An unusual case of trace amounts of a contaminating protein facilitating formation of a heterotrimeric protein complex.
Keywords: cortactin, Arg, protein–protein complexes, cytoskeleton
Abstract
Crystallization of contaminating proteins is a frequently encountered problem for macromolecular crystallographers. In this study, an attempt was made to obtain a binary cocrystal structure of the SH3 domain of cortactin and a 17-residue peptide from the Arg nonreceptor tyrosine kinase encompassing a PxxPxxPxxP (PxxP1) motif. However, cocrystals could only be obtained in the presence of trace amounts of a contaminating protein. A structure solution obtained by molecular replacement followed by ARP/wARP automatic model building allowed a ‘sequence-by-crystallography’ approach to discover that the contaminating protein was lysozyme. This 1.65 Å resolution crystal structure determination of a 1:1:1 heterotrimeric complex of Arg, cortactin and lysozyme thus provides an unusual ‘caveat emptor’ warning of the dangers that underpurified proteins harbor for macromolecular crystallographers.
1. Introduction
The crystallization of impurities is a well known problem for macromolecular crystallographers. This problem is often seen when the target protein(s) is not purified to sufficient homogeneity. Commonly seen by-product crystals of incomplete purifications include lysozyme, the affinity tag (e.g. GST) or the protease used to cleave the affinity tag (e.g. TEV). Usually, when by-product crystals are observed they occur instead of the target protein of interest. In this manuscript, we describe an unusual case in which we obtained by-product crystals that included both the target protein of interest (cortactin SH3 domain), a peptide that cortactin binds (derived from the Arg nonreceptor tyrosine kinase) and lysozyme. We reproducibly obtained these heterotrimeric crystals only on addition of the Arg peptide; crystallization trials of cortactin SH3 domain alone produced only cortactin SH3 crystals. We believe this to be the first observed case in which the addition of a peptide to a target-protein crystallization trial has resulted in the formation of heterotrimeric crystals with a nonphysiologically relevant protein.
The expression of recombinant protein from bacterial host cells (Escherichia coli) is the most popular and simple avenue to obtain sufficient quantities of protein for macromolecular crystallization. Lysozyme is commonly used to aid lysis of the bacterial cells by specifically catalyzing the hydrolysis of 1,4-glycosidic bonds in the peptidoglycan cell wall of bacteria. Lysozyme has the advantage of providing a gentle way to break bacteria cell walls without damaging the target protein (Lesley, 2001 ▶). We, and many other structural biology laboratories, therefore commonly use lysozyme to aid cell lysis in our protein-purification protocols.
Cortactin is a key regulator of actin polymerization in response to tyrosine kinase signaling. This protein contains an N-terminal acidic (NTA) domain that binds the Arp2/3 complex, a repeat region that binds F-actin and a C-terminal Src homology 3 (SH3) domain that interacts with N-WASp, Arg and WIP (MacGrath & Koleske, 2012 ▶). Cortactin binds and activates the Arp2/3 complex via its NTA region and regulates actin polymerization within cell-edge protrusions, including invadopodia (Lapetina et al., 2009 ▶). Additionally, Arg and cortactin bind and activate N-WASp, a strong Arp2/3 complex activator. The interaction of the cortactin SH3 domain with a PxxP1 motif in Arg may provide an initial step in assembly and function of an Arg–cortactin–N-WASp complex in cell-edge protrusions (Lapetina et al., 2009 ▶).
To provide a structural basis for this first step of Arg–cortactin–N-WASp complex formation, we initiated cocrystallization trials of cortactin SH3 domain (UniProt Q60598) with an Arg peptide encompassing the PxxP1 motif (UniProt P42684). To our surprise, while conducting initial crystallization trials with what we expected to be crystallization-quality cortactin SH3 domain incubated with the Arg peptide, we reproducibly obtained heterotrimeric crystals of cortactin SH3, Arg peptide and lysozyme (UniProt P00698). In this manuscript, we discuss the purification of cortactin SH3 domain and show how contamination by trace amounts of lysozyme resulted in cocrystallization with our target proteins of interest. We discuss our crystallization and structure determination for this complex to 1.65 Å resolution and describe a difficult molecular-replacement solution. Finally, we describe the interactions between cortactin and Arg, between cortactin and lysozyme and between lysozyme and Arg. Overall, these findings provide an unusual ‘caveat emptor’ warning of the dangers that underpurified proteins harbor for macromolecular crystallographers.
2. Materials and methods
2.1. Protein purification for crystallization
Murine cortactin (gene ID 13043) SH3 domain (Ser487CTN–Gln546CTN; 60 amino acids; 6.8 kDa) was subcloned into pGEX-6p-1 using BamHI and XhoI restriction-enzyme sites (we have used the superscript nomenclature CTN throughout for cortactin residues, ARG for Arg residues and LYS for lysozyme residues). Initial protein expression and purification were carried out in E. coli BL21 (DE3) cells induced at 293 K overnight with 0.2 mM IPTG. After induction, the cells were pelleted, resuspended and co-lysed using a freeze–thaw protocol and sonication in the presence/absence of 1 mg ml−1 lysozyme. Following sonication and centrifugation (20 000g) at 277 K for 30 min, the resulting supernatant was incubated with glutathione Sepharose 4B beads (GE) for 4 h at 277 K. 20 bed volumes of PBS buffer were then used to wash the resin prior to incubation with PreScission protease at 277 K overnight to remove the GST tag, leaving a five-residue Gly-Pro-Leu-Gly-Ser non-native N-terminal sequence. Cortactin SH3 protein was further purified on a Superdex 75 column in 20 mM HEPES pH 7.5 buffer containing 100 mM NaCl, 1 mM EDTA and 1 mM DTT. The protein was concentrated to 10.0 mg ml−1 for crystallization.
2.2. Crystallization, data collection and refinement
Screening for crystallization conditions was conducted using a Matrix Hydra II eDrop crystallization robot (Murthy et al., 2007 ▶) and the Qiagen JCSG+ crystallization screening kit. Following the discovery of initial hits, the vapor-diffusion methodology was used to optimize the conditions. Before crystallization, murine cortactin SH3 (10 mg ml−1) was incubated with the Arg PxxP1 motif (S563SVVPYLPRLPILPSKT; 17 amino acids; 1.9 kDa) at 40 mM to give a 1:2 protein:peptide ratio. Crystals grew within one week at room temperature (298 K) by hanging-drop vapor diffusion with a drop size of 0.8 µl protein plus 0.8 µl precipitant. The best crystals grew using precipitant conditions consisting of 1.0 to 1.2 M sodium citrate pH 6.0. The crystals were flash-cooled in liquid nitrogen using the crystallization conditions prior to data collection. Crystallographic data were collected on beamline X6A of the National Synchrotron Light Source (NSLS) (Table 1 ▶) and the data were processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). Initial structure solution was obtained using Phaser (McCoy et al., 2005 ▶) by molecular replacement using the full cortactin SH3 domain (PDB entry 2d1x; Hashimoto et al., 2006 ▶) as a search model. Following complete structure determination as described in §3, the model was refined and built using REFMAC5 (Murshudov et al., 2011 ▶) and Coot (Emsley & Cowtan, 2004 ▶). The final model was validated using MolProbity (Chen et al., 2010 ▶). Structure analyses were performed using PISA (Krissinel & Henrick, 2007 ▶) and PDBsum (Laskowski, 2001 ▶). The refined model has been deposited in the PDB with accession code 3ulr.
Table 1. Crystallographic data-collection and refinement statistics.
Values in parentheses are for the highest resolution bin.
| Data collection | |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 38.7, b = 57.8, c = 95.5 |
| X-ray source | NSLS X6A |
| Wavelength (Å) | 1.0789 |
| Resolution (Å) | 20–1.65 (1.71–1.65) |
| Total reflections | 143160 |
| Unique reflections | 25122 |
| Completeness (%) | 95.0 (65.8) |
| Rmerge (%) | 8.7 (35.0) |
| 〈I/σ(I)〉 | 14.9 (2.6) |
| Multiplicity | 5.7 (2.9) |
| Wilson B factor (Å2) | 20.7 |
| Refinement statistics | |
| Resolution (Å) | 20–1.65 (1.69–1.65) |
| R factor (%) | 18.4 (32.2) |
| Free R factor (%) | 22.5 (33.3) |
| Residues built | |
| Lysozyme (chain A) | 129 [Lys19–Leu147] |
| Cortactin SH3 (chain B) | 64 [Pro483–Gln546] |
| Arg (chain C) | 13 [Ser563–Leu575] |
| Free R reflections (%) | 5.1 |
| No of free R reflections | 1277 |
| No. of non-H atoms | 1920 |
| No. of water molecules | 292 |
| Model quality | |
| R.m.s.d. bond lengths (Å) | 0.019 |
| R.m.s.d. bond angles (°) | 1.77 |
| Mean B factors (Å2) | |
| Overall | 24.2 |
| Protein atoms (chain A/B/C) | 21.5/22.1/29.4 |
| Water | 35.2 |
| Ramachandran plot (%) | |
| Favored | 99.0 |
| Allowed | 1.0 |
| Disallowed | 0.0 |
| PDB code | 3ulr |
3. Results
3.1. Structure determination
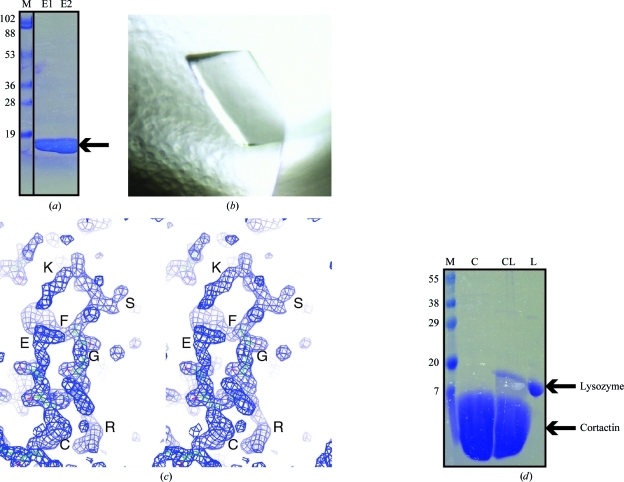
To determine the structure of cortactin SH3 domain in complex with the Arg PxxP1 motif, we began by expressing and purifying cortactin SH3 domain using standard protocols. The bacterial culture yielded around 2.0 mg protein per litre. We followed a simple purification scheme consisting of three steps: (i) affinity purification on glutathione Sepharose 4B beads, (ii) overnight cleavage on the beads by PreScission protease and (iii) size-exclusion chromatography. We were easily able to obtain cortactin SH3 domain that eluted as a monodisperse peak from size-exclusion chromatography. We concentrated this sample to 10 mg ml−1 and analyzed the purity of the sample by SDS–PAGE (Fig. 1 ▶ a).
Figure 1.
Identification of a contaminating protein. (a) SDS–PAGE (12%) analysis of cortactin SH3 purified by SEC. 10 µl of cortactin SH3 domain at 2 mg ml−1 concentration (a total of 20 µg) was loaded onto the gel. (b) Crystals of the heterotrimeric cortactin SH3–Arg peptide–lysozyme complex that formed in 1.0 M sodium citrate pH 6.0. These crystals only formed on pre-incubation of Arg PxxP1 peptide with cortactin SH3 domain. (c) Stereoview showing electron density for the initial ARP/wARP solution, allowing us to ‘sequence by crystallogaphy’. Residues KSFGRCE can clearly be identified. (d) Overloaded SDS–PAGE (15%) analysis of cortactin SH3. Lane L, lysozyme control; lane CL, 10 µl cortactin SH3 domain at 20 mg ml−1 concentration (a total of 200 µg); this is the same protein crystallization sample shown in (a); lane C, 10 µl cortactin SH3 domain at 20 mg ml−1 concentration (a total of 200 µg) obtained by following an optimized purification protocol; lane M, molecular-weight markers (labelled in kDa).
Our initial analysis of protein purity led us to believe that the sample was sufficiently pure for protein crystallization trials, so we conducted trials with cortactin SH3 domain alone. These crystallization trials yielded cortactin SH3 crystals (to be published elsewhere). We therefore decided to conduct cocrystallization trials of cortactin SH3 domain with a synthesized PxxP1 peptide derived from its binding partner, Arg. To our satisfaction, we were able to obtain a new cortactin SH3-domain crystal form only in the presence of Arg peptide; these crystals did not grow in the absence of the Arg peptide (Fig. 1 ▶ b). We therefore considered this to be a complexed crystal and optimized this crystal form for synchrotron data collection at the NSLS (beamline X6A). These complex crystals diffracted X-rays to 1.65 Å resolution and belonged to space group P212121, with unit-cell parameters a = 38.7, b = 57.8, c = 95.5 Å, α = β = γ = 90°, allowing a full data set to be collected.
We decided to use molecular replacement to solve the crystal structure using a previous structure of cortactin SH3 domain in complex with an AMAP peptide (PDB entry 2d1x; Hashimoto et al., 2006 ▶). The Matthews coefficient suggested the presence of three SH3 molecules per asymmetric unit; however, Phaser was only able to find one molecule. This first solution yielded a translation Z score of 8.9, but no second or third cortactin solutions could be determined.
Since this was a high-resolution data set, we decided to conduct automatic model building using ARP/wARP (Perrakis et al., 2001 ▶). We input the sequences for the cortactin SH3 domain and the Arg peptide and ran ARP/wARP to search for these sequences three times in the asymmetric unit. Following 200 cycles of autobuilding, ARP/wARP had built 192 residues and placed 66 in sequence. On visual inspection of the electron-density map, we found that only the first SH3 domain of cortactin (59 residues) and a portion of the Arg peptide (seven residues) fitted the map. The remaining 126 residues were built as a polyglycine trace by the sequence-assignment protocol of ARP/wARP (Cohen et al., 2004 ▶). We noted that the R and R free factors following ARP/wARP were 24.2% and 30.9%, respectively.
After multiple rounds of automated model building using ARP/wARP, both in parallel with varying parameters and sequentially, we were unable to obtain a sequence trace for more than one cortactin SH3 domain and one Arg peptide. We therefore conducted a more detailed visual inspection of the resulting electron-density map. We observed that although ARP/wARP had traced the backbone correctly in some parts of the map, it had been unable to fit side chains and so had left the trace as a polyglycine sequence. We noticed that there were some easily identifiable residues within the electron density of these stretches (Fig. 1 ▶ c). Post-ARP/wARP electron-density map sequencing has previously been conducted (Hilge et al., 2001 ▶) and we therefore decided to ‘sequence by crystallography’. We traced a peptide that looked to have the sequence KSFGRCELAAAML and then conducted a BLAST search to see if we could determine the protein that contained this sequence. BLAST returned a clear indication that this protein was lysozyme, with 11/12 identical residues to chicken egg-white lysozyme. We therefore ran Phaser to search for two molecules: the SH3 domain of cortactin (PDB entry 2d1x) and lysozyme (PDB entry 3m3u; N. K. Varshney & V. Sitaramam, unpublished work). This yielded translation Z scores of 8.7 and 32.5 for the first molecule (cortactin SH3) and second molecule (lysozyme), respectively. ARP/wARP autobuilding was able to build 193 residues. The R and R free factors following ARP/wARP were now 23.8% and 28.9%, respectively. On inspection of the resulting electron-density maps, we were able to build the bound Arg peptide, which was sandwiched between the cortactin SH3 domain and lysozyme (described below). Multiple rounds of manual refinement and model building yielded final R and R free factors of 18.4% and 22.5%, respectively. The final model was validated using MolProbity (Chen et al., 2010 ▶) and PROCHECK (Laskowski et al., 1993 ▶).
3.2. The impact of impurities
We were attempting to obtain a cocrystal of the cortactin SH3 domain and a peptide encompassing the Arg PxxP1 motif. Therefore, when we obtained crystals only once peptide had been added to the crystallization conditions, we rapidly moved to optimization and synchrotron-radiation data collection. Following the discovery that the crystals contained a contaminating lysozyme bound in a 1:1:1 fashion with cortactin SH3 and the Arg peptide, we went back to our protein-purification preparations. We conducted an overloaded SDS–PAGE analysis of the crystallization-quality protein to investigate whether we could in fact observe the contaminating lysozyme. We found that a very small amount of contaminating lysozyme had co-purified with cortactin SH3 domain through GST-affinity, tag-cleavage and size-exclusion chromatography (Fig. 1 ▶ d, left lane). Furthermore, this small amount of contamination was not observed by SDS–PAGE when the samples were not overloaded (Fig. 1 ▶ a). We then conducted a further purification of cortactin SH3 domain, this time without including lysozyme in the cell-lysis buffer. For this modified preparation, lysozyme was not observed in the final protein sample (Fig. 1 ▶ d, right lane); furthermore, this new cortactin preparation failed to cocrystallize with the Arg peptide, presumably because of the lack of lysozyme contamination. We again note that an extremely small amount of lysozyme contamination results in the cocrystallization of a 1:1:1 complex.
3.3. Structure and interface analysis
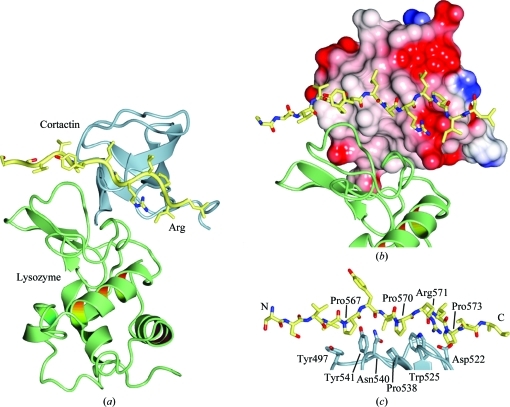
The SH3 domain of cortactin maintains a typical SH3 fold with five β-strands (βA–βE; Kaneko et al., 2008 ▶; Li, 2005 ▶) and has an r.m.s.d. of 0.8–1.1 Å over 59–61 Cα atoms when compared with the four peptide chains of the previously determined crystal structure of cortactin SH3 domain (PDB entry 2d1x; Hashimoto et al., 2006 ▶; Figs. 2 ▶ a and 2 ▶ b). Lysozyme adopts an experimentally identical conformation to that observed previously (Diamond, 1974 ▶; Diamond & Levitt, 1971 ▶; Vaney et al., 1996 ▶; Dong et al., 1999 ▶; e.g. an r.m.s.d. of 0.3 Å over 129 Cα atoms when compared with PDB entry 193l). The interfaces of the heterotrimeric cortactin–Arg–lysozyme complex were analyzed using the PISA server (Krissinel & Henrick, 2007 ▶) and using SC (Lawrence & Colman, 1993 ▶). A brief summary follows.
Figure 2.
Cortactin SH3–Arg PxxP–lysozyme complex structure. (a) Overall view of the cortactin SH–Arg PxxP1–lysozyme heterotrimeric complex structure. Cortactin is shown in blue, Arg in yellow and lysozyme in green. (b) Electrostatic potential of the surface of cortactin. Arg binds to the hydrophobic PxxP1 binding surface of the SH3 domain. Lysozyme packs against both a hydrophobic patch on cortactin and Arg PxxP1. (c) Close-up view of the interaction between Arg PxxP1 and the cortactin SH3 domain, highlighting residues discussed in the text. The images were generated with CCP4mg (Potterton et al., 2003 ▶).
3.3.1. Cortactin–Arg
Previous work has indicated that this interaction is the physiologically relevant interaction in the cortactin–Arg complex (Lapetina et al., 2009 ▶). The interaction between cortactin and the Arg peptide adopts a type II-like SH3–PPII helix interaction, buries a total of 789 Å2 and encompasses 12 residues from cortactin and 11 residues from Arg. Interestingly, three Arg proline residues are involved in the interaction with cortactin: Pro567ARG, Pro570ARG and Pro573ARG. Pro567ARG stacks against cortactin tyrosines Tyr497CTN and Tyr541CTN, Pro570ARG stacks against Asn540CTN, Pro538CTN and Trp525CTN, and Pro573ARG stacks against Trp525CTN and Asp522CTN (Fig. 2 ▶). A salt bridge is made between cortactin residue Asp522CTN and Arg residue Arg571ARG (Fig. 2 ▶ c). A total of seven hydrogen bonds are also made in the interaction. The shape complementarity for this interface is 0.85 and the PISA complexation significance score (CSS), a measure of predicted physiological relevance, is 1.000, indicating that the interface should be physiologically relevant.
3.3.2. Lysozyme–cortactin
We built the major interaction between lysozyme and cortactin SH3 domain as the asymmetric unit interaction. This interaction buries a total of 973 Å2 and encompasses 14 residues from cortactin and 16 residues from lysozyme that make an interface with five hydrogen bonds. The shape complementarity for this interaction is 0.59. The PISA CSS score for this interface is 0.031. Overall, we believe this interaction reflects a crystal-packing artifact.
3.3.3. Lysozyme–Arg
Analysis of the interaction between lysozyme and the Arg peptide in the asymmetric unit reveals an interaction that buries 393 Å2 and that yields a PISA CSS score of 0.017. For analytical purposes, when we treat the cortactin–Arg complex as a single protein (i.e. a cortactin–Arg chimera) we find that the buried surface area for the interaction with lysozyme is 1257 Å2, encompassing 21 residues from the cortactin–Arg chimera and 18 from lysozyme. This interface yields a PISA CSS score of 0.100 and a shape complementarity of 0.61. When compared with previous analyses of protein–protein interactions, this surface falls within the range of predominantly crystallographic interactions (Bahadur et al., 2004 ▶).
4. Discussion
All crystallographers know that contaminating proteins (e.g. GST, TEV, lysozyme etc.) can result in misleading crystallization hits. This manuscript, however, describes to our knowledge the first case of a contaminating lysozyme facilitating the cocrystallization of a heterotrimeric complex with the target proteins of interest.
One of the first tasks that a crystallographer should undertake when a new crystal form is obtained is a search of the PDB for structures with similar unit-cell parameters. This can sometimes save a significant amount of time and effort if the crystallized protein is in fact a common contaminant. In the case presented in this manuscript, however, the typical search of the PDB did not yield a positive hit. Furthermore, molecular replacement found what we thought was the major component of this crystal (cortactin SH3 domain).
We were aided in our discovery that the contaminating protein was lysozyme by two factors. Firstly, the resolution was high enough to use the ARP/wARP autobuilding software to ‘bootstrap’ from our initial molecular-replacement solution of a single SH3 domain. Secondly, the high-resolution data and reasonable phases from ARP/wARP enabled us to ‘sequence by crystallography’ and discover the identity of the contaminating protein.
So what lessons should one learn from this example? Firstly, an overloaded gel should be run for crystallization samples to highlight the presence of contaminating proteins; as shown in Fig. 1 ▶, samples that can look clean may contain co-purifying contaminants. Secondly, lysozyme, or potentially other contaminating proteins, may be able to co-purify through multiple steps of purification. Again, adequate diagnostics at the SDS–PAGE level should be conducted. Thirdly, these heterotrimeric crystals were only obtained when both binding partners (cortactin SH3 and Arg peptide) were present, leading us to assume the presence of a complex containing both proteins. As these crystals diffracted well, we neglected to run a diagnostic gel of the crystals; however, this would have indicated the presence of the contamination. Finally, and unsurprisingly in this often serendipitous field where molecules can sometimes ‘come along for the ride’ (Evdokimov et al., 2001 ▶; Chen et al., 2002 ▶), structure determination frequently requires the rational intervention of a crystallographer. Overall, this study provides both an unusual ‘caveat emptor’ warning of the dangers that underpurified proteins harbor for macromolecular crystallographers and an optimistic example of ‘contra felicem vix deus vires habet’.
Supplementary Material
PDB reference: cortactin–Arg–lysozyme complex complex, 3ulr
Acknowledgments
Vivian Stojanoff and Jean Jakoncic of NSLS beamline X6A are thanked. This research was supported in part by a Pilot Grant from the Yale Comprehensive Cancer Center and pilot grants from the Women’s Health Research at Yale program and the Connecticut Breast Health Initiative.
References
- Bahadur, R. P., Chakrabarti, P., Rodier, F. & Janin, J. (2004). J. Mol. Biol. 336, 943–955. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L. & Hughson, F. M. (2002). Nature (London), 415, 545–549. [DOI] [PubMed]
- Cohen, S. X., Morris, R. J., Fernandez, F. J., Ben Jelloul, M., Kakaris, M., Parthasarathy, V., Lamzin, V. S., Kleywegt, G. J. & Perrakis, A. (2004). Acta Cryst. D60, 2222–2229. [DOI] [PubMed]
- Diamond, R. (1974). J. Mol. Biol. 82, 371–391. [DOI] [PubMed]
- Diamond, R. & Levitt, M. (1971). Biochem. J. 125, 92P. [DOI] [PMC free article] [PubMed]
- Dong, J., Boggon, T. J., Chayen, N. E., Raftery, J., Bi, R.-C. & Helliwell, J. R. (1999). Acta Cryst. D55, 745–752. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evdokimov, A. G., Anderson, D. E., Routzahn, K. M. & Waugh, D. S. (2001). J. Mol. Biol. 305, 891–904. [DOI] [PubMed]
- Hashimoto, S. et al. (2006). Proc. Natl Acad. Sci. USA, 103, 7036–7041.
- Hilge, M., Perrakis, A., Abrahams, J. P., Winterhalter, K., Piontek, K. & Gloor, S. M. (2001). Acta Cryst. D57, 37–43. [DOI] [PubMed]
- Kaneko, T., Li, L. & Li, S. S.-C. (2008). Front. Biosci. 13, 4938–4952. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Lapetina, S., Mader, C. C., Machida, K., Mayer, B. J. & Koleske, A. J. (2009). J. Cell Biol. 185, 503–519. [DOI] [PMC free article] [PubMed]
- Laskowski, R. A. (2001). Nucleic Acids Res. 29, 221–222. [DOI] [PMC free article] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Lawrence, M. C. & Colman, P. M. (1993). J. Mol. Biol. 234, 946–950. [DOI] [PubMed]
- Lesley, S. A. (2001). Protein Expr. Purif. 22, 159–164. [DOI] [PubMed]
- Li, S. S.-C. (2005). Biochem. J. 390, 641–653. [DOI] [PMC free article] [PubMed]
- MacGrath, S. M. & Koleske, A. J. (2012). In the press.
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Murthy, T., Wang, Y., Reynolds, C. & Boggon, T. J. (2007). J. Assoc. Lab. Autom. 12, 213–218.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Perrakis, A., Harkiolaki, M., Wilson, K. S. & Lamzin, V. S. (2001). Acta Cryst. D57, 1445–1450. [DOI] [PubMed]
- Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. (2003). Acta Cryst. D59, 1131–1137. [DOI] [PubMed]
- Vaney, M. C., Maignan, S., Riès-Kautt, M. & Ducruix, A. (1996). Acta Cryst. D52, 505–517. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: cortactin–Arg–lysozyme complex complex, 3ulr