The first crystal structure of a pectin methylesterase from an enteropathogen is presented. This enzyme from Y. enterocolitica has biological significance for the evolution of pectin-metabolic pathways within pectinolytic bacteria and related agents of foodborne illness.
Keywords: carbohydrate esterases, pectins, methylesterification, enteropathogens
Abstract
Pectin methylesterases (PMEs) are family 8 carbohydrate esterases (CE8s) which remove the methyl group from methylesterified galacturonic acid (GalA) residues within pectin. Although the role of pectinases such as PMEs within dedicated phytopathogens has been well established, the significance of homologous enzymes found within the genomes of human enteropathogens remains to be determined. Presented here is the low-resolution (3.5 Å) structure of the CE8 from Yersinia enterocolitica (YeCE8). The high degree of structural conservation in the topology of the active-site cleft and catalytic apparatus that is shared with a characterized PME from a bacterial phytopathogen (i) indicates that YeCE8 is active on methylated pectin and (ii) highlights a more prominent role for pectin utilization in Yersinia than in other enteropathogenic species.
1. Introduction
Family 8 carbohydrate esterases (CE8s) are a group of sequence-related enzymes found within the domains Archaea, Bacteria and Eukarya (http://www.cazy.org; Cantarel et al., 2009 ▶). Currently, 40 different CE8s have been characterized as pectin methylesterases (PMEs), enzymes which remove the methyl group from methyl-esterified galacturonic acid (GalA) residues within pectin. This is a significant biological event in processes such as cell-wall turnover, fruit ripening and pathogenesis. Removal of methanol from a methyl-esterified GalA (i.e. demethoxylation, referred to below as demethylation) restores the inherent negative charge on the uronate group of GalA, which loosens the higher order structure of the pectic network through electrorepulsive effects (Pelloux et al., 2007 ▶). In bacteria, two different isozymes have been described, PmeA/PemA (pectin methylesterase A) and PmeB/PemB (pectin methylesterase B), both of which are required for optimal growth of the phytopathogen Dickeya dadantii 3937 (formerly Erwinia chrysanthemi 3937) on highly methylated pectic substrates (Shevchik et al., 1996 ▶). PmeA is a secreted PME that is active on highly polymerized methylated pectin and is closely related to plant PMEs, whereas PmeB belongs to a smaller subfamily of PMEs that are specific for methylated oligogalacturonides and are lipid-anchored to the periplasmic face of the outer membrane (Shevchik et al., 1996 ▶; Kazemi-Pour et al., 2004 ▶). These two distinct profiles suggest unique roles for each isozyme along the pectinolytic cascade. In eukaryotes, a separate nomenclature has been adopted in which pectin methylesterase isozymes are delineated numerically (i.e. Pme1, Pme2). Below, enzymes will be identified using the CAZy classification system, which consists of the italicized initials of the host species followed by the enzyme family (i.e. the PME from Yersinia enterocolitica is YeCE8).
Recently, a role other than demethylesterification of pectin has been suggested for a small subclade of CE8s (Eklöf et al., 2009 ▶). The YbhC gene product from Escherichia coli K-12 MG1655 (EcCE8) does not display any esterase activity on highly methylated polygalacturonic acid, nor does it interact with this polymer in binding studies. Structural analysis indicated that EcCE8 contains several unique motifs: a remodelled active-site cleft with several loop insertions and mutations to key catalytic residues, including the primary general acid–base catalyst (see Supplementary Material1). Intriguingly, phylogenetic analysis of EcCE8 indicated that a cluster of enzymes belonging to enteric bacteria, such as Enterobacter, Klebsiella, Salmonella and Shigella, appear to have evolved from a common PmeB ancestor (Eklöf et al., 2009 ▶). Previously, the exclusive role of CE8s in pectin degradation suggested that this enzyme was a highly conserved member of a pectin-utilization pathway found in a number of foodborne pathogens from Enterobacteriaceae (Rodionov et al., 2004 ▶; Abbott & Boraston, 2008 ▶). The relationship now however appears to be more biologically complex. Therefore, in order to further explore the role of CE8s in Enterobacteriaceae, the homologue from Y. enterocolitica, YeCE8, has been cloned and structurally characterized at low resolution (3.5 Å). The complex evolutionary history of the emergence of pectin-utilization systems within enteropathogens is discussed.
2. Materials and methods
2.1. Cloning, production and purification of recombinant YeCE8
The gene sequence of YeCE8 (locus tag Ye0549), without the N-terminal 25-amino-acid signal peptide, was amplified from the genomic DNA of biotype O:8, serotype 1B Y. enterocolitica subsp. enterocolitica 8081 (ATCC ID No. 27729D-5). An in-frame 5′ NheI restriction site, encoded within the primer sequence ATATGCT AGCGCGCAGTATAATGCAGTTGTTTCT, and 3′ XhoI restriction site, encoded within the primer sequence ATATCTCGAGTCAATGCACGGCCCAATCAGGGAA, were introduced into the PCR product for directional cloning. The amplicon was purified, digested and ligated into pET28a plasmid vector (Novagen, catalogue No. 69864) treated with complementary enzymes. The generated expression construct, pYECE8, contained a thrombin-cleavable N-terminal His6 tag (MGSSHHHHHHSSGLVPR–GSHM). Ligated vectors were subsequently transformed into DH5α competent cells for propagation. Positive clones were purified and verified by restriction digest and DNA sequencing.
Recombinant YeCE8 protein was produced in E. coli BL21 (DE3) (Novagen, catalogue No. 69041). Transformed cells were grown at 310 K with shaking at 180 rev min−1 in LB supplemented with 50 µg ml−1 kanamycin to an OD of ∼0.8 at 600 nm. At this density, the temperature was decreased to 289 K for 1 h with shaking; gene expression was then induced using 0.2 mM isopropyl β-d-1-thiogalactopyranoside with shaking overnight. The following morning, the cells were harvested by centrifugation at 6000g and disrupted by chemical lysis. Following clarification by centrifugation at 15 000g, YeCE8 was purified by immobilized metal-affinity chromatography using Ni-Sepharose resin (GE Healthcare, catalogue No. 17-5318-06) equilibrated in 500 mM sodium chloride, 20 mM Tris–HCl pH 8.0. Bound protein was eluted with a stepwise gradient from 0 to 500 mM imidazole in the aforementioned buffer. Protein fractions were analyzed for purity by SDS–PAGE and samples containing appreciable amounts of pure YeCE8 were pooled and buffer-exchanged to remove imidazole using a stirred-cell ultrafiltration device with a 5000 MWCO membrane (Millipore, catalogue No. PLCC02510). Calcium ions were added to a final concentration of 2 mM and the N-terminal six-histidine tag was removed using restriction-grade thrombin (Novagen, catalogue No. 69671). After complete digestion, YeCE8 was further purified by gel-filtration chromatography using a Sephacryl S-200 column (GE Biosciences, catalogue No. 17-0584-05) in 20 mM Tris–HCl pH 8.0. Purified polypeptides were concentrated to 20 mg ml−1 for crystallization trials, as determined using a calculated extinction coefficient of 1.20 mg ml−1 (Gasteiger et al., 2005 ▶).
2.2. YeCE8 crystallization conditions and structure solution
Repeated attempts to crystallize YeCE8 were performed using commercially available crystal screens and the hanging-drop vapour-diffusion method at 291 K. Despite these efforts, no promising crystallization conditions were detected within a period of three months with routine surveillance. After a period of approximately one year the plates were rechecked and a single crystal was observed in a well containing unbuffered 4 M sodium formate (condition No. 33 of Crystal Screen; Hampton Research, catalogue No. HR2-110). The crystal was flash-cooled directly in a cryostream at 113 K as the high salt concentration acted as an effective cryoprotectant. Crystallized YeCE8 was verified for X-ray diffraction and data were collected on beamline X8-C at the National Synchrotron Light Source, Brookhaven National Laboratories. Data were processed with MOSLFM (Battye et al., 2011 ▶), POINTLESS (Evans, 2006 ▶) and SCALA (Evans, 2006 ▶) to a resolution of 3.5 Å (Table 1 ▶).
Table 1. Data-collection and structure statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Wavelength (Å) | 1.1 |
| Space group | P4332 |
| Molecules per asymmetric unit | 1 |
| Solvent content (%) | 76.3 |
| Unit-cell parameters (Å) | a = b = c = 170.6 |
| Resolution (Å) | 40.00–3.50 (3.69–3.50) |
| Rmerge | 0.17 (0.60) |
| 〈I/σ(I)〉 | 21.2 (5.9) |
| Completeness (%) | 99.9 (99.9) |
| Multiplicity | 24.7 (21.1) |
| Mosaicity (°) | 0.89 |
| No. of crystals | 1 |
| Temperature (K) | 100 |
| Oscillation (°) | 1 |
| Rotation range (°) | 129 |
| Exposure per frame (s) | 20 |
| Crystal-to-detector distance (mm) | 200 |
| Detector | CCD |
| Refinement | |
| No. of reflections | 10653 |
| Rwork/Rfree | 0.20/0.25 (0.18/0.26) |
| No. of atoms | |
| Protein | 2409 [chain A] |
| Water | 13 [chain B] |
| B factors (Å2) | |
| Protein | 59.9 |
| Water | 36.2 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.0048 |
| Bond angles (°) | 0.92 |
| Ramachandran plot (%) | |
| Favoured | 95.6 |
| Allowed | 4.4 |
| Outliers | 0.0 |
The structure of YeCE8 was solved using the MOLREP automated molecular-replacement program (Vagin & Teplyakov, 2010 ▶) with PmeA from D. dadantii 3937 (referred to as DdCE8 below; PDB entry 1qjv; 59% identity over 335 amino acids; Jenkins et al., 2001 ▶) as a search model. One molecule was found in the asymmetric unit. Successive rounds of rigid-body and restrained refinement using REFMAC (Murshudov et al., 2011 ▶) and manual model building in Coot (Emsley & Cowtan, 2004 ▶) were performed to complete the model. 5% of the reflections were flagged as ‘free’ and were used to monitor refinement progress. All waters were added manually based upon difference maps and acceptable distances from local amino acids (2.5–3.5 Å). Validation of the YeCE8 structure was performed using MolProbity (Chen et al., 2010 ▶) and the resulting statistics are provided in Table 1 ▶. Structural alignments were performed with Coot (Emsley & Cowtan, 2004 ▶). PyMOL (DeLano, 2002 ▶) was used for figure generation and secondary-structure analysis.
3. Results and discussion
3.1. The crystal structure of YeCE8
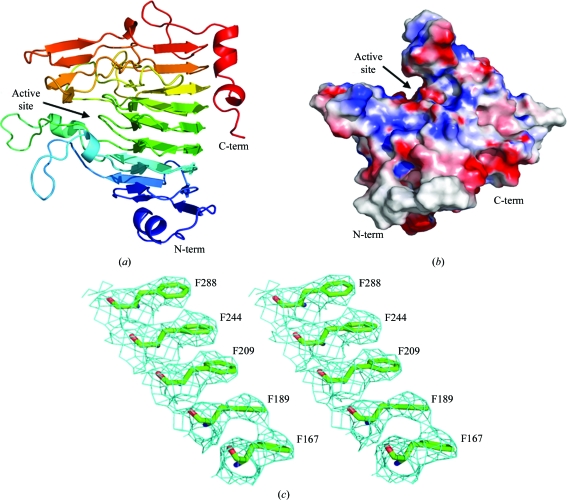
Family 8 carbohydrate esterases adopt a right-handed β-helix (Fig. 1 ▶ a), which is a common scaffold for a variety of unrelated pectinase families, including polygalacturonases and rhamnogalacturonases (GH28) and pectin and pectate lyases (PL1, PL3 and PL9) (Abbott & Boraston, 2008 ▶; Jenkins & Pickersgill, 2001 ▶). A prominent cleft is visible on the surface of YeCE8 which is conserved in all structurally characterized PmeAs (Fig. 1 ▶ b; Fries et al., 2007 ▶; Jenkins et al., 2001 ▶). The open cleft provides the enzyme with access to internal methylations along the length of pectic polymers. The model includes residues 41–360 of the YeCE8 primary structure; a portion of the N-terminus was too disordered to model accurately. In addition to the 2409 protein atoms (chain A), 13 water atoms (chain B) were added manually on the basis of their proximity to neighbouring side chains (2.5–3.5 Å). Although this structure was solved to a low resolution (3.5 Å), the high redundancy improved the electron-density quality (Fig. 1 ▶ c) and enabled the majority of side-chain fitting to be performed using automated refinement. However, some regions required careful inspection and side-chain truncations were introduced where required to improve the refinement statistics. In areas of ambiguous electron density, manual model building was directed by superimposition with the DdCE8 structure (Jenkins et al., 2001 ▶).
Figure 1.
The crystal structure of YeCE8. (a) Cartoon representation of the YeCE8 β-helix fold coloured in a rainbow gradient from blue (N-terminus) to red (C-terminus). (b) Electrostatic surface potential of YeCE8, highlighting the prominent active-site cleft. (c) Wall-eyed representation of the electron density for YeCE8 at 3.5 Å resolution. The maximum-likelihood (Murshudov et al., 2011 ▶) σA-weighted (Read, 1986 ▶) 2F obs – F calc map (coloured in cyan) is contoured at 1.0σ (0.11 e− Å−3). The internal aromatic stack of phenylalanine residues is a structural feature found in other β-helix proteins that serves to anchor neighbouring β-strands and stabilize the overall fold (Jenkins & Pickersgill, 2001 ▶).
A DaliLite search (v.3; Holm & Rosenström, 2010 ▶) for YeCE8 produced the highest matches with a series of DdCE8 complexes and its apo structure (Fries et al., 2007 ▶; Jenkins et al., 2001 ▶). Consistent Z values were observed in the range 50.2–50.7 and r.m.s.d.s were in the range 0.7–0.9 Å over 319 aligned Cα atoms for each chain of these deposited structures. The similar values observed for the complexed and apo forms underlines that these enzymes are rigid and do not display significant conformational changes in the active-site cleft or along the β-helix backbone following substrate binding. The two eukaryotic PMEs from Solanum lycopersicum (PDB entry 1xg2; Di Matteo et al., 2005 ▶; 25% identity) and Daucus carota (PDB entry 1gq8; Johansson et al., 2002 ▶; 28% identity) were the next two closest results, with Z values of 35.0 and 33.8 and r.m.s.d. values of 1.8 Å over 317 and 319 Cα atoms, respectively. YbhC, a PmeB-like homologue from E. coli K-12 with unknown activity (PDB entry 3grh; Eklöf et al., 2009 ▶; 25% identity), followed the PMEs with a Z value of 31.5 and an r.m.s.d. of 1.6 Å over 326 Cα atoms. Alignments with pectinases that display distinct activities and low primary-structure identities (<15%) were also observed (r.m.s.d. values of 2.8–3.7 Å), highlighting the flexibility of the β-helix scaffold in diverse pectinolytic functions.
3.2. The YeCE8 active site
A closer look at the core active site of YeCE8 superimposed with the DdCE8–GalA6 complex (PDB entry 2ntb; Fries et al., 2007 ▶) indicates that YeCE8 is a PME (Fig. 2 ▶). YeCE8 side chains make direct sugar contacts with the oligogalacturonide in five different subsites (labelled −2 to +3 from the nonreducing end; Fig. 2 ▶ a). The scissile GalA methyl group is positioned in subsite +1. The terminal sugar residue at the nonreducing end does not display any direct interactions with the enzyme. Every key residue identified in the DdCE8 enzyme is absolutely conserved; the only subtle difference detected is that the uronate group of GalA in subsite −2 is positioned to make a hydrogen bond to the peptidyl N atom of Ser109 in YeCE8, which replaces the interaction with the peptidyl N atom of Ala110 observed in DdCE8. Significantly, the catalytic triad consisting of Gln176, Asp177 and Asp199 is strictly conserved, with the putative acid–base catalyst Asp177 within hydrogen-bonding distance of O6 of the substrate (Asp177 Oδ1 and Oδ2 are 2.5 and 3.5 Å from the O6A and O6B atoms of the +1 GalA, respectively). These observations strongly support the contention that YeCE8 and DdCE8 have analogous biological functions in the demethylesterification of pectin.
Figure 2.
Comparison of the functional groups involved in direct carbohydrate interactions within the active sites of YeCE8 (green) and DdCE8 (grey). The amino acids are shown as sticks and labelled as in YeCE8/DdCE8. Five of the six sugar residues of the GalA6 product (PDB entry 2ntb) are displayed as yellow sticks; the GalA residue with the scissile methyl group is coloured cyan. Subsites are indicated by black dashed ellipses and are numbered in red. The putative YeCE8 nucleophile (Asp199) is not shown, but is structurally conserved with the homologous DdCE8 residue (Asp199). (b) Ribbon representation of superimposed YeCE8 (green; PDB entry 3uw0) and DdCE8 (grey; PDB entry 2ntb) displaying the open active site of the PmeA-type PMEs.
The recently reported structure of the PmeB-like homologue YbhC from E. coli K12 (PDB entry 3grh) provides further clues to the functional specificity of YeCE8. Comparing the global folds of the two subclasses of PMEs previously revealed that the ‘closing off’ of one end of the active site by loop insertions defines the structural basis for the distinct activities of the polysaccharide-specific PmeA and the oligosaccharide-specific PmeB (Eklöf et al., 2009 ▶). The open active-site cleft of YeCE8 presented here indicates that YeCE8 is also a PmeA-type PME and therefore active on large polymeric substrates (Fig. 2 ▶ b).
3.3. Pectin utilization within Yersinia
The presence of pectin-utilization pathways within the genomes of enteropathogenic species from Enterobacteriaceae has been described previously (Rodionov et al., 2004 ▶; Abbott & Boraston, 2008 ▶). The biological significance of the utilization of a structural plant cell-wall polysaccharide as an energy source by these microbes, however, remains to be determined. Although subtle differences in the complement of biocatalysts and their predicted cellular locations are noted between species such as Salmonella, Shigella, E. coli, Klebsiella and Yersinia, several features are absolutely conserved, including a KdgR repressor protein, a member of the KdgM outer membrane porin family, a CE8 and an intact cytoplasmic pathway (Hugouvieux-Cotte-Pattat et al., 1996 ▶; Rodionov et al., 2004 ▶; Abbott & Boraston, 2008 ▶). Determining when pectin-utilization pathways are activated during enteropathogenic life cycles may be a key discovery for understanding the role of pectin utilization in environmental persistence, food-crop contamination or colonization of the host gut (Abbott et al., 2010 ▶). In this light, the presence of a conserved pathway within the zoonotic Y. pestis, which is not known to pass through the host digestive system, is intriguing.
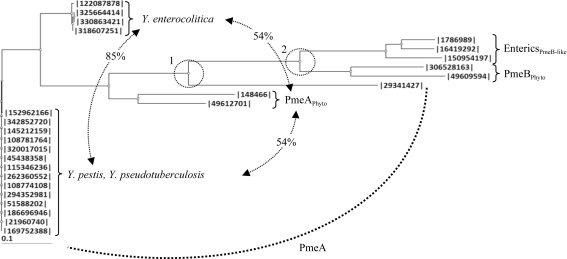
The potential role of a subclade of CE8s in a biological process other than pectin demethylesterification suggests that the CE8 homologue is not essential for pectin harvesting in enteric bacteria (Eklöf et al., 2009 ▶). Insights into the evolutionary history of CE8s found within a group of enteropathogens from Enterobacteriaceae, including Klebsiella, Erythrobacter, Citrobacter, Salmonella and Shigella, revealed that this group of CE8s arose from the PmeB isozyme (Fig. 3 ▶) and display notable functional differences from PmeAs, including (i) a lipidated anchor, (ii) a remodelled active-site cleft and (iii) an acid–base catalyst that is mutated from an aspartate to an asparagine (see Supplementary Material1; Eklöf et al., 2009 ▶). The phylogenetic analysis presented here indicates that CE8s found within the genus Yersinia contain highly conserved PmeA-like homologues, as opposed to the PmeB-like homologues in other enteric bacteria, and retain the structural signatures required for polymeric pectin demethylesterification by DdCE8 (Fig. 3 ▶; Supplementary Material1). This observation is consistent with the genomes of Yersinia displaying the most developed pectin-utilization pathways of any enteropathogen from the Enterobacteriaceae. Indeed, Y. pestis, Y. pseudotuberculosis and Y. enterocolitica each contain an exopolygalacturonase (YeGH28) and two pectate lyases (YePL2A and YePL2B) that are also found in plant-cell macerating phytopathogens (Abbott & Boraston, 2008 ▶; Rodionov et al., 2004 ▶; Hugouvieux-Cotte-Pattat et al., 1996 ▶). Importantly, the presence of a bone fide pectin methylesterase supports the idea that pectin metabolism may play a more significant role in the biological fitness or transmission of Yersinia than in other related enteric bacteria.
Figure 3.
Phylogram of CE8 enzymes from Yersinia and bacterial phytopathogens and enteropathogens. Neighboring-joining phylogram of CE8s from Y. pestis (342852720, A1122; 145212159, Pestoides F; 108781764, Antiqua; 320017015, biovar Mediaevalis str. Harbin 35; 5438358, biovar Microtus str. 91001; 115346236, CO92; 262360552, D106004; 21960740, KIM 10; 108774108, Nepal516; 294352981, Z176003), Y. pseudotuberculosis (152962166, IP_31758; 51588202, IP_32953; 186696946, PB1/+; 169752388, YPIII) and Y. enterocolitica [122087878, subsp. enterocolitica 8081; 325664414, subsp. palearctica 105.5R(r); 318607251, subsp. palearctica Y11; 330863421, W22703] strains with (1) PmeA from B. thetaiotaomicron (29341427) and the phytopathogens D. dadantii 3937 (148466) and Pectobacterium atrosepticum SCRI1043 (49612701), (2) PmeB from D. dadantii 3937 (306528163) and P. atrosepticum SCRI1043 (49609594) and (3) PmeB-like enzymes from the enteropathogens K. pneumonia subsp. pneumonia MGH78578 (150954197), S. enterica subsp. enterica serovar Typhimurium str. LT2 (16419292) and E. coli str. K-12 substr. MG1655 (1786989). The relatedness (represented as percentage identity) between the Y. enterocolitica, PmeAPhyto and Y. pestis/Y. psudotuberculosis subclades are labelled. Two significant divergences separating the Yersinia enzymes from the other PMEs and the PmeAs from the PmeBs are indicated by dashed circles and labelled.
Supplementary Material
PDB reference: pectin methylesterase, 3uw0
Supplementary material file. DOI: 10.1107/S1744309111055400/hv5208sup1.pdf
Acknowledgments
This work was funded by a grant from the Natural Sciences and Engineering Research Council of Canada. ABB is a Canada Research Chair in Molecular Interactions and a Michael Smith Foundation for Health Research Scholar.
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: HV5208).
References
- Abbott, D. W. & Boraston, A. B. (2008). Microbiol. Mol. Biol. Rev. 72, 301–316. [DOI] [PMC free article] [PubMed]
- Abbott, D. W., Gilbert, H. J. & Boraston, A. B. (2010). J. Biol. Chem. 285, 39029–39038. [DOI] [PMC free article] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V. & Henrissat, B. (2009). Nucleic Acids Res. 37, D233–D238. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Di Matteo, A., Giovane, A., Raiola, A., Camardella, L., Bonivento, D., De Lorenzo, G., Cervone, F., Bellincampi, D. & Tsernoglou, D. (2005). Plant Cell, 17, 849–858. [DOI] [PMC free article] [PubMed]
- Eklöf, J. M., Tan, T.-C., Divne, C. & Brumer, H. (2009). Proteins, 76, 1029–1036. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Fries, M., Ihrig, J., Brocklehurst, K., Shevchik, V. E. & Pickersgill, R. W. (2007). EMBO J. 26, 3879–3887. [DOI] [PMC free article] [PubMed]
- Gasteiger, E. H. C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D. & Bairoch, A. (2005). The Proteomics Protocols Handbook, edited by J. M. Walker, pp. 571–607. Totowa: Humana Press.
- Holm, L. & Rosenström, P. (2010). Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed]
- Hugouvieux-Cotte-Pattat, N., Condemine, G., Nasser, W. & Reverchon, S. (1996). Annu. Rev. Microbiol. 50, 213–257. [DOI] [PubMed]
- Jenkins, J., Mayans, O., Smith, D., Worboys, K. & Pickersgill, R. W. (2001). J. Mol. Biol. 305, 951–960. [DOI] [PubMed]
- Jenkins, J. & Pickersgill, R. (2001). Prog. Biophys. Mol. Biol. 77, 111–175. [DOI] [PubMed]
- Johansson, K., El-Ahmad, M., Friemann, R., Jörnvall, H., Markovic, O. & Eklund, H. (2002). FEBS Lett. 514, 243–249. [DOI] [PubMed]
- Kazemi-Pour, N., Condemine, G. & Hugouvieux-Cotte-Pattat, N. (2004). Proteomics, 4, 3177–3186. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Pelloux, J., Rustérucci, C. & Mellerowicz, E. J. (2007). Trends Plant Sci. 12, 267–277. [DOI] [PubMed]
- Read, R. J. (1986). Acta Cryst. A42, 140–149.
- Rodionov, D. A., Gelfand, M. S. & Hugouvieux-Cotte-Pattat, N. (2004). Microbiology, 150, 3571–3590. [DOI] [PubMed]
- Shevchik, V. E., Condemine, G., Hugouvieux-Cotte-Pattat, N. & Robert-Baudouy, J. (1996). Mol. Microbiol. 19, 455–466. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: pectin methylesterase, 3uw0
Supplementary material file. DOI: 10.1107/S1744309111055400/hv5208sup1.pdf