S-Ribosylhomocysteinase (LuxS) encoded by the LuxS gene from Streptococcus mutans was solubly expressed in Escherichia coli, purified and crystallized. Diffraction by the crystal extended to 2.4 Å resolution.
Keywords: Streptococcus mutans, LuxS
Abstract
S-Ribosylhomocysteinase (LuxS) encoded by the luxS gene from Streptococcus mutans plays a crucial role in the quorum-sensing system. LuxS was solubly expressed in Escherichia coli with high yield. The purity of the purified target protein, which was identified by SDS–PAGE and MALDI–TOF MS analysis, was >95%. The protein was crystallized using the hanging-drop vapour-diffusion method with PEG 3350 as the primary precipitant. X-ray diffraction data were collected at Beijing Synchrotron Radiation Facility (BSRF). Diffraction by the crystal extended to 2.4 Å resolution and the crystal belonged to space group C2221, with unit-cell parameters a = 55.3, b = 148.7, c = 82.8 Å.
1. Introduction
Dental caries is a bacterial infection affecting dental hard tissue and is recognized as one of the most common diseases in humans, especially in children. Accumulating evidence from bacteriological studies and animal models has confirmed that Streptococcus mutans is the principal cariogenic bacterium (Loesche, 1986 ▶; Loesche et al., 1982 ▶). The virulence properties of S. mutans can be attributed to three major mechanisms: adhesion, acidogenicity and aciduricity (Banas, 2004 ▶). This primary oral pathogen can metabolize dietary carbohydrates and secrete acids. The acids (mainly lactate) reduce the plaque micro-environmental pH (Loesche, 1986 ▶). The resulting perennial low pH in the oral environment leads to carious lesions. At the same time, this accumulation of acidic metabolites would be lethal if S. mutans had no powerful ability to withstand them. The fact that S. mutans is able to survive in a sustained acidic microenvironment signifies that acid resistance is one virulence property (Hamilton & Buckley, 1991 ▶). The acid-resistance mechanisms focus on the protection and repair of macromolecules, including DNA, alterations of metabolic pathways, secondary metabolism, cell density, biofilm formation and regulatory systems, and intracellular pH homeostasis (Matsui & Cvitkovitch, 2010 ▶).
Bacteria often survive in nature as sessile communities called biofilms. Bacterial biofilm formation is regulated by various gene-regulation systems, including the quorum-sensing system (Davies et al., 1998 ▶). Quorum sensing is a process of bacterial cell-to-cell communication that involves a small-molecule signal called autoinducer-2 (AI-2), which is the only common quorum-sensing system that exists in both Gram-positive and Gram-negative bacteria (Xavier & Bassler, 2003 ▶; Miller et al., 2004 ▶). LuxS is the AI-2 synthase and functions as an S-ribosylhomocysteine (SRH) cleavage enzyme. This pathway has been found in over 55 species, indicating that AI-2 provides a universal language for interspecies communication (Xavier & Bassler, 2003 ▶). AI-2 is produced from S-adenosylmethionine (SAM) in at least three enzymatic steps (Schauder et al., 2001 ▶). SAM is the main methyl donor in archaebacterial, eubacterial and eukaryotic cells. Consumption of SAM produces S-adenosylhomocysteine (SAH); SAH is then detoxified by the Pfs enzyme to yield adenine and S-ribosylhomocysteine (SRH), the conversion of which to 4,5-dihydroxy-2,3-pentanedione (DPD) and homocysteine is catalyzed by LuxS. DPD can be rearranged to AI-2 (Lewis et al., 2001 ▶; Pei & Zhu, 2004 ▶; Wnuk et al., 2009 ▶).
Studies have shown that mutations in LuxS affect the structure of the bacterial biofilm formed by S. mutans. Microscopic analysis of the structure of in vitro-grown biofilms revealed that LuxS-mutant biofilms adopted a much more granular appearance compared with the relatively smooth confluent layer observed for the wild type (Merritt et al., 2003 ▶). Wen and Burne reported similar observations, namely that inactivation of LuxS resulted in fewer, loose and hive-like biofilms on hydroxylapatite discs, especially in the presence of sucrose. The LuxS-deficient strain was more sensitive to acid killing, but could still undergo acid adaptation (Wen & Burne, 2004 ▶). The LuxS mutant also resulted in down-regulation of the ffh gene (encoding the subunits of the F1F0-ATPase, which pumps protons to maintain a neutral cytosolic pH which is linked to the ability of an organism to survive under acidic conditions; Kuhnert et al., 2004 ▶; Bender et al., 1986 ▶), the brpA gene (a transcriptional regulator that is involved in biofilm formation, autolysis and cell division; Wen et al., 2006 ▶), the recA gene, the smnA gene (AP endonuclease) and the nth gene (endonuclease) (Wen & Burne, 2004 ▶).
To date, structures of LuxS from four species (Deinococcus radiodurans, Helicobacter pylori, Haemophilus influenzae and Bacillus subtilis) have been solved (Ruzheinikov et al., 2001 ▶; Lewis et al., 2001 ▶). S. mutans LuxS shares 42, 38, 37 and 37% sequence identity to LuxS from D. radiodurans, H. pylori, H. influenzae and B. subtilis, respectively. All of the structural studies provide evidence for a Lewis acid function of the metal ion in the LuxS-catalyzed reaction and identified the catalytic function of the active-site residues. Nevertheless, the detailed structure of S. mutans LuxS remains unknown. Its three-dimensional structural information will help us to further understand its functional role in acid-resistance mechanisms.
In this study, we report the expression, crystallization and preliminary X-ray analysis of S. mutans LuxS as the first step towards determining its three-dimensional structure.
2. Materials and methods
2.1. Materials
Enzymes for recombinant DNA technology, such as Taq polymerase, T4p DNA ligase, BamHI and XhoI, were obtained from New England Biolabs. A PCR Amplification Kit (including PCR buffer and dNTP mix) was also from New England Biolabs. Plasmid Mini Kit I and DNA Quick Purify/Recover Kit were purchased from Omega Bio-Tek. Crystallization screening kits were purchased from Hampton Research.
S. mutans strain UA159 was a gift from Professor Lihong Guo of Peking University.
2.2. Primer design and PCR amplification
The primers were designed according to the nucleotide sequence of S. mutans strain UA159 (GenBank Accession No. AE014133). In order to facilitate subsequent cloning, BamHI and XhoI restriction-endonuclease sites (bold) were attached to the 5′-termini of the upstream and downstream primers, respectively: forward, 5′-CGCG GATCCATGACAAAAGAAGTTACTGTTG-3′; reverse, 5′-CCGCTCGAGTTACACTAGATGACGCTCAAAAG-3′. The polymerase chain reaction was carried out using the genomic DNA of S. mutans as a template. The PCR mixture was subjected to 35 cycles of denaturation (45 s, 367 K), annealing (45 s, 328 K) and extension (60 s, 345 K) using a DNA Thermal Cycler (Eppendorf). The PCR product was separated on an agarose gel containing 1% agarose, purified using the DNA Quick Purify/Recover Kit and digested with BamHI and XhoI overnight.
2.3. Cloning, expression and purification
The digested PCR product was recovered and cloned into the prokaryotic expression vector pGEX-6P-I. The recombinant plasmid was transformed into E. coli strain BL21 (DE3). The cells were cultured in Luria–Bertani medium containing 100 µg ml−1 ampicillin. Protein expression was induced by the addition of IPTG (0.1 mM final concentration) at 289 K when the OD600 of the culture reached approximately 0.8 and was maintained at 289 K for 16 h. The cells were harvested by centrifugation at 5000 rev min−1 for 10 min at 277 K. The pellet was resuspended in lysis buffer (1×PBS pH 7.4, 1 mM DTT and 1 mM PMSF) and homogenized by sonication. The cell lysate was cleared by centrifugation at 15 000 rev min−1 for 45 min at 277 K.
The supernatant was loaded onto a self-packaged GST affinity column (3 ml Glutathione Sepharose 4B resin; GE Healthcare) equilibrated with lysis buffer. The contaminant proteins were washed out with wash buffer (lysis buffer plus 200 mM NaCl) and monitored using Coomassie Brilliant Blue G250 solution. The fusion protein was then digested with PreScission protease at 277 K overnight. The target protein was eluted with lysis buffer. The eluant was then concentrated and desalted using buffer A (25 mM Tris–HCl pH 8.1, 1 mM DTT) on a Sephadex G-25 gel-filtration column. The desalted protein was further purified using a HiTrap Q HP (Pharmacia) column (buffer A, 25 mM Tris–HCl pH 8.1, 1 mM DTT; buffer B, 25 mM Tris–HCl pH 8.1, 500 mM NaCl, 1 mM DTT). The fractions containing the target protein were pooled and concentrated using an Ultrafree 5000 molecular-weight cutoff filter unit (Millipore) and were further purified using a Superdex-75 (Pharmacia) column (using buffer C: 10 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT). The fractions containing the target protein were pooled and concentrated to 10 mg ml−1. All purification steps were carried out at 289 K.
The purified protein was analyzed by SDS–PAGE and MALDI–TOF MS as reported elsewhere (Zhang et al., 2010 ▶). Total proteins were measured using the Bradford method (Bradford, 1976 ▶).
2.4. Crystallization
Crystallization experiments were carried out at 291 K. Initial screening was performed at 291 K in 24-well plates by the hanging-drop vapour-diffusion method using commercial screening kits from Hampton Research, including Crystal Screen, Crystal Screen 2, PEG/Ion, PEG/Ion 2 and Index. Optimization was followed by refinement of the conditions through variation of the precipitants, pH, protein concentration and additives. Typically, droplets consisting of 1 µl protein solution and an equivalent volume of reservoir solution were equilibrated against 200 µl reservoir solution.
2.5. X-ray diffraction, data collection and processing
X-ray diffraction data sets were collected using a MAR165 CCD detector (MAR Research) at Beijing Synchrotron Radiation Facility (BSRF) with a wavelength of 1.0000 Å. The crystals were immersed in a cryoprotectant solution (reservoir solution supplemented with 12% glycerol) for 5–10 s, picked up in a loop and flash-cooled in a nitrogen-gas stream at 100 K. A data set consisting of 200 frames was collected. The exposure time per frame was 0.8 s, the crystal-to-detector distance was 150 mm and the oscillation range per frame was 0.5°. All intensity data were indexed, integrated and scaled using the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
A 483 bp DNA fragment was obtained by PCR amplification. The DNA fragment was digested with BamHI and XhoI and cloned into pGEX-6P-1. The bacteria harbouring the recombinant plasmid were identified by PCR and plasmid digestion and confirmed by DNA sequencing.
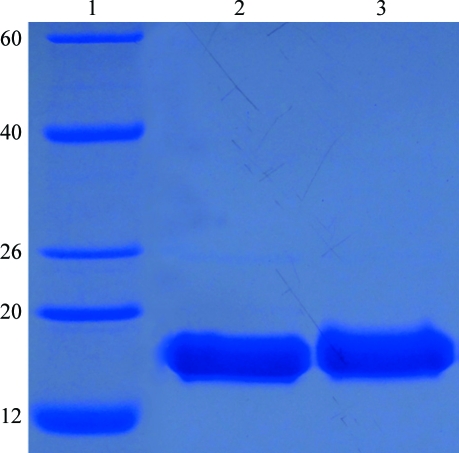
LuxS fused with an N-terminal GST tag was solubly expressed in E. coli. The fusion protein was cleaved with PreScission protease, producing the target protein with an additional five amino-acid residues (GPLGS) at the N-terminus. After a series of purification steps, the purified target protein was >95% pure on SDS–PAGE stained with Coomassie Brilliant Blue (Fig. 1 ▶). MALDI–TOF MS analysis of trypsin-digested purified protein provided convincing evidence that this protein was S. mutans LuxS (data not shown). The precise sequence of the protein that was crystallized consisted of amino-acid residues 1–160 of S. mutans LuxS and an additional five amino-acid residues (GPLGS) at the N-terminus.
Figure 1.
SDS–PAGE of purified S. mutans LuxS. Lane 1, molecular-weight markers (labelled in kDa); lanes 2 and 3, purified S. mutans LuxS used for crystallization.
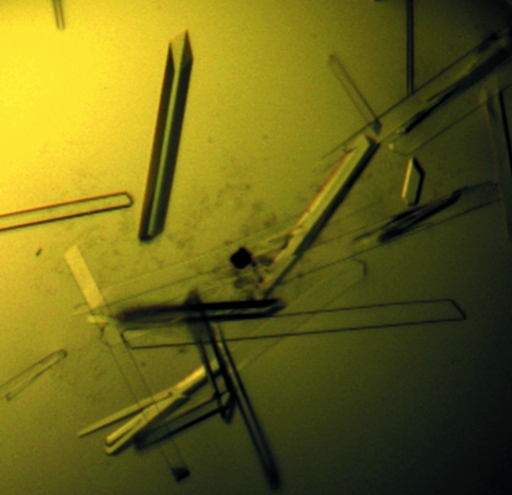
Microcrystals appeared after 3 d from one condition of PEG/Ion 2 [0.03 M citric acid, 0.07 M bis-tris propane pH 7.6, 20%(w/v) PEG 3350]. This condition was further optimized by variation of the precipitants, buffer pH and protein concentration, and larger crystals (Fig. 2 ▶) were obtained at 291 K using the vapour-diffusion method in hanging-drop mode by mixing 1 µl protein with 1 µl reservoir solution [0.1 M Tris–HCl pH 8.0, 20%(w/v) PEG 3350] and equilibrating against 200 µl reservoir solution. The larger crystals were reproducible and suitable for X-ray diffraction. The crystals were incubated in reservoir solution supplemented with 12%(v/v) glycerol.
Figure 2.
Crystals of S. mutans LuxS. The dimensions of the typical crystal used for data collection were about 30 × 30 × 550 µm.
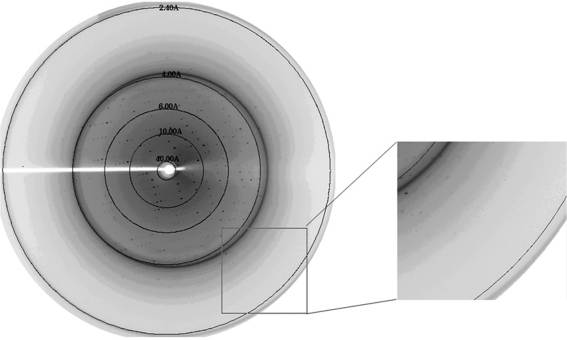
An X-ray diffraction data set was collected from a single crystal to 2.4 Å resolution (Fig. 3 ▶). The crystal belonged to space group C2221, with unit-cell parameters a = 55.3, b = 148.7, c = 82.8 Å (see Table 1 ▶). There was estimated to be one molecule per asymmetric unit, giving a Matthews coefficient V M of 2.4 Å3 Da−1 and a solvent content of 46.5%.
Figure 3.
X-ray diffraction pattern from a crystal of S. mutans LuxS. The crystal diffracted to 2.4 Å resolution.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Space group | C2221 |
| Unit-cell parameters (Å) | a = 55.3, b = 148.7, c = 82.8 |
| Wavelength (Å) | 1.0000 |
| Resolution (Å) | 50.00–2.40 (2.44–2.40) |
| No. of observed reflections | 42012 (2288) |
| No. of unique reflections | 12834 (636) |
| Multiplicity | 3.3 (3.6) |
| Completeness (%) | 96.5 (99.1) |
| 〈I/σ(I)〉 | 9.1 (2.7) |
| Rmerge† (%) | 10.2 (63.2) |
R
merge = 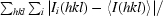
 , where 〈I
(hkl)〉 is the mean of the observations Ii(hkl) of reflection hkl.
, where 〈I
(hkl)〉 is the mean of the observations Ii(hkl) of reflection hkl.
S. mutans LuxS shares 42 and 38% sequence identity with D. radiodurans autoinducer-2 synthesis protein and H. pylori LuxS, respectively. We are now attempting to determine the structure of S. mutans LuxS by the molecular-replacement method with Phaser (McCoy et al., 2007 ▶) using D. radiodurans autoinducer-2 synthesis protein (PDB entry 1vh2 chain A; Badger et al., 2005 ▶) and H. pylori LuxS (PDB entry 1j6x chain A; Lewis et al., 2001 ▶) as models. Correct solutions are indicated by clear rotation and translation Z-scores, with initial R work and R free values of 0.38 and 0.42, respectively. Further model building and refinement are under way.
The three-dimensional structure of S. mutans LuxS will help us to further understand its specific functions in caries and may also be a good target for the rational design of anti-S. mutans drugs.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (81170945) and by the Development Program of the Science and Technology Department, Jilin Province (200905177). The diffraction data were collected at Beijing Synchrotron Radiation Facility (BSRF). We are grateful to Professor Zhiyong Lou of Tsinghua University for his technical assistance with data collection and processing, valuable comments and critical discussion.
References
- Badger, J. et al. (2005). Proteins, 60, 787–796. [DOI] [PubMed]
- Banas, J. A. (2004). Front. Biosci. 9, 1267–1277. [DOI] [PubMed]
- Bender, G. R., Sutton, S. V. & Marquis, R. E. (1986). Infect. Immun. 53, 331–338. [DOI] [PMC free article] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H., Costerton, J. W. & Greenberg, E. P. (1998). Science, 280, 295–298. [DOI] [PubMed]
- Hamilton, I. R. & Buckley, N. D. (1991). Oral Microbiol. Immunol. 6, 65–71. [DOI] [PubMed]
- Kuhnert, W. L., Zheng, G., Faustoferri, R. C. & Quivey, R. G. (2004). J. Bacteriol. 186, 8524–8528. [DOI] [PMC free article] [PubMed]
- Lewis, H. A., Furlong, E. B., Laubert, B., Eroshkina, G. A., Batiyenko, Y., Adams, J. M., Bergseid, M. G., Marsh, C. D., Peat, T. S., Sanderson, W. E., Sauder, J. M. & Buchanan, S. G. (2001). Structure, 9, 527–537. [DOI] [PubMed]
- Loesche, W. J. (1986). Microbiol. Rev. 50, 353–380. [DOI] [PMC free article] [PubMed]
- Loesche, W. J., Syed, S. A., Laughon, B. E. & Stoll, J. (1982). J. Periodontol. 53, 223–230. [DOI] [PubMed]
- Matsui, R. & Cvitkovitch, D. (2010). Future Microbiol. 5, 403–417. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Merritt, J., Qi, F., Goodman, S. D., Anderson, M. H. & Shi, W. (2003). Infect. Immun. 71, 1972–1979. [DOI] [PMC free article] [PubMed]
- Miller, S. T., Xavier, K. B., Campagna, S. R., Taga, M. E., Semmelhack, M. F., Bassler, B. L. & Hughson, F. M. (2004). Mol. Cell, 15, 677–687. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pei, D. & Zhu, J. (2004). Curr. Opin. Chem. Biol. 8, 492–497. [DOI] [PubMed]
- Ruzheinikov, S. N., Das, S. K., Sedelnikova, S. E., Hartley, A., Foster, S. J., Horsburgh, M. J., Cox, A. G., McCleod, C. W., Mekhalfia, A., Blackburn, G. M., Rice, D. W. & Baker, P. J. (2001). J. Mol. Biol. 313, 111–122. [DOI] [PubMed]
- Schauder, S., Shokat, K., Surette, M. G. & Bassler, B. L. (2001). Mol. Microbiol. 41, 463–476. [DOI] [PubMed]
- Wen, Z. T., Baker, H. V. & Burne, R. A. (2006). J. Bacteriol. 188, 2983–2992. [DOI] [PMC free article] [PubMed]
- Wen, Z. T. & Burne, R. A. (2004). J. Bacteriol. 186, 2682–2691. [DOI] [PMC free article] [PubMed]
- Wnuk, S. F., Robert, J., Sobczak, A. J., Meyers, B. P., Malladi, V. L., Zhu, J., Gopishetty, B. & Pei, D. (2009). Bioorg. Med. Chem. 17, 6699–6706. [DOI] [PMC free article] [PubMed]
- Xavier, K. B. & Bassler, B. L. (2003). Curr. Opin. Microbiol. 6, 191–197. [DOI] [PubMed]
- Zhang, L., Xiang, H., Gao, J., Hu, J., Miao, S., Wang, L., Deng, X. & Li, S. (2010). Protein Expr. Purif. 69, 204–208. [DOI] [PubMed]