H; 118015: Fig. 3K,L,O,P; 239910: Fig. 3S,T,W,X; 127924: Fig. 3AA,BB,EE,FF; 51544: Fig. 4C,D,G,H; 242283: Fig. 4K,L,O,P), whereas injection of a control MO did not result in significant change in gene expression (supplementary material Fig. S2). In addition, there appeared to be a reduction in both the number of cells expressing the target and in the level of expression observed in the remaining cells (12533: Fig. 3C,G; 118015: Fig. 3K,O; 239910: Fig. 3S,W; 127924: Fig. 3AA,EE; 51544: Fig. 4C,G; 242283: Fig. 4K,O). The reduced expression levels were reversed or alleviated by co-injection of the NtruncNvashA:venus mRNA with the NvashA MO (Fig. 3D,L,T,BB and Fig. 4D,L; MO_Res).
Conversely, injection of NvashA:venus in vitro transcribed mRNA resulted in an increase in expression levels for each gene and appeared to expand the total number of cells expressing each target (12533: Fig. 3B,D,F,H; 118015: Fig. 3J,L,N,P; 239910: Fig. 3R,T,V,X; 127924: Fig. 3Z,BB,DD,FF; 51544: Fig. 4B,D,F,H; 242283: Fig. 4J,L,N,P). The overexpression phenotype can be reversed by co-injection of the translation-blocking NvashA morpholino (Fig. 3D,L,T,BB and Fig. 4D,L; OE_Res). Based on both knockdown and overexpression, we conclude that NvashA is necessary for proper expression and sufficient to increase expression of each target. This supports the hypothesis that we have identified six novel NvashA targets, which are likely to have roles in neural functions. Additionally, we verified the feasibility of more extensive array experiments to identify targets of NvashA during Nematostella development.
NvashA overexpression confirms the presence of distinct domains along the oral-aboral axis
Overexpression NvashA phenotypes confirms molecular differences along oral-aboral axis of Nematostella. Previous reports described molecularly distinct domains demarcated by broad expression of genes, such as Nvwnts (Kusserow et al., 2005), Nvdlx (Ryan et al., 2007) and Nvfgfs (Matus et al., 2007; Rentzsch et al., 2008) along the oral-aboral axis of Nematostella. Ubiquitous overexpression of NvashA (supplementary material Fig. S1) increases the number of cells expressing and level of expression of neural genes within their normal expression domains but does not result in ectopic expression of neural genes elsewhere in the embryo (Fig. 2H; Fig. 3B,F,J,N,R,V,Z,DD; Fig. 4B,F,J,N). Furthermore, for targets with broader expression patterns, such as Nvelav (Fig. 2G), 51544 and 242283 (Fig. 4A and 4I, respectively), only the most aboral domain showed a robust response to overexpression of NvashA (Fig. 2H; Fig. 4B,J). However, the Nvelav upregulated domain appears to encompass a larger percentage of the aboral half of the embryo (compare Fig. 2H with Fig. 4B,J). Additionally, for NvanthoRFamide, which is predominantly expressed in a more oral domain and is mostly excluded from the aboral region, a much weaker response to NvashA overexpression was observed. Taken together, the overexpression of NvashA phenotypes support the functional significance of the previously described molecular domains along the oral-aboral axis.
NvashA is co-expressed with 242283 and 12533 neural markers
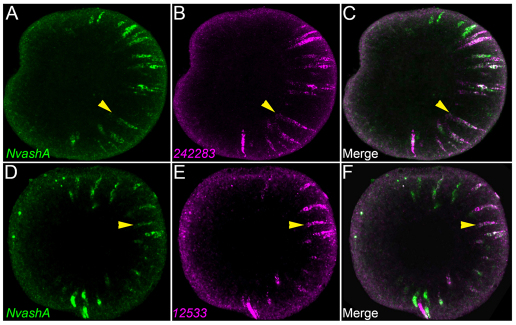
To determine whether NvashA is co-expressed with neural cell markers, we performed double fluorescent mRNA in situ hybridization. We were only able to reproducibly detect the LWamide-like (242283) (Fig. 5B) and the canalicular multispecific organic anion transporter (12533) (Fig. 5D) neural markers when performing double fluorescent detection of neural markers and NvashA. Thus, our analysis is limited to these two neural genes. We detected NvashA co-expression in ∼70% of both the 242283- and the 12533-positive cells (Fig. 5, yellow arrowheads for examples of co-expressing cells). The widespread co-expression of NvashA with 242283 and 12533 suggests that, at least for some neural cell types, NvashA is co-expressed with genes associated with neural differentiation. This supports a role for NvashA in differentiating neural cells.
Fig. 5.
NvashA co-expression with 242283 and 12533 neural markers. (A-C) z-projection (15 μm) from confocal z-series of double fluorescent in situ hybridization showing expression of NvashA (A,C; green) and 242283 (B,C; magenta). In each animal, 69% of cells showed co-expression of NvashA and 242283 (n=10 animals with>50 cells scored per animal). (D-F) z-projection (16.5 μm) from confocal z-series of double fluorescent in situ hybridization showing expression of NvashA (D,F; green) and 12533 (E,F; magenta). In each animal, 64% of cells showed co-expression of NvashA and 12533 (n=6 animals with >30 cells scored per animal). All images are lateral view with oral to the left. Yellow arrows indicate examples of double-positive cells.
NvashB-D overexpression suggests that they are likely to regulate neural gene expression
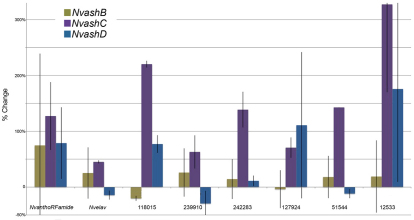
To determine whether the remaining Nvash genes are also likely regulators of cnidarian neurogenesis, in vitro transcribed mRNA for each gene fused to the venus coding sequence was microinjected. Expression of NvanthoRFamide, Nvelav and the six identified NvashA target genes was assayed by qPCR at 24 hpf. NvashB-D showed differential abilities to promote the NvashA target genes. NvashB did not reproducibly induce expression of any of the NvashA target genes as indicated by the large error bars (Fig. 6, olive bars). NvashC was able to positively regulate expression of all NvashA targets in biological replicates (Fig. 6, purple bars). NvashD was able to promote expression of NvanthoRFamide, 118015 and 12533, but does not show reproducible changes to expression levels of the remaining genes (Fig. 6, blue bars). Thus, NvashC and NvashD are able to differentially regulate subsets of the NvashA target genes. We conclude that, based on sufficiency of each NvashC-D genes to promote at least a subset neural gene expression at 24 hpf and expression in neurogenic domains of NvashB-D, each Nvash gene probably has some role in Nematostella neural development.
Fig. 6.
NvashB-D genes also regulate neural target genes. Percent change of gene expression levels for NvashA target genes in 24 hpf animals injected with NvashB:venus (olive bars), NvashC:venus (purple bars), and NvashD:venus (blue bars) compared with control animals. Gene expression was assayed in two biological replicates for each treatment by qPCR. Error bars represent s.e.m.
DISCUSSION
Achaete-scute proneural genes have a conserved role in promoting neural development
Our work demonstrates that manipulating NvashA levels affects the gene expression of the NvanthoRFamide neurotransmitter, the Nvelav neural marker, the post-synaptic glycine receptor (PrtID=127924) and the LWamide-like neural peptide (PrtID=242283), as well as two ATP-binding cassette membrane transporter genes (PrtIDs=118015, 12533) associated with neural expression and function (Gohlke et al., 2008). Additionally, we observe co-expression of NvashA with the 242283 and 12533 neural genes by double fluorescent in situ hybridization, suggesting a role for NvashA in differentiating neural cells. Together, these data indicate that at least one function of NvashA during cnidarian development is promotion of neurogenesis.
The remaining three Nvash genes are not expressed as early as NvashA; however, two sets of data suggest that they also regulate neurogenesis. First, they are all expressed in the developing lateral regions of the directive mesenteries where some adult cnidocyte containing nematosomes originate (Williams, 1975; Williams, 1979). This expression suggests that there might be a link between achaete-scute homolog expression in the developing cnidocytes of adult Hydra (Grens et al., 1995; Lindgens et al., 2004) and the cnidocytes in forming nematosomes of Nematostella (Williams, 1975; Williams, 1979). Also, in animals microinjected with NvashC or NvashD:venus mRNA, a subset of NvashA targets were upregulated (Fig. 6; purple and blue bars). One explanation for the observed variability in the capacity of different Nvash genes to induce NvashA target genes is that there are individual Nvash-specific neural target genes, which is consistent with their differential expression during development. It is not surprising that when there is a shared expression domain, NvashA-D in the oral ectodermal ring, for example, that the NvashA target gene expressed in that domain (NvanthoRFamide in the oral ectoderm) (Marlow et al., 2009) is regulated by multiple family members. However, future studies characterizing each gene individually throughout Nematostella development will be required to verify further the targets of each gene and their putative contributions to cnidarian neurogenesis. Taken together, our functional and expression data and the previously described achaete-scute homolog (cnash) expression in the differentiating cnidocytes (Grens et al., 1995; Lindgens et al., 2004) and in differentiated sensory neurons of adult Hydra vulgaris (Hayakawa et al., 2004) suggest that ash genes represent a conserved component of neurogenesis between cnidarians and bilaterians.
Nvash genes probably only promote development of a subset of the nervous system
It is not clear whether NvashA has a pan-neural role in cnidarian neurogenesis. Although the two definitive known neural markers are regulated by NvashA, it is still unknown how many neurons are formed in the embryonic ectoderm. Other genes known to regulate neurogenesis, such as sox family transcription factors, display ‘salt and pepper’ patterns that are temporally similar to NvashA, but are expressed in many more cells (Magie et al., 2005). Interestingly, one of the putative atonal family proneural transcription factors is expressed in a ‘salt and pepper’ pattern in the Nematostella embryo (data not shown). achaete-scute and atonal genes both regulate generic neural properties but also function to promote distinct neural subtypes (Bertrand et al., 2002; Gohlke et al., 2008). Additionally, neural genes associated with specific neural subtypes (i.e. specific neurotransmitters and neurotransmitter receptors) are downregulated by overexpression of NvashA (data not shown), indicating that NvashA might suppress one neural subtype in favor of promoting another. Lastly, we also observed no regulation of minicollagens, which are expressed in developing cnidocytes in late gastrula stages (Zenkert et al., 2011), in the NvashA overexpression array (data not shown). Together, these data indicate that NvashA does not have a pan-neural role during embryonic development.
Co-factors are likely to be important for providing context to regulate specific neural markers
One interesting observation that comes from this work is the degree to which regional patterning and co-factors could be influencing neural fates in early Nematostella embryos. The broad expression pattern of Nvelav (Fig. 2G) (Marlow et al., 2009), 242283 and 51544 (Fig. 4A and 4I, respectively) argues that neural development is not restricted to any particular domain along the oral-aboral axis. However, we did identify four genes with restricted expression in an aboral domain of the embryo (Fig. 3). Ubiquitous ectopic NvashA did not expand the domain of these targets, but rather increased expression only within their normal expression domains (Figs 2, 3, 4). Moreover, in the case of NvanthoRFamide, at most only a few cells were added to the expression pattern, implying that increased levels occurred within already NvanthoRFamide-positive cells. These data suggest that NvashA requires additional inputs to produce distinct neural cell types in distinct domains of the Nematostella embryonic ectoderm.
The idea of regional neural patterning is not novel for cnidarians as there are examples of regional differences in molecular marker expression in the nerve nets of cnidarians (Galliot et al., 2009; Marlow et al., 2009). However, our data provide the first functional link of a neurogenic transcription factor with a broad expression pattern, which has regionally specific neural target genes. Additional microarray analysis and characterization of target genes for NvashA and other neural promoting transcription factors coupled with analysis of upstream inputs into these pathways in the early embryo are likely to provide a better understanding of the various regional boundaries in the Nematostella embryo. Current data suggests that Nvwnt- (Kusserow et al., 2005), Nvdlx- (Ryan et al., 2007) and Nvfgf- (Matus et al., 2007; Rentzsch et al., 2008) expressing domains might play a role in patterning the oral-aboral axis of Nematostella embryos and, thus, are good candidates for possible factors that regulate distinct neural subtypes domains. Of particular interest is fibroblast growth factor (FGF) signaling because previous work identified it as a key regulator of the aboral domain and larval apical tuft sensory organ (Rentzsch et al., 2008). Additionally, the aboral region of the 24 hpf embryonic ectoderm appears to be more sensitive to ectopic NvashA indicating that this region is more neurogenic at this time. Additionally, FGF signaling and the closely related epidermal growth factor (EGF) signaling provide crucial instructive cues during bilaterian central nervous system development (Stern, 2005; Watanabe et al., 2009). Thus, study of these pathways is interesting for investigating the origins of the bilaterian central nervous systems.
NvashB expression in presumptive endoderm
Expression of NvashB in the endodermal tissue prior to and during the initiation of gastrulation is similar to achaete-scute complex genes expression and function in promoting the formation of interstitial cell and adult midgut precursors during endodermal specification and patterning in Drosophila melanogaster (Tepass and Hartenstein, 1995). Interestingly, the early expression of achaete-scute genes in Drosophila is widespread but the cell types ultimately regulated by achaete-scute in the fly endoderm arise in a ‘salt and pepper’ pattern and not in uniform domains (Tepass and Hartenstein, 1995). Unfortunately, there are no established molecular markers for distinct cell types in Nematostella endomesoderm and further investigations of achaete-scute class proneural gene functions during formation of distinct endodermal fates cannot be carried out in detail at this time. It is interesting to speculate whether the NvashB expression in the presumptive endoderm is the first indication of proneural gene function during Nematostella endodermal neural development or if it represents an ancestral role for proneural genes regulating distinct endodermal cell types.
Conclusion
We have identified a definitive role for, at least, the NvashA gene in regulating cnidarian embryonic ectodermal neurogenesis, which suggests that achaete-scute class proneural genes represent an ancient component of a conserved metazoan neurogenic pathway. These findings provide a foundation for investigating the mechanisms of neural development in cnidarians, the bilaterian sister clade. Future comparisons of neurogenic programs between distinct bilaterian taxa and Nematostella as well as other cnidarians will provide a better understanding of the mechanisms that led to the evolution of central nervous systems. Lastly, the identification and additional study of this conserved component of metazoan neural development in the highly regenerative Nematostella vectensis might provide insight about mechanisms of neural regeneration.
Supplementary Material
Acknowledgments
We thank Dr Robert E. Paull (University of Hawaii, Manoa) and Dr Gernot Presting (University of Hawaii, Manoa) for use of their facilities to carry out the microarray hybridizations and scanning of the microarray slides. We also thank Dr Steven DiNardo (University of Pennsylvania, Philadelphia, PA) for his critical reading and comments on this manuscript.
Footnotes
Funding
This work was made possible by funding from The National Institutes of Health Eunice Kennedy Shriver National Institute of Child Health and Human Development [F32HD055000]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.073221/-/DC1
References
- Bertrand N., Castro D. S., Guillemot F. (2002). Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–530 [DOI] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- Collins A. G., Schuchert P., Marques A. C., Jankowski T., Medina M., Schierwater B. (2006). Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Syst. Biol. 55, 97–115 [DOI] [PubMed] [Google Scholar]
- Dunn C. W., Hejnol A., Matus D. Q., Pang K., Browne W. E., Smith S. A., Seaver E., Rouse G. W., Obst M., Edgecombe G. D., et al. (2008). Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745–749 [DOI] [PubMed] [Google Scholar]
- Galliot B., Quiquand M., Ghila L., de Rosa R., Milijkovic-Licina M., Chera S. (2009). Origins of neurogenesis, a cnidarian view. Dev. Biol. 332, 2–24 [DOI] [PubMed] [Google Scholar]
- Gohlke J. M., Armant O., Parham F. M., Smith M. V., Zimmer C., Castro D. S., Nguyen L., Parker J. S., Gradwohl G., Portier C. J., Guillemot F. (2008). Characterization of the proneural gene regulatory network during mouse telencephalon development. BMC Biol. 6, 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grens A., Mason E., Marsh J. L., Bode H. (1995). Evolutionary conservation of a cell fate specification gene: the Hydra achaete-scute homolog has proneural activity in Drosophila. Development 121, 4027–4035 [DOI] [PubMed] [Google Scholar]
- Hayakawa E., Fujisawa C., Fujisawa T. (2004). Involvement of Hydra achaete-scute gene CnASH in the differentiation pathway of sensory neurons in the tentacles. Dev. Genes Evol. 214, 486–492 [DOI] [PubMed] [Google Scholar]
- Hejnol A., Obst M., Stamatakis A., Ott M., Rouse G. W., Edgecombe G. D., Martinez P., Baguna J., Bailly X., Jondelius U., et al. (2009). Assessing the root of the bilaterian animals with scalable phylogenomic methods. Proc. R. Soc. Biol. Sci. 476, 4261–4270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- Jager M., Chiori R., Alie A., Dayraud C., Queinnec E., Manuel M. (2010). New insights on ctenophore neural anatomy: immunofluorescence study in Pleurobrachia pileus (Muller, 1776). J. Exp. Zool. Mol. Dev. Evol. 314B, 1–17 [DOI] [PubMed] [Google Scholar]
- Kass-Simon G., Scappaticci A. A. (2002). The behavioral and developmental physiology of nematocysts. Can. J. Zool. 80, 1772–1794 [Google Scholar]
- Kunisch M., Haenlin M., Campos-Ortega J. A. (1994). Lateral inhibition mediated by the Drosophila neurogenic gene Delta is enhanced by proneural proteins. Proc. Natl. Acad. Sci. USA 91, 10139–10143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusserow A., Pang K., Sturm C., Hrouda M., Lentfer J., Schmidt H. A., Technau U., von Haeseler A., Hobmayer B., Martindale M. Q., Holstein T. W. (2005). Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433, 156–160 [DOI] [PubMed] [Google Scholar]
- Layden M. J., Meyer N. P., Pang K., Seaver E. C., Martindale M. Q. (2010). Expression and phylogenetic analysis of the zic family in the evolution and development of metazoans. EvoDevo 1, 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgens D., Holstein T. W., Technau U. (2004). Hyzic, the Hydra homolog of the zic/odd-paired gene, is involved in the early specification of the sensory nematocytes. Development 131, 191–201 [DOI] [PubMed] [Google Scholar]
- Lynch J. W. (2004). Molecular structure and function of the glycine receptor chloride channel. Physiol. Rev. 84, 1051–1095 [DOI] [PubMed] [Google Scholar]
- Magie C. R., Pang K., Martindale M. Q. (2005). Genomic inventory and expression of the Sox and Fox genes in the cnidarian Nematostella vectensis. Dev. Genes Evol. 96, 239–257 [DOI] [PubMed] [Google Scholar]
- Marlow H. Q., Srivastava M., Matus D. Q., Rokhsar D., Martindale M. Q. (2009). Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev. Neurobiol. 69, 235–254 [DOI] [PubMed] [Google Scholar]
- Matus D. Q., Thomsen G. H., Martindale M. Q. (2007). FGF signaling and gastrulation and neural development in Nematostella vectensis, an anthozoan cnidarian. Dev. Genes Evol. 217, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam N. H., Srivastava M., Hellsten U., Dirks B., Chapman J., Salamov A., Terry A., Shapiro H., Lindquist E., Kapitonov V. V., et al. (2007). Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94 [DOI] [PubMed] [Google Scholar]
- Rentzsch F., Fritzenwanker J. H., Scholz C. B., Technau U. (2008). FGF signaling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis. Development 135, 1761–1769 [DOI] [PubMed] [Google Scholar]
- Richards G. S., Simionato E., Perron M., Adamska M., Vervoort M., Degnan B. M. (2008). Sponge Genes provide New Insight into the evolutionary origin of the neurogenic circuit. Curr. Biol. 18, 1156–1161 [DOI] [PubMed] [Google Scholar]
- Roffers-Agarwal J., Hutt K., Gammill L. S. (2009). Regulation of neural crest migration by the putative phosphatase, paladin. Dev. Biol. 331, 471 [Google Scholar]
- Ryan J. F., Mazza M. E., Pang K., Matus D. Q., Baxevanis A. D., Martindale M. Q., Finnerty J. R. (2007). Pre-bilaterian origins of the hox cluster and the hox code: evidence for the sea anemone, Nematostella vectensis. PLoS ONE 2, e153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmich J., Rudolf R., Trepel S., Leitz T. (1998). Immunohistochemical studies of GLWamides in cnidaria. Cell Tissue Res. 294, 169–177 [DOI] [PubMed] [Google Scholar]
- Seipel K., Yanze N., Schmid V. (2004). Development and evolutionary aspects of the basic helix-loop-helix transcription factors Atonal-like 1 and Achaete-scute homolog 2 in the jellyfish. Dev. Biol. 269, 331–345 [DOI] [PubMed] [Google Scholar]
- Simionato E., Ledent V., Richards G., Thomas-Chollier M., Kerner P., Coornaert D., Degnan B. M., Vervoort M. (2007). Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol. Biol. 7, 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern C. D. (2005). Neural induction: old problem, new findings, yet more questions. Development 132, 2007–2021 [DOI] [PubMed] [Google Scholar]
- Tepass U., Hartenstein V. (1995). Neurogenic and proneural genes control cell fate specification in the Drosophila endoderm. Development 121, 393–405 [DOI] [PubMed] [Google Scholar]
- Watanabe H., Fujisawa T., Holstein T. W. (2009). Cnidarians and the evolutionary origin of the nervous system. Dev. Growth Differ. 51, 167–183 [DOI] [PubMed] [Google Scholar]
- Williams R. B. (1975). A redescription of the brackish-water sea anemone Nematostella vectensis Stephenson, with an appraisal of congeneric species. J. Nat. Hist. 9, 51–64 [Google Scholar]
- Williams R. B. (1979). Studies on nematosomes of Nematostella vectensis Stephenson (Coelenterata: Actiniaria). J. Nat. Hist. 13, 69–80 [Google Scholar]
- Zenkert C., Takahashi T., Diesner M., Ozbek S. (2011). Morphological and molecular analysis of the Nematostella vectensis cnidom. PLoS ONE 6, e22725 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.