Abstract
Amyloid fibrils are important in diverse cellular functions, feature in many human diseases and have potential applications in nanotechnology. Here we developed methods that combine optical trapping and fluorescent imaging to characterize the forces that govern the integrity of amyloid fibrils formed by a yeast prion protein. A critical advance was to employ the self-templating properties of amyloidogenic proteins to tether prion fibrils, enabling their manipulation in the optical trap. At normal pulling forces the fibrils were impervious to disruption. At much higher forces (up to 250 pN), discontinuities occurred in force-extension traces prior to fibril rupture. Selective amyloid disrupting agents and mutations demonstrated that such discontinuities resulted from the unfolding of individual subdomains. Thus, our results reveal unusually strong non-covalent intermolecular contacts that maintain fibril integrity, even when individual monomers partially unfold and extend fibril length.
Amyloid fibrils are self-associating protein aggregates with a common cross-β-sheet structure 1. Formation of amyloids under normal physiological conditions has long been associated with devastating human diseases, with amyloids sometimes causing pathology 2 and sometimes protecting against it 3,4. More recently it has been appreciated that some amyloidogenic proteins serve an astonishing variety of normal biological functions, including protein scaffolding, environmental adaptation, long-term memory and biofilm formation 5. Moreover, the capacity to acquire an amyloid conformation is an inherent property of most proteins 6, suggesting it is one of the most ancient folds in protein chemistry. These discoveries have produced an explosion of interest in the nature of amyloids but the fibrous nature of these proteins generally prohibits standard structural analyses. Thus, the molecular forces that govern their structures have been difficult to elucidate.
Amongst the amyloidogenic proteins, fibrils formed by the Sup35 prion protein of Saccharomyces cerevisiae are of particular interest because they constitute a protein-based mechanism for the inheritance of biological traits. In its normal soluble state, Sup35 is a translation-termination factor. When it switches into the prion (amyloid-forming) state, less soluble protein is available to function in translation, causing the read-through of nonsense codons. Associated changes in gene expression create a wide variety of novel phenotypes, enhancing survival in diverse environments 5. These phenotypes are heritable by a “protein-only” mechanism: mother cells faithfully pass prion conformers on to their daughters, perpetuating the cycle of conformational conversion.
Intense study of Sup35 prions has already provided many insights. First and foremost, these studies have shown that the basis of prion replication is the ability of prion fibrils to template the conformational conversion of protein in the non-prion conformation into the prion conformation 7–9. The fibril-severing activity of Hsp104 then provides fragments for inheritance in daughter cells 10–13. Further, like most amyloid proteins, Sup35 forms a variety of structurally related but distinct fibril types 13. Biologically these variants lead to phenotypically distinct transmissible prion states that are readily assayed in vivo. When different amyloid variants are taken up by prion-minus yeast cells after cell wall removal (protein-only transformation), they create distinct stably propagating prion phenotypes, or “strains” 14. Amyloid variants that are more easily fragmented generate a greater number of seeds 13 and leave a lower concentration of soluble Sup35 available for translation 15. Thus, somewhat paradoxically, structurally weak fibrils create phenotypically stronger prions.
Here, we develop optical trapping as a technique to manipulate individual prion fibrils and directly characterize their fragmentation. Although optical trapping has provided a wealth of information on DNA-enzyme interactions 16, motor protein movements 17, and protein folding landscapes 18–20, it has not been applied to amyloids because a suitable experimental geometry has not been available. Manipulation of amyloid fibrils has, to date, been achieved only by atomic force spectroscopy (AFM). In these experiments fibrils were laid onto a supporting surface and forces were applied perpendicular to the fibril axis 21–23. To directly assess the molecular forces that maintain fibril integrity, we sought to stretch fibrils parallel to the fibril axis. We also wished to keep them free in solution, to eliminate the complications of surface interactions. By taking advantage of the self-assembly properties of prion fibrils, we devised a method to attach one end of the fibrils to a glass surface and the other end to a polystyrene bead. This allowed us to optically trap and pull on the beads. The advances in our instrumentation allowed for application of much higher forces than previously employed in trapping experiments and for direct fluorescent visualization of fibrils during force application.
RESULTS
Prion seeds produce fluorescently labeled fibrils with uniform biological properties
Sup35 has three distinct regions. The carboxy-terminal region contains the translation termination domain but is completely extraneous to prion behavior per se 8,9. The ability of Sup35 to exist stably and heritably in one of two different states, the prion state or the non-prion state, derives from the divergent properties of the amino-terminal region (N) and the middle region (M) 24. N is extremely rich in glutamine and asparagine residues and drives the protein into the prion amyloid. M is highly charged (lysines and glutamates) and promotes the soluble non-prion state (Fig. 1a). To allow visualization of fibrils in optical trapping experiments, we employed a variant of NM with a single cysteine substitution at amino acid 184. The NM variant was labeled with an alexa-555 fluorophore and mixed with an equal concentration of wild-type NM for polymerization. This substitution is in the M domain and lies outside the amyloid fibril core 25–27, having no effect on fibril formation 25, 28.
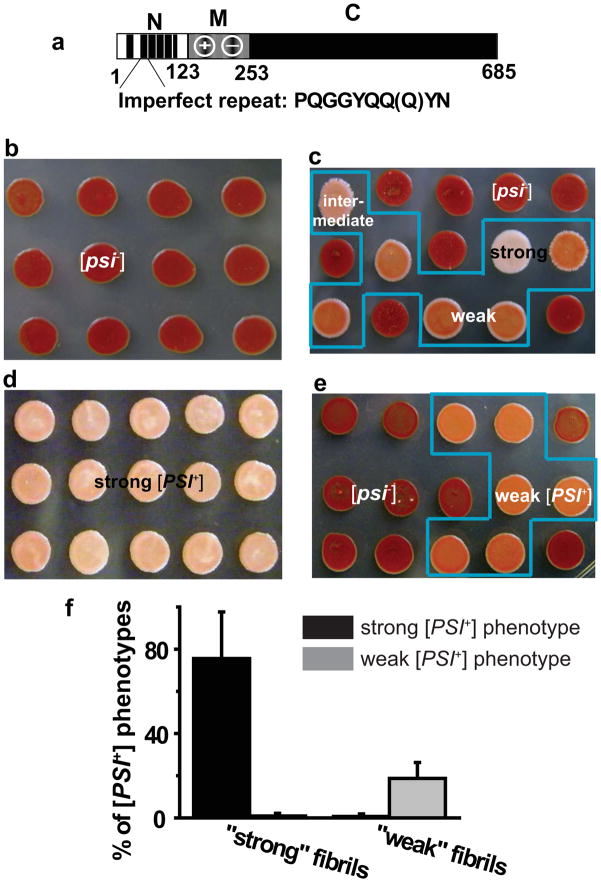
Figure 1.
The biological homogeneity of NM fibrils was confirmed by protein-only transformation. (a) Diagrams of wild-type Sup35. The black segments in the N-terminal region of Sup35 (N) represent oligopeptide repeats. The highly charged nature of the middle region (M) is indicated. The C-terminal region (C) functions in translation termination. (b–f) Biologically distinct prion states were induced when prion-minus cells were transformed with fluorescently labeled fibrils formed under different conditions. (b) Colonies transformed with buffer alone. (c) Colonies transformed with spontaneously assembled NM fibrils. A mixture of prion phenotypes is highlighted in the blue box. (d) Colonies after “protein-only” transformation at 4°C with NM fibrils that were seeded by lysates of yeast cells that carried a strong prion element. The uniform strong prion phenotypes are indicated by the light pink-colored colonies in the blue box. (e) Colonies after “protein-only” transformation at 37°C with NM fibrils that were seeded by lysates from cells carrying a weak prion element. The uniform weak prion phenotypes are indicated by the dark salmon-colored colonies in the blue box. (f) Quantification of transformation efficiency and specificity of distinct prion fibrils. Strong fibrils refer to the fibrils used in (d), while weak fibrils refer to the fibrils used in (e).
Initially, the proteins were allowed to assemble into amyloid spontaneously. However, such fibrils produced a spectrum of prion strains with different levels of nonsense suppression when they were assayed by protein-only transformation of prion-minus cells (Fig. 1b and c). To ensure that fibrils were both biologically homogenous and of clear relevance to the in vivo prion state, we seeded polymerization with lysates of cells carrying phenotypically strong or weak prion elements. For strong prions, polymerization was performed at 4°C, a condition which itself favors assembly of amyloids that confer the “strong” prion phenotype 14. Indeed, the resulting fibrils produced a uniform and strong nonsense suppression phenotype when analyzed by protein-only transformation (Fig. 1d and f). For weak prion elements, polymerization reactions were seeded at 37°C, which favors assembly of amyloids conferring a “weak” prion phenotype. When assayed by protein-only transformation of prion-minus cells, these fibrils produced only weak biological phenotypes (Fig. 1e and f). Thus, these procedures resulted in the preparation of fluorescently labeled protein fibrils that carried true prion states, but of single subtypes.
Prion assembly properties provide a strategy for optical manipulation
To manipulate single fibrils with the optical trap, we attempted to attach one end of the fibrils to a glass surface and the other end to a polystyrene bead that would provide “handles” for optical pulling experiments stretching the fibrils from their ends. Several strategies reported in the literature for the attachment of other proteins for optical trapping experiments were tested but all failed to achieve the desired geometries 18,19,29. As an alternative, we sought to take advantage of a quintessential property of yeast prions, their ability to serve as a template for the conformational conversion of solublemonomers. This recruitment of monomers occurs exclusively at fibril ends 30,31. We asked if we could invert this process and employ unstructured monomers bound to surfaces to recruit and capture preformed fibrils from solution.
NM monomers were adsorbed onto the glass surface of a flow chamber, with the expectation that the highly charged glass surface would interact with the highly charged M domain, leaving the amyloidogenic N domain free to interact with fibrils (Fig. 2a, Supplementary methods). The remaining exposed glass surface was then blocked against non-specific protein interactions with a solution of casein. Next, fluorescently labeled NM fibrils were introduced to the chamber for capture by the pre-affixed monomers. Finally, polystyrene beads (0.8 μm), which had themselves been pre-bound with NM monomers, were introduced for recruitment to the remaining free ends of the fibrils.
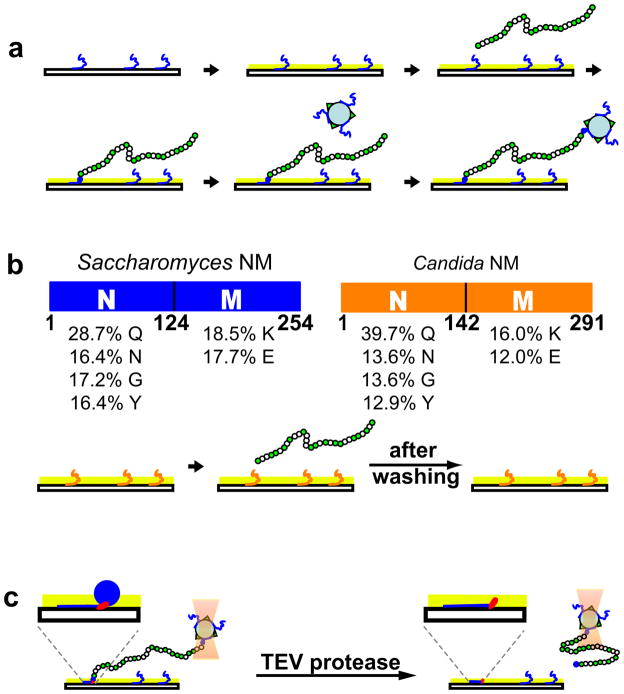
Figure 2.
NM monomers effectively and specifically bind individual NM fibrils to a glass surface on one end and a polystyrene bead on the other. (a) Schematic diagram of the method for the attachment of NM fibrils. Monomers (blue) are adhered to the glass surface and the remaining surface is coated with casein (light green). Green circles, fluorescently labeled NM monomers; white circles, wild-type NM monomers. Polystyrene beads (light blue circles) labeled with Alexa488 (green triangle) and Alexa555-NM (blue) are introduced and captured by the other free ends of the fibrils. (b) Top, The molecular architectures and amino-acid compositions of Saccharomyces cerevisiae NM (blue) and Candida albicans NM (orange). Bottom, schematic illustration of the experiment demonstrating the specificity between pre-bound monomers and fibrils. From left to right, deposition of Candida NM on the glass surface; incubation of Saccharomyces NM fibrils in the flow channel; imaged after washing. (c) Schematic diagram of the detachment of NM fibrils from the glass surface with a treatment of TEV protease. The TEV recognition site is shown in red and highlighted in the inserts. The M domain (blue line) binds with the glass surface and the N domain (blue circle) interacts with the end of NM fibrils. The bead was held in position by the optical trapping (light pink).
We employed fluorescent imaging to visualize the fibrils in order to optimize conditions and validate the nature of their attachments (Supplementary movie 1 and 2; see methods for experimental details). Casein proved very effective in preventing non-specific interactions between prion fibrils and the glass surface. The self-recognition properties of pre-attached monomers were necessary and sufficient to tether one end of the fibrils to the glass surface and capture the other end with the polystyrene bead.
To determine if NM monomers captured the NM fibrils via bona fide prion interactions, we took advantage of the “prion species barrier”. This barrier inhibits prion transmission between Sup35 proteins that are derived from evolutionarily divergent protein sequences 32. We asked if the NM domain from the Candida albicans Sup35 protein could capture the NM fibrils of Saccharomyces cerevisiae. The Candida protein has the same highly unusual amino acid composition as the Saccharomyces protein, rich in asparagine and glutamine residues (Fig. 2b), but the sequence is distinct and the two prions do not cross seed each other 32. When Candida monomers were pre-deposited on the glass surface followed by casein blocking, fibrils of the Saccharomyces protein were very rarely tethered to the surface (Fig. 2b and Supplementary movie 3). Thus, the attachment between the surface-bound NM monomers and NM fibril ends exhibits the species-specific interactions characteristic of the bona fide prion fold.
The M domain of the capturing monomer provides the anchor for fibril attachment
To determine if the M domain of the individual monomers pre-attached to the surface was solely responsible for anchoring the fibrils to the glass surface, we engineered NM monomers to carry a TEV protease cleavage site (ENLYFQG) at the junction between N and M (aa 128–134). These monomers, pre-bound to the glass surface, were used to recruit fibrils that were entirely composed of NM proteins that did not contain a TEV site. We captured the fibril-tethered bead in the trap and then introduced TEV protease. This released the entire fibril from the glass surface (Fig. 2c and Supplementary movie 4). We conclude that, except for the M domain of the single anchoring subunit, no interactions exist between the glass surface and the prion fibrils. Thus, individual prion monomers can function as a highly specific amyloid “glue”, with the M domain providing an anchor to the surface and the N domain capturing a preformed fibril at its end.
Prion fibrils are remarkably resistant to disruption by mechanical forces
We first pulled fibrils that create phenotypically strong prions in the optical trapping experiments, since, as discussed in the introduction, they are structurally weaker and more likely to be fragmented. We pulled fibrils with forces up to ~100 pN, which is sufficient to unfold many globular proteins at low loading rates typically achieved in optical trapping force spectroscopy (1–100 pN/s) 18–20, 33. (Note: the non-equilibrium nature of mechanically unfolding proteins is strongly dependent on loading rate. Unfolding proteins at higher loading rates, such as with an AFM (~104–106 pN/s), generally requires larger force magnitudes, 100–500pN.) Fibril tethers were subjected to force-extension measurements by focusing the laser beam on the beads to trap them and then moving the flow chamber with attached NM fibrils on a piezoelectric stage with nanometer resolution (Supplementary movie 5). Individual NM fibrils were first centered directly above the surface attachment point and then pulled from the center position to full extension (stretching phase). After being held at the extended position at high force for a prolonged period (holding phase–40–80s), the fibrils were finally relaxed (relaxation phase) back to the center position. The extension-relaxation had no effect on NM fibrils as the curves were overlapped (data not shown).
To achieve much higher forces, we used an optical trapping instrument that was configured to include a second stationary trap (Fig. 3a). A lower force primary branch was guided through acousto-optic deflectors to enable precise calibration of the bead’s position 34. The second stationary branch, which had minimal optical components to reduce power loss, was then switched on to perform the pulling experiments. This allowed us to achieve forces of up to 250 pN, employing the same typical loading rate for optical trap experiments (Fig. 3b and c; Supplementary methods). Again, pulling had no effect on fibril integrity (Fig. 3c and d). Force-extension curves from the stretching and relaxation cycles were superimposable (Fig. 3d, left), even when individual fibrils were subjected to multiple stretching cycles (Fig. 3d, right), indicating that no rupture or permanent deformation had occurred. Thus, although NM fibrils are assembled via non-covalent monomer interactions, they are remarkably resistant to disruption by mechanical forces.
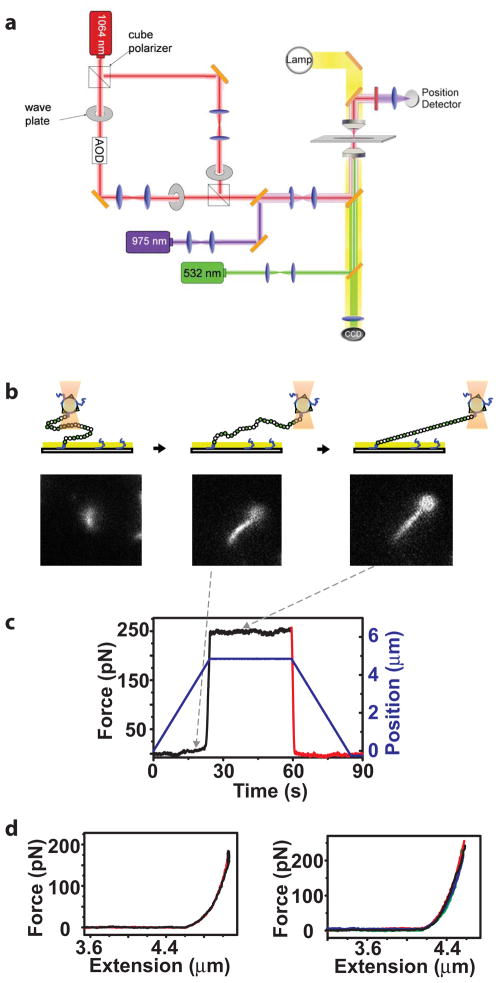
Figure 3.
Stretching NM fibrils by optical trapping at high forces with simultaneous fluorescent imaging. (a) Schematic of the trapping instrument with dual trapping capabilities used for high force experiments (see Methods for details). 1024, nm, trapping laser; 975 nm, position detection laser; 532 nm, fluorescence imaging. (b) Schematic representation and fluorescence snapshots of the experiments. From left to right, the bead was first centered on top of the attachment point, and then the stage was moved along the x-axis until the fibril was fully extended. (c) Representative pulling and relaxation traces for NM fibrils in the presence of normal assembly buffer. The force and the position of the stage were plotted as a function of time. Black line, the stretching and holding phase; red line, the relaxation phase; blue line, position of the bead relative to the attachment point of the tether on the glass surface. (d) Left, force applied vs. fibril extension length (force-extension curve) for the tethered fibril experiencing one stretching-relaxation cycle. Black line, stretching and holding phase; red line, relaxation phase. The x-axis is expanded to illustrate the behavior in the fully extended position. Right, the force-extension curves for a single tethered fibril that experienced four sequential stretching-relaxation cycles superimpose upon each other. For clarity, the holding phase is not shown. Stretching and relaxation curves from the same cycle are shown in the same color, with subsequent curves in a different color.
Complex pulling traces are produced with more extreme conditions
To increase the probability of detecting rupture events, we repeated fibril stretching in the presence of guanidine hydrochloride (GdHCl), a chaotropic reagent that decreases protein stability 25,35. NM fibrils with a strong prion phenotype unfold in the presence of GdHCl at D1/2~1.5 M 25. Under the highest pulling forces we could achieve (250 pN), 0.4 and 0.8 M GdHCl had no effect. To our surprise, at 1.2 M GdHCl, instead of simple rupture most pulling trials yielded complex traces characterized by sudden force-dropping events. These occurred during both the stretching and holding phases. Several examples are shown in Fig. 4a, with force-dropping events indicated by the black arrows and final rupture indicated by the red arrows. Most fibrils could be subjected to multiple cycles of stretching and relaxation, producing many associated force-dropping events before rupture (two examples in Fig. 4b). In total, we recorded 50 stretching-relaxation cycles from 15 tethered NM fibrils and observed 116 force-dropping events prior to rupture.
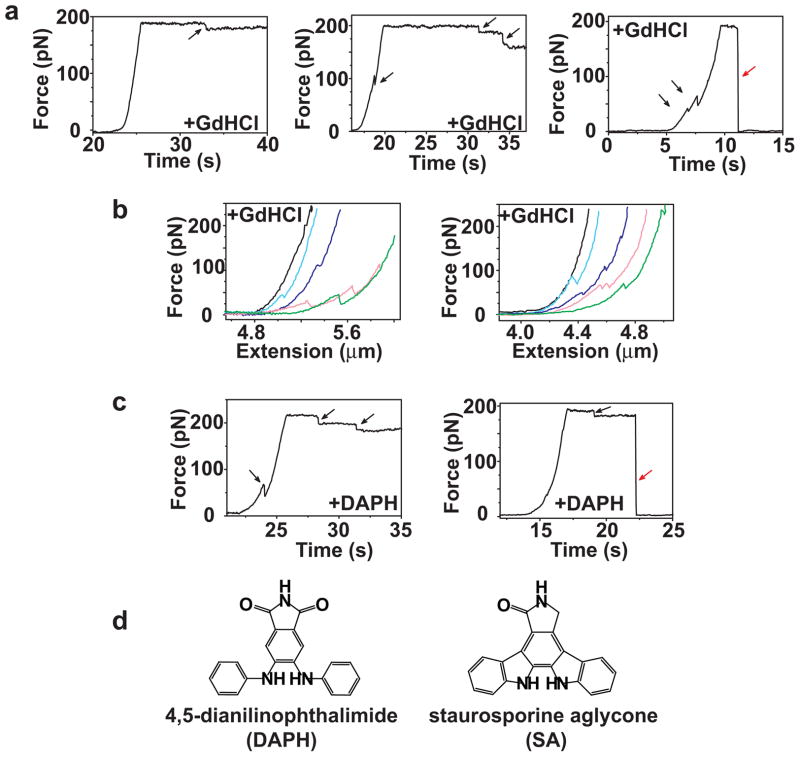
Figure 4.
Representative pulling traces (force vs. time) for NM fibrils under different experimental condition. (a) Force-extension curves of NM fibrils in the presence of 1.2 M GdHCl. Black arrows highlight sudden force changes in the stretching and holding traces. Red arrows mark rupture of the tethered fibril with a concomitant drop to zero force. (b) Representative force-extension curves for two NM fibrils that experienced multiple stretching-holding-relaxation cycles in the presence of 1.2 M GdHCl. Black, light blue, blue, light pink and green correspond to cycles 1–5 for each fibril. For clarity, only stretching phases are shown. (c) Force-extension curves in the presence of 50 μM DAPH. (d) The structures of DAPH and SA.
When individual fibrils were subjected to multiple stretching-relaxation cycles, the stretching curves from subsequent cycles were not superimposable (Fig. 4b), in contrast to those produced in the absence of GdHCl (Fig. 3d, right). This was because discontinuities in force curves increased fibril length. These length extensions were generally retained in the relaxation phase, likely because the high concentration of GdHCl prevented the protein from returning to its pre-existing state (see below and Supplementary Fig. 1).
Excluding artificial origins of the force-dropping events
As occurred with other proteins, they can form branched or kinked amyloid fibrils 36, and such geometries could give rise to complex mechanical behavior. The use of simultaneous fluorescent imaging in our optical trapping experiments allowed us to eliminate this concern. We were able to focus exclusively on uniform, unbranched fibrils with a single swivel point of attachment to the glass surface. It is possible, however, fibrils adhere to the glass surface at multiple points that are too close to the surface to be resolved by fluorescence. If so, abrupt elongations and sudden force drops would then be due to these attachments sequentially being “peeled” from the surface. The ability of TEV to release the entire fibril from the surface when the only molecule containing a TEV site was the anchoring monomer (see above) made this extremely unlikely, but it was possible that the addition of GdHCl allowed additional attachments to form. We found, however, 1.2 M GdHCl prevented pre-deposited NM monomers from recruiting NM fibrils to the glass surface and very rarely produced additional points of attachment (Supplementary Fig. 2).
Another explanation for the force discontinuities is that they reflect sequential release of small segments of the M domain from the individual anchoring monomer. This seemed extremely unlikely because the total length extensions we obtained were often much longer than an individual M domain (see below). To investigate this possibility more directly, we visualized the position of rupture by fluorescence imaging. If the sequential force dropping events were due to the release of anchoring attachments, rupture events would simply represent the final release of the last anchoring M domain segment. To visualize the position of fibril rupture, we trapped the bead with fibrils in the extended position and manually aligned the fibril with the x-axis of the piezo stage without the prolonged process of centering. Fluorescent images could then be stably recorded before and after rupture. (Without the prolonged process of centering, photobleaching that occurs due to simultaneous exposure to the trapping and visualizing laser beams were minimized, allowing direct visualization.) In the recorded movies (Supplementary movie 6), fragments representing various fractions of the initial fibril could be seen still attached to the surface after rupture. Thus, rupture was indeed not due to detachment of the anchoring M domain from the glass surface (or, for that matter, of the streptavidin linkage to the bead), but to severing of the fibril itself.
The complex pulling traces reflect intrinsic properties of prion fibrils
Do discontinuities in the pulling traces derive from features integral to the structural and biological properties of the prion fibrils? We employed several different types of experiment to investigate this question. First, we pulled fibrils in the presence of DAPH (4,5-dianilinophthalimide), rather than GdHCl. DAPH is a highly specific disrupter of particular types of amyloid fibrils and, in particular, disrupts molecular contacts in the N domain of NM fibrils. It does not affect other types of protein folds 37. We recorded 51 stretching-relaxation cycles from 24 tethered NM fibrils in the presence of 50 μM DAPH and observed 146 force-dropping events prior to rupture. These were similar in character to those observed in 1.2 M GdHCl (Fig. 4c).
We then performed the experiments in the presence of staurosporine aglycone (SA). SA is a very close analog of DAPH, containing only two extra carbon–carbon bonds that impose planarity on the structure (Fig. 4d). In contrast to DAPH, however, SA does not dissociate NM fibrils and has no effect on the amyloid fold 37. In the presence of SA, we observed no discontinuities in force extension curves. Thus, the force-dropping events that occurred in the presence of DAPH were specifically due to intrinsic structural properties of amyloid fold.
Second, we stretched an amyloid variant made from the same NM protein, but embodying a weak prion element. As previously described 13, such fibrils are more resistant to chemical denaturation and more resistant to physical fragmentation by agitation. When stretched by optical pulling in the presence of DAPH, only two isolated force-dropping events were observed in 76 stretching-relaxation cycles with 16 fibrils. The two fibrils that experienced such an event were stretched multiple times thereafter with no further drops in force. These “weak phenotype” NM fibrils also resisted unfolding in GdHCl at the highest technically feasible concentration (1.4 M; further increases in GdHCl caused the polystyrene beads to stick to the glass surface).
Mutations in the repeat region dramatically change mechanical behaviors of fibrils
Force dropping events were induced by the specific amyloid disrupting agent DAPH (but not by SA) and were profoundly altered when fibrils embodied a different prion subtype (with the same M domain). Taken together, these results establish that discontinuities in the force-extension curves reflect intrinsic structural and biological properties of the prion fibrils and are not due to extrinsic surface attachments. We therefore considered a rather remarkable explanation: individual subdomains of the fibrils have the capacity to sequentially unfold under mechanical force, while non-covalent intermolecular contacts maintain fibril integrity.
If so, which segment of the protein might unfold to produce fibril-lengthening events? Although the atomic level structure of NM fibrils is unknown, much evidence indicates that the M domain is not involved in the amyloid core 25–27. Indeed, we found that the M domain was readily cleaved from the fibrils and released as soluble fragments. The cleaved N-domain-only fibrils maintained their fibril integrity as confirmed by electron microscopy. Further, they had the same biochemical properties as wild-type NM fibrils (Supplementary Fig. 3).
Since the N domain is responsible for fibril integrity, how might force-dropping unfolding events in this domain be associated with fibril lengthening? In a previously proposed β-helix model for NM fibrils 25,38, a continuous segment of the N domain forms the amyloid core, with the termini from neighboring monomers in self-contact, head-to-head and tail-to-tail (Fig. 5a). The β-helical region between them, largely composed of the repeat region, is sequestered from intermolecular interactions 38. In such a structure, the repeat sequences might successively unfold, increasing the length of the fibril, while the integrity of the fibrils is maintained by head-to-head and tail-to-tail contacts.
Figure 5.
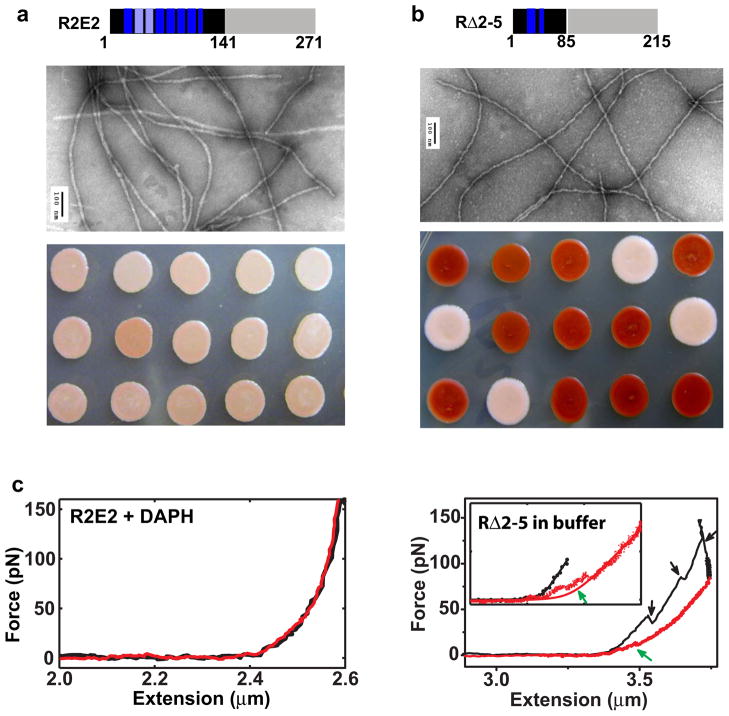
Expansion or deletion of the repeat region dramatically changed mechanical behavior of prion fibrils. (a) A previously proposed β-helix model for NM fibrils. The M domain is not involved in the amyloid core and is not shown. The color scheme for the N domain is demonstrated on the top left. An expansion of two monomers is shown on the top right. (b) and (c), top, schematic illustration of R2E2 and RΔ2-5. The blue segments in the N-terminal region represent oligopeptide repeats. Middle, R2E2 (left) and RΔ2-5 (right) fibrils imaged by transmission electron microscopy. Scale bar, 100 nm. The R2E2 fibrils have an average diameter of 16 ± 2 nm, slightly wider than that of wild-type “strong” NM fibrils, 12±2 nm. The RΔ2-5 fibrils have a diameter comparable to wild-type fibrils, but many showed a distinct wavy morphology. Bottom, protein-only transformation of R2E2 (left) and RΔ2-5 (right) fibrils. (c) Representative force-extension curves for R2E2 (left) and RΔ2-5 (right) fibrils. The black traces, stretching phase; the red traces, relaxation phase. Black arrows, force-dropping events in the stretching curves; green arrows, force plateaus in the relaxation curves at low forces. Insert, expansion at the low force regime. The stretching and relaxation curves were fit to a modified WLC model, shown as black and red curves, respectively, in the inserts. The deviation of the relaxation curves from the WLC fitting is highlighted by the green arrow.
To investigate how the repeat region influences the mechanical behavior of prion fibrils, we stretched fibrils assembled from NM protein carrying two additional repeats, R2E2 39. We first determined that R2E2 fibrils seeded by cellular wild-type prion seeds uniformly produced strong or weak prion phenotypes (depending upon conditions described above) when assayed by protein-only transformation (Fig. 5b and Supplementary Fig. 4). This established that R2E2 proteins could faithfully receive strain-specific templating information from wild-type protein and transmit that information back to wild-type protein. In the presence of 50 μM DAPH, we stretched eleven R2E2 fibrils of the strong prion subtype with 5 stretching-relaxation cycles for each. All fibrils remained intact without unfolding or rupture, except for one fibril that ruptured without prior unfolding. The elimination of unfolding events by expansion of the repeat region, when the intermolecular contacts that define prion strains remain intact, supports the repeat region as the origin of unfolding.
Since expansion of the repeat region added mechanical stability, we asked if deleting this region might destabilize fibrils sufficiently to achieve unfolding and rupture in the absence of denaturant. We employed an NM protein carrying a deletion of repeats two through five, RΔ2-5. As confirmed by protein only transformation, this protein faithfully received strain-specific templating information from the wild-type protein and transmitted it back to wild-type protein (Fig. 5c and Supplementary Fig. 4). Thus, for this protein, too, the strain-determining intermolecular contacts remained unaltered. Next, we stretched NM fibrils carrying the RΔ2-5 deletion in the optical trap. Under the same conditions used for the other fibril variants, unfolding and rupture occurred under physiological conditions, without the need for GdHCl or DAPH (Fig. 5d, black curves). We also observed plateaus in the relaxation curves at very low forces (red curve, Fig 5d) that suggested refolding could occur in the absence of denaturant (Supplementary Fig. 1). In contrast to wild-type fibrils, which sustained many unfolding events before rupture (Fig. 4b), many RΔ2-5 fibrils ruptured after one unfolding event and no fibrils were able to sustain more than three events without rupturing. Again, the profound changes in the mechanical behavior of the prion fibrils when the repeat region was contracted support this region as the origin of unfolding.
Characterization of length extensions and rupture events
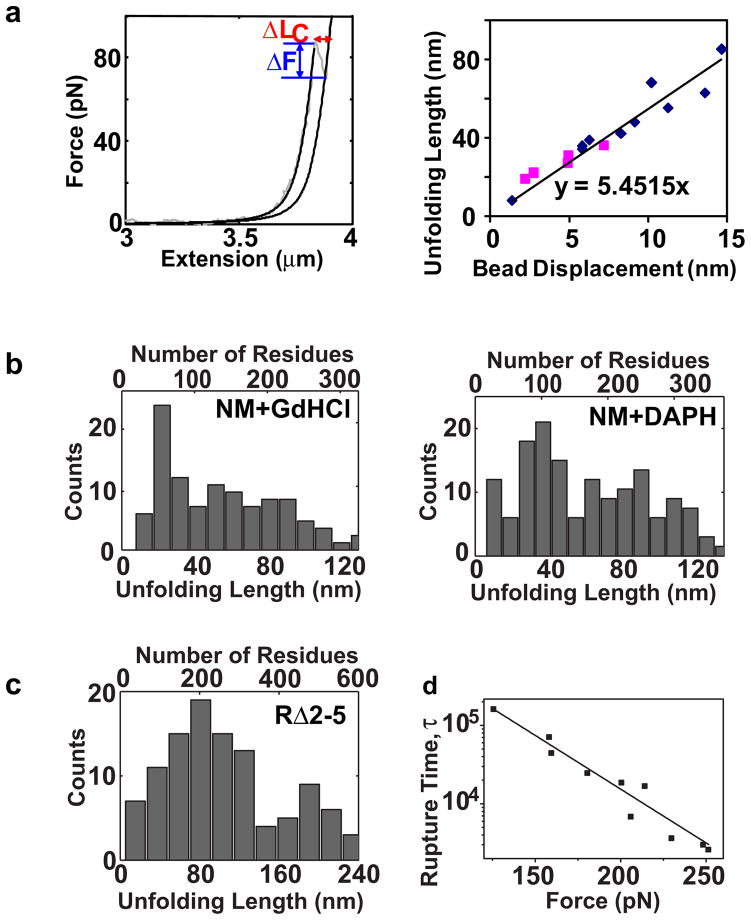
In order to calculate the length of extension (additional contour length added to the fibril as a result of unfolding, ΔLc), the force-extension curves with force-dropping events in the enthalpic stretching phase were fit with a modified WLC model to determine the contour lengths (Lc) before and after unfolding (Fig. 6a, left). A linear correlation was obtained between the length of extension of individual force-dropping events and the instantaneous bead displacement that occurred upon unfolding (determined by dividing the change in force by the trap stiffness; Fig. 6a, right). This provided a calibration factor to calculate the extension length of each force-dropping event occurring during the holding phase based on the bead displacements obtained from the experiments.
Figure 6.
The distribution of the length of extension is broad. (a) Left, An example of WLC fits used to determine the length added to the fibril at each force-dropping event that occurred during the stretching phase. Right, a linear correlation was obtained between the length of extension determined from WLC fits and the instantaneous bead displacement that occurred upon the unfolding. Black Points, length of extension from WLC fits to wild-type NM fibrils; gray points, length of extension from WLC fits to RΔ2-5 fibrils. (b) Histogram of the length of extension for NM fibrils at each force- dropping event in the presence of 1.2 M GdHCl (left) or 50 μM DAPH (right). (c) Histogram of the extension lengths obtained with RΔ2-5 fibrils. A 12nm and a 24nm bin width were used for (b) and (c) respectively (Supplementary methods). (d) Rupture time vs. force for the RΔ2-5 fibrils. The rupture events that occurred during the holding phase were fit to a Bell model as described in the methods.
In the presence of GdHCl or DAPH, the extension of the unfolding events for NM fibrils from both the stretching and holding phases revealed a broad distribution (Fig. 6b). Such a distribution contrasts with the unfolding of protein polymers formed by multiple covalently linked globular domains strung together end to end with a flexible linker between them, which often show discrete and precise extension lengths by unfolding well-defined domains 40–42. The smallest population of extensions was approximately 15 to 20 nm, roughly matching the length of repeat region in the N domain (~9 residues per repeat × 0.34 to 0.4 nm per residue × 5 repeats = 15.5 to 18 nm). Individual extensions that are longer than the total length of the repeat region presumably derive from near simultaneous unfolding of multiple repeat regions. The extensions of RΔ2-5 fibrils were much larger than with wild-type NM, often longer than the calculated length of the entire protein (Fig. 6c). They must also represent events in multiple amyloid monomers along the length of the fibril occurring virtually simultaneously.
Unfolding and rupture events with RΔ2-5 fibrils were not complicated by the presence of denaturants. This allowed us to use data obtained in the constant force regime (holding phase) to assess the strength of the intermolecular interactions that maintain fibril integrity and are broken upon rupture. The lifetime of an interaction, τ, under a constant applied force, F, can be described by Bell’s equation, τ (F) = τ0 exp(−βFx‡) where τ0 is the intrinsic lifetime under no load, β−1 = kBT 43, and x‡ is the characteristic distance along the reaction coordinate required to achieve rupture (transition distance) (Supplementary methods). The lifetime-prior-to-rupture showed a clear relationship to applied force (τ0 = 99±28 days and x‡= 0.13±0.01 nm; Fig. 6d). This indicates that the intermolecular contacts have extremely high mechanical stability.
DISCUSSION
Unexpected behaviors of prion fibrils under mechanical forces
We have described a method to tether amyloid fibrils to a glass surface on one end and a polystyrene bead on the other, using monomers of the same protein as a remarkably strong and highly specific molecular glue. This allowed us to apply optical trapping with simultaneous fluorescence imaging, a technique that has never been reported for any amyloid, to characterize the forces that govern the integrity of prion fibrils. Under unusually high pulling forces, we discovered unexpected properties of prion fibrils. First, multiple force dropping events were observed prior to fibril rupture. These force-dropping events 1) occurred in the presence of a highly selective amyloid-disrupting agent but not in the presence of the inactive analog; 2) occurred with phenotypically strong prion fibrils but not phenotypically weak prions; and 3) were highly sensitive to mutations in the N domain, in proteins that carried the same strain-specific prion determining monomer: monomer interface and identical M domains. These results collectively establish that the force-dropping events reflect properties intrinsic to the prion fold and are not a result of extrinsic aspects of surface attachments. Moreover, they establish that fibrils of a single prion type contain two regions with distinct mechanical stabilities. Unfolding, presumably of the repeat region, results in increased fibril length. Fibrils can sustain multiple unfolding events while maintaining fibril integrity, until mechanical failure of the stronger region causes rupture. These properties were completely unexpected because these prion fibrils exhibit simple cooperative unfolding kinetics by chemical and thermal denaturation 14,25.
Second, a broad distribution of unfolding lengths was observed for prion fibrils. This is in contrast to the unfolding of protein polymers, which have multiple identical well-folded globular domains linked with flexible peptide linkers. When individual domains of such polymers unfold, discrete extension lengths are observed 44–46. In contrast, in our prion polymers, a continuous amyloid core runs the entire length of the fibril joining thousands of molecules, via both inter and intermolecular interactions, into a rigid, highly cooperative structure 7,25,47–49. The mechanical pulling forces is likely distributed to individual monomers in a very different way than it would be when flexible linkers intervene between folded domains, making it more likely for simultaneous unfolding events to occur. Further, the amyloidogenic N domain of the prion protein displays low sequence complexity, is extremely Gln/Asn rich, and contains multiple tandem nine-mer repeats. We speculate that these sequence features may allow slightly different lengths of the repeat region to unfold in each event, cumulatively resulting in a broad distribution of fibril-lengthening events. This may also explain the different extension-length profiles observed with DAPH and GdHCl, since these compounds likely destabilize prion fibrils in somewhat different ways. And finally, although we have made substantial efforts to generate uniform fibrils with biologically pure phenotypes, it is entirely possible that fibrils are not completely homogeneous, but instead contain local structural fluctuations and imperfections.
Implications for the structure of prion fibrils
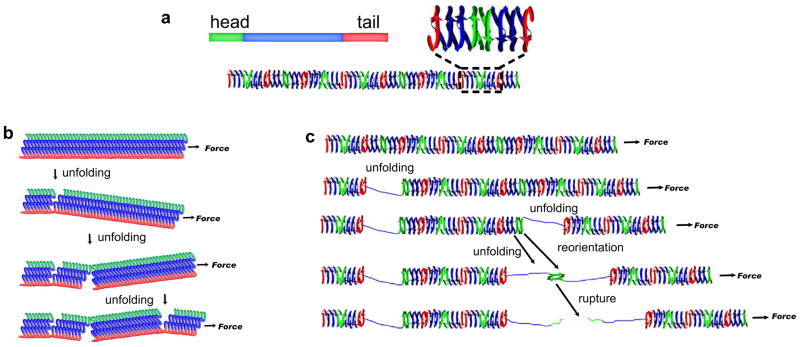
The molecular structure of NM fibrils has not been determined, but two general models have been proposed 25,50–52, a β-pleated sheet model and a β-helix model (Figs. 5a and 7). Our observations cannot be reconciled with the β-pleated sheet model. In that model, each monomer is stacked on top of the next along its entire length, one monomer per rung and each amino acid on top of the same amino acid in the adjacent monomers (Fig. 7a) 52. To achieve an increase in fibril length in such an arrangement, both termini of the interfaces have to split open, alternating from side to side, with each split extending more than half of the fibril width but not so far as to cause rupture (Fig. 7a). This would require that the two termini of the interface have very similar mechanical integrities. But then the entire interface should dissociate and rupture the fibril immediately.
Figure 7.
Cartoon representation of the mechanical unfolding of NM amyloid fibrils with different structures. (a) A possible mechanism with the β-pleated sheet model. (b) A possible mechanism with the β-helix model. See discussion for details.
In contrast, the β-helix model does provide a framework for interpreting our data (Figs. 5a and 7b). First, in this model the repeat region is largely sequestered from intermolecular contacts. Multiple repeat regions could unfold, with fibril length increasing at each event, while fibril integrity is maintained by intermolecular contacts. Indeed, mutations of the repeat region dramatically alter the unfolding behavior. Fibrils with repeat expansions are mechanically stronger and fibrils with repeat deletions are mechanically weaker. This suggests that repeat expansions and deletions do not simply add or remove additional identical β-strands in the helix but slightly change the molecular geometry in some way. This is consistent with the fact that R2E2 and RΔ2-5 fibrils showed different diameters and morphologies from wild-type fibrils (Fig. 5b and c).
Second, in the β-helix model, the termini from adjacent monomers contact each other in a “head-to-head” and “tail-to-tail” manner (Fig. 5a) 25. Simple geometry dictates that the β-strands at monomer interfaces are antiparallel (inset, Fig. 5a), while the β-helical strands within are parallel 38. Antiparallel and parallel and β-strands might easily have somewhat different mechanical stabilities, constituting the mechanically stronger and mechanically weaker domains we observe. But even more interestingly, if two neighboring repeat regions were to unfold, it would cause the intermolecular contact region between them to reorient relative to the pulling force (from perpendicular to parallel; Fig. 7b) 53. Breaking this interface would then necessitate the shearing of β-strands. The simultaneous shearing of all of the hydrogen bonds requires much stronger mechanical forces than the simple stepwise unzipping of strands in a β-helix 54,55. Thus, even if the stabilities of inter- and intra-molecular contacts are rather similar under the influence of chemical denaturants, under the influence of mechanical forces unfolding of the intramolecular domains would actually strengthen the intermolecular contact through reorientation to a geometry requiring simultaneous shearing of the entire interface. Supporting this concept, computational simulation of β-helical proteins has predicted a reorientation of the β-strands after partial unfolding 53. Moreover, the distance along the reaction coordinate required to achieve rupture of our prion fibrils (x‡ of 0.13 nm) is very similar to previously determined β-strand shearing transitions determined by optical trapping for globular proteins of known structure 55.
The same basic unfolding schema might be obtained with NM fibrils composed of more complex protein arrangements, which have been suggested for several other amyloidogenic proteins. For example, a domain-swapped trimeric β-helix model has been proposed for the mammalian prion protein 56, which presents additional inter-molecular contacts and more intrinsic entanglements between the monomers. A cryo-electron microscopy study suggested that β-(2)-microglobulin fibrils consist of multiple β-sheet-rich globular subunits with multiple hierarchical intra- and inter-subunit contacts 57. In any case, whether NM fibrils have a structure that builds upon an underlying β-helix or not, our data establish that there must be distinct subdomains, which can sustain multiple lengthening events while a mechanically stronger monomer: monomer interface maintains fibril integrity.
Applications of the new methodology
The methods we have developed should provide a broadly applicable tool for investigating many interesting questions in the amyloid field. The mechanically resistant prion fibrils we described are fragmented in the yeast cytoplasm by the protein-remodeling factor Hsp104 12,58. The fibril-severing activity of Hsp104, providing fragments for inheritance in daughter cells, is key to the extraordinary ability of prions to serve as protein-based elements of inheritance 13. Our methods offer an opportunity to investigate the mechanism by which Hsp104 deploys 12 ATP sites to convert chemical activity into mechanical rupture of prion fibrils. Moreover, our method for tethering amyloid fibrils should also be applicable to investigating forces that govern the integrity of other amyloids, since most such proteins template polymerization by recruiting soluble monomers at their ends, acting as strong and specific “molecular glue”. Several functional amyloids whose functions rely on their mechanical properties could be interesting candidates for investigation. For example, the amyloids in biofilms provide an extremely strong mechanism for organismal adhesion. If a β-strand reorientation, such as we have suggested, occurs with these fibrils it would provide a compelling explanation for their essential biological properties. The method we have developed allows for testing this possibility. The properties of the fibrils studied here, with their highly efficient and specific tethering capacity and an ability to sustain multiple length-extending unfolding events prior to rupture, should also find utility in the design of protein-based nanostructures.
METHODS
Protein purification, labeling and fibril assembly
Wild-type NM and NM mutants were cloned, expressed, purified and labeled as previously described (1) (see Supplementary Methods for details).
Validation of fibril attachment and optimizing conditions
We validated the success of the fibril attachment method by fluorescence visualization. Fibrils were visualized either with a commercially available inverted microscope (Axiovert, Carl Zeiss microscope, field of observation 60 μm × 60 μm) or with a customized Nikon TE2000 microscope on the optical trapping instrument. With the customized microscope, images were recorded on a back-thinned EMCCD camera (ANDOR, field of observation 15 μm × 15 μm).
When casein was not employed to block non-specific adsorption, fibrils littered the glass surface, whereas when casein was employed without pre-depositing NM monomers, fibrils were rarely detected. Thus, the casein successfully prevented non-specific interactions between prion fibrils and the glass surface. When NM monomers were pre-bound, followed by casein blocking, many fibrils were tethered in the field of observation (~60μm × 60μm, Supplementary movie 1). This confirmed that in the presence of casein the self-recognition properties of pre-bound monomers were required to capture the fibrils. In the final step – incubation of NM-coated beads with affixed fibrils – beads only attached efficiently if they had been pre-decorated with NM monomers and if the incubation was long enough to allow for specific capture.
Because of variations in experimental conditions, it proved necessary to test different concentrations of surface monomers and incubation times for each experiment, surveying the flow chambers to identify the best samples for pulling experiments. We typically worked with samples in which 5 to 10 fibrils were tethered in the optical trapping visual field (which is smaller than the viewing field in a standard inverted microscope, ~15μm × 15μm, Supplementary movie 2a). With such a density, most fibrils were tethered to the glass slide at only one end, and carried a single bead at the other end for optical manipulation. When fibril density was higher than this, many fibrils were captured on the glass surface at both ends although the main body of the fibril remained mobile in solution (Supplementary movie 2b).
Cleavage of NM fibrils from the glass surface by TEV protease
A NM variant was engineered with a TEV protease recognition sequence (ENLYFQG) substituting for amino acids 128 to 135 of wild-type NM. This region is located at the beginning of the M domain, and is not involved in the formation of the amyloid core. These TEV-cleavable monomers were introduced into the flow channel and allowed to attach to the glass surface exactly as was done with wild-type protein. They were then used to tether wild-type NM fibrils as described in the main text. The polystyrene bead was coated with wild-type NM and attached to the other end of the fibril. The bead that attached to the end of a fibril was trapped and held in a fully extended position prior to introducing the protease to the chamber by flow. After transient fluorescence imaging of the intact fibrils, 20 μl of 20-unit/μl TEV protease was introduced into the chamber by flow. An additional fluorescent image was then recorded after 10-minute incubation of TEV protease. To better resolve the fibrils that were released from the surface but still attached to the beads, a constant flow of 1×CRBB buffer was introduced through the flow channel to extend the fibrils along the flow direction.
Stretching NM fibrils at high forces with a double trap instrument
The instrument was designed for dual trapping capabilities by splitting the trapping laser (1064 nm 10 W; IPG Photonics, Oxford, MA) into two branches of approximately equal power using a cube polarizer (ThorLabs; Newton, NJ). One branch (the primary branch) was guided through acousto-optic deflectors (AODs; IntraAction, Bellwood, IL) enabling precise steering of the beam; however, the AODs also impart ~50% power loss. The second branch, which experienced minimal power losses along its path, was used to apply maximum possible force. Half-wave plates (CVI; Albuquerque, NM), which can rotate the polarization of incoming light, were placed in each path to ensure maximum power throughput at the second cube polarizer. The maximum force capacity of our trapping instrument was determined by using the force-extension behavior of the fibrils as an internal control (See Supplementary methods for details).
Prior to running an experiment, the secondary branch was aligned with the primary branch and the position detection laser (975 nm, Avanex, San Jose, CA) in the image plane by adjusting the position of the first lens in the telescope pair of the second branch using a 3-axis micrometer stage. The bead was initially trapped with the primary branch and centered over the surface attachment point of the tethered fibril. The bead was subjected to position calibration as previously described 59. Briefly, the bead is raster scanned with the AODs through a grid of positions to map detector signal in 2 dimensions 34. The bead is then held in the center of the detector zone while position is registered to determine trap stiffness via the equipartition method 59. Then the bead was exchanged between the primary branch and the secondary branch to perform a stiffness calibration. After performing the stiffness calibration, the pulling experiments were executed as described in the main text by using the secondary branch.
Fitting the force-extension curves by a modified worm-like chain (WLC) model
Under no externally applied forces, NM fibrils exhibited end-to-end distances comparable to their fully extended length (contour length, Lc), indicating that persistence length (Lp) is of the same order at Lc. Therefore, the Palmer-Boyce WLC model, which is appropriate when Lp ~ Lc, was used to model the force-extension data (Supplementary Methods) 60. To fit the force-extension curves with discontinuities in the stretching phases, force-extension data prior to unfolding was fit first and then LC was adjusted in order to fit the data after unfolding. ΔLC was determined by the LC prior and after the discontinuities occurred.
Supplementary Material
Acknowledgments
We thank members of the Lindquist laboratory for comments on the manuscript. We are grateful to Dr. Steven Block, Dr. Wonmuk Hwang and Dr. Karen Allendoerfer for their critical reading. SL is an investigator of the Howard Hughes Medical Institute (HHMI). This work was supported by a NIH grant GM025874 to SL, NSF Career Award (0643745) to MJL, American Heart Association fellowship to JD (0725849T). The project described was also supported by National Institute of Biomedical Imaging and Bioengineering grant (T32EB006348). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Biomedical Imaging and Bioengineering or the National Institutes of Health.
Footnotes
CONTRIBUTIONS
J.D. initiated the project. J.D. carried out sample preparation and yeast biology; J.D. and C.E.C. designed and carried out the optical trapping experiments and data analysis; M.C.B, M.J.L. and S.L. supervised the projects and interpreted the results. J.D. and S.L. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Nelson R, Eisenberg D. Structural models of amyloid-like fibrils. Fibrous Proteins: Amyloids, Prions and Beta Proteins. 2006;73:235. doi: 10.1016/S0065-3233(06)73008-X. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 3.Cheng IH, et al. Accelerating amyloid-beta fibrillization reduces oligomer levels and functional deficits in Alzheimer disease mouse models. J Biol Chem. 2007;282:23818–23828. doi: 10.1074/jbc.M701078200. [DOI] [PubMed] [Google Scholar]
- 4.Douglas PM, et al. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci U S A. 2008;105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–50. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 6.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 7.Serio TR, et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–21. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 8.Glover JR, et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–9. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 9.Paushkin SV, Kushnirov VV, Smirnov VN, TerAvanesyan MD. In vitro propagation of the prion-like state of yeast Sup35 protein. Science. 1997;277:381–383. doi: 10.1126/science.277.5324.381. [DOI] [PubMed] [Google Scholar]
- 10.Satpute-Krishnan P, Langseth SX, Serio TR. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. Plos Biol. 2007;5:251–262. doi: 10.1371/journal.pbio.0050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 12.Chernoff YO, Lindquist SL, Ono B, Ingevechtomov SG, Liebman SW. Role of the Chaperone Protein Hsp104 in Propagation of the Yeast Prion-Like Factor [Psi(+)] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 15.Uptain SM, Sawicki GJ, Caughey B, Lindquist S. Strains of [PSI(+)] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J. 2001;20:6236–45. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodside MT, Garcia-Garcia C, Block SM. Folding and unfolding single RNA molecules under tension. Curr Opin Chem Biol. 2008;12:640–646. doi: 10.1016/j.cbpa.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Block SM. Kinesin motor mechanics: Binding, stepping, tracking, gating, and limping. Biophys J. 2007;92:2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bechtluft P, et al. Direct observation of chaperone-induced changes in a protein folding pathway. Science. 2007;318:1458–1461. doi: 10.1126/science.1144972. [DOI] [PubMed] [Google Scholar]
- 19.Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 20.Kellermayer MSZ, Smith SB, Granzier HL, Bustamante C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 21.Karsai A, et al. Mechanical manipulation of Alzheimer’s amyloid beta 1–42 fibrils. J Struct Biol. 2006;155:316–326. doi: 10.1016/j.jsb.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Kellermayer MSZ, et al. Reversible mechanical unzipping of amyloid beta-fibrils. J Biol Chem. 2005;280:8464–8470. doi: 10.1074/jbc.M411556200. [DOI] [PubMed] [Google Scholar]
- 23.Raman EP, Takeda T, Barsegov V, Klimov DK. Mechanical unbinding of A beta peptides from amyloid fibrils. J Mol Biol. 2007;373:785–800. doi: 10.1016/j.jmb.2007.08.034. [DOI] [PubMed] [Google Scholar]
- 24.Liu JJ, Sondheimer N, Lindquist SL. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+] Proc Natl Acad Sci U S A. 2002;99 (Suppl 4):16446–53. doi: 10.1073/pnas.252652099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–72. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tessier PM, Lindquist S. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature. 2007;447:556. doi: 10.1038/nature05848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–7. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 28.Scheibel T, et al. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc Natl Acad Sci U S A. 2003;100:4527–32. doi: 10.1073/pnas.0431081100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tskhovrebova L, Trinick J, Sleep JA, Simmons RM. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997;387:308–12. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 30.Collins SR, Douglass A, Vale RD, Weissman JS. Mechanism of prion propagation: Amyloid growth occurs by monomer addition. Plos Biol. 2004;2:1582–1590. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DePace AH, Weissman JS. Origins and kinetic consequences of diversity in Sup35 yeast prion fibers. Nat Struct Biol. 2002;9:389–396. doi: 10.1038/nsb786. [DOI] [PubMed] [Google Scholar]
- 32.Santoso A, Chien P, Osherovich LZ, Weissman JS. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–88. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- 33.Dudko OK, Hummer G, Szabo A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc Natl Acad Sci U S A. 2008;105:15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lang MJ, Asbury CL, Shaevitz JW, Block SM. An automated two-dimensional optical force clamp for single molecule studies. Biophys J. 2002;83:491–501. doi: 10.1016/S0006-3495(02)75185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao Y, Li H. How do chemical denaturants affect the mechanical folding and unfolding of proteins? J Mol Biol. 2008;375:316–324. doi: 10.1016/j.jmb.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 36.Andersen CB, et al. Branching in Amyloid Fibril Growth. Biophys J. 2009;96:1529–1536. doi: 10.1016/j.bpj.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, et al. Direct and selective elimination of specific prions and amyloids by 4,5-dianilinophthalimide and analogs. Proc Natl Acad Sci U S A. 2008;105:7159–7164. doi: 10.1073/pnas.0801934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tessier PM, Lindquist S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+] Nat Struct Mol Biol. 2009;16:598–605. doi: 10.1038/nsmb.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu JJ, Lindquist S. Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature. 1999;400:573–6. doi: 10.1038/23048. [DOI] [PubMed] [Google Scholar]
- 40.Greene DN, et al. Single-molecule force spectroscopy reveals a stepwise unfolding of Caenorhabditis elegans giant protein kinase domains. Biophys J. 2008;95:1360–1370. doi: 10.1529/biophysj.108.130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oberhauser AF, Hansma PK, Carrion-Vazquez M, Fernandez JM. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc Natl Acad Sci U S A. 2001;98:468–472. doi: 10.1073/pnas.021321798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao Y, Li HB. Polyprotein of GB1 is an ideal artificial elastomeric protein. Nature Materials. 2007;6:109–114. doi: 10.1038/nmat1825. [DOI] [PubMed] [Google Scholar]
- 43.Bell GI. Models for Specific Adhesion of Cells to Cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 44.Carrion-Vazquez M, et al. Mechanical and chemical unfolding of a single protein: A comparison. Proc Natl Acad Sci U S A. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gebhardt JCM, Bornschlogla T, Rief M. Full distance-resolved folding energy landscape of one single protein molecule. Proc Natl Acad Sci U S A. 2010;107:2013–2018. doi: 10.1073/pnas.0909854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kellermayer MSZ, Smith S, Bustamante C, Granzier HL. Mechanical manipulation of single titin molecules with laser tweezers. Adv Exp Med Biol. 2000;481:111–128. doi: 10.1007/978-1-4615-4267-4_7. [DOI] [PubMed] [Google Scholar]
- 47.Zhang JN, Muthukumar M. Simulations of nucleation and elongation of amyloid fibrils. J Chem Phys. 2009;130 doi: 10.1063/1.3050295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pallitto MM, Murphy RM. A mathematical model of the kinetics of beta-amyloid fibril growth from the denatured state. Biophys J. 2001;81:1805–1822. doi: 10.1016/S0006-3495(01)75831-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee CC, Nayak A, Sethuraman A, Belfort G, Mcrae GJ. A three-stage kinetic model of amyloid fibrillation. Biophys J. 2007;92:3448–3458. doi: 10.1529/biophysj.106.098608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kishimoto A, et al. beta-Helix is a likely core structure of yeast prion Sup35 amyloid fibers. Biochem BiophysRes Commun. 2004;315:739–745. doi: 10.1016/j.bbrc.2004.01.117. [DOI] [PubMed] [Google Scholar]
- 51.Lazo ND, Downing DT. Amyloid fibrils may be assembled from beta-helical protofibrils. Biochem. 1998;37:1731–1735. doi: 10.1021/bi971016d. [DOI] [PubMed] [Google Scholar]
- 52.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keten S, Buehler MJ. Large deformation and fracture mechanics of a beta-helical protein nanotube: Atomistic and continuum modeling. Comput Methods Appl Mech Engrg. 2008;197:3203–3214. [Google Scholar]
- 54.Brockwell DJ, et al. Pulling geometry defines the mechanical resistance of a beta-sheet protein. Nat Struct Biol. 2003;10:731–737. doi: 10.1038/nsb968. [DOI] [PubMed] [Google Scholar]
- 55.Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu RevBiochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 56.Yang SC, Levine H, Onuchic JN, Cox DL. Structure of infectious prions: stabilization by domain swapping. Faseb J. 2005;19:1778–1782. doi: 10.1096/fj.05-4067hyp. [DOI] [PubMed] [Google Scholar]
- 57.White HE, et al. Globular Tetramers of beta(2)-Microglobulin Assemble into Elaborate Amyloid Fibrils. J Mol Biol. 2009;389:48–57. doi: 10.1016/j.jmb.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sondheimer N, Lindquist S. Rnq1: An epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 59.Neuman KC, Block SM. Optical trapping. Rev Sci Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palmer JS, Boyce MC. Constitutive modeling of the stress-strain behavior of F-actin filament networks. Acta Biomaterialia. 2008;4:597–612. doi: 10.1016/j.actbio.2007.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.