Native and selenomethionine-derivatized (SeMet) crystals of Bacillus subtilis YwfE in the presence of ADP, MgCl2 and the dipeptide l-Ala-l-Gln were obtained using the hanging-drop vapour-diffusion method.
Keywords: l-amino-acid ligases, ATP-grasp fold
Abstract
Bacillus subtilis YwfE, an l-amino-acid ligase, catalyzes the formation of an α-dipeptide from l-amino acids in an ATP-dependent manner. In order to elucidate the substrate-recognition mode and the reaction mechanism of this ligase, native and selenomethionine-derivatized (SeMet) crystals of YwfE in the presence of ADP, MgCl2 and the dipeptide l-Ala-l-Gln were obtained using the hanging-drop vapour-diffusion method. These crystals diffracted to 1.9 and 2.8 Å resolution, respectively. Preliminary SAD phase calculations using the data set from the SeMet crystal suggested that the crystal belonged to the hexagonal space group P6522, with unit-cell parameters a = b = 90.85, c = 250.31 Å, and contained one molecule in the asymmetric unit with a solvent content of 57.3%.
1. Introduction
Peptide biosynthesis is carried out by the ribosome in all living organisms. In some bacteria and fungi, nonribosomal peptide synthetase (NRPS) produces a variety of bioactive peptides such as antibiotics and siderophores. In addition to these machineries, there are other methods by which enzymes catalyze the formation of a peptide bond. d-Alanyl-d-alanine ligase (Ddl; EC 6.3.2.4), one of the key enzymes in the peptidoglycan biosynthetic pathway, ligates two d-Ala molecules to produce a d-Ala-d-Ala dipeptide in an ATP-dependent manner (Fan et al., 1994 ▶). Glutathione synthetase (EC 6.3.2.3) catalyzes the second step in the biosynthesis of glutathione from γ-glutamylcysteine and glycine by the cleavage of ATP to ADP and phosphate (Polekhina et al., 1999 ▶). Glutamine synthetase (EC 6.3.1.2) catalyzes a condensation reaction of glutamate and ammonia to form glutamine, with concomitant hydrolysis of ATP (Eisenberg et al., 2000 ▶). These enzymes contain a unique nucleotide-binding fold referred to as an ATP-grasp fold. In addition, there are enzymes that contain the ATP-grasp fold that catalyze reactions other than amino-acid and peptide biosynthesis reactions: for example, carbamoyl-phosphate synthetase in the arginine and pyrimidine biosynthetic pathway (Holden et al., 1999 ▶), four of the 15 purine-biosynthetic enzymes (PurD, PurT, PurK and PurP; Zhang et al., 2008 ▶) and some others. These members of the ATP-grasp superfamily of enzymes catalyze the ATP-dependent ligation of a carboxylate-containing molecule to an amino or thiol group-containing molecule in a number of unrelated biosynthetic pathways, accepting a variety of substrates (Galperin & Koonin, 1997 ▶). The formation of an acyl-phosphate intermediate to activate the carboxyl substrate is thought to be a common feature of this superfamily. Recently, a new member of this superfamily was identified by in silico screening using the ATP-grasp motif and classified into a new enzyme category as an l-amino-acid ligase (EC 6.3.2.28) which catalyzes the formation of an α-peptide bond from l-amino acids (Tabata et al., 2005 ▶).
YwfE from Bacillus subtilis, the first reported l-amino-acid ligase, catalyzes the formation of the dipeptide antibiotic bacilysin, which consists of an l-Ala at the N-terminus and a non-proteinogenic amino acid, l-anticapsin, at the C-terminus. Its enzymatic properties have been investigated using recombinant protein and it has been reported that YwfE can produce 44 kinds of dipeptides, such as l-Ala-l-Gln, from proteinogenic l-amino acids, while it did not react with highly charged amino acids or d-amino acids (Tabata et al., 2005 ▶). In contrast, RizA and RizB from B. subtilis NBRC3134 have been reported to produce dipeptides with a charged amino acid, l-Arg, at the N-terminus (Kino et al., 2009 ▶) and various oligopeptides consisting of 2–5 l-amino acids (Kino et al., 2010 ▶). Both enzymes were thought to be involved in the biosynthetic pathway of the peptide antibiotic rhizocticin. Most recently, several l-amino-acid ligases have been identified from bacterial genomes and showed different substrate specificities (Arai & Kino, 2010 ▶). However, little is known regarding the distinct substrate-specificity mechanisms of l-amino-acid ligases owing to a lack of structural information.
In order to obtain insight into the structural properties of the substrate-binding mode of l-amino-acid ligase, we aimed to determine the crystal structure of YwfE, the best studied l-amino-acid ligase, with bound ligands. Structural comparison of YwfE with other ATP-grasp enzymes, especially Ddl and its homologues, will provide detailed information about the substrate recognition and reaction mechanism of l-amino-acid ligases. Furthermore, l-amino-acid ligase could become an attractive tool for enzymatic synthesis of commercially useful peptides consisting of l-amino acids. For example, the artificial sweetener aspartame is composed of a dipepteide of l-amino acids (l-aspartyl-l-phenylalanine methyl ester). l-Ala-l-Gln is used as a component of patient infusions because of its higher solubility compared with l-Gln alone. In this communication, we describe the overproduction, purification, crystallization and preliminary X-ray analysis of YwfE. We also report data suggesting that YwfE forms a dimer in solution.
2. Materials and methods
2.1. Protein preparation
The genes encoding full-length YwfE (Met1–Val472) and a deletion mutant (Lys4–Tyr468) were PCR-amplified from genomic DNA of B. subtilis, which was obtained from the Riken Bioresource Center (Tsukuba, Japan). The amplified PCR products were cloned into the pGEX-6P-1 expression vector (GE Healthcare) with BamHI and XhoI sites to produce an N-terminal GST-fusion protein. Escherichia coli strain ArcticExpress RIL cells (DE3) (Stratagene) containing the expression plasmid were grown in Luria–Bertani medium containing 50 µg ml−1 ampicillin at 303 K. When the optical density at 600 nm of the culture reached ∼0.5, expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.3 mM. After cultivation for 18 h at 298 K, the cells were harvested by centrifugation at 5000g for 15 min, suspended in phosphate-buffered saline (PBS) containing 1 mM dithiothreitol (DTT) and lysed by sonication. The lysate was centrifuged at 10 000g for 20 min and the supernatant obtained was loaded onto a Glutathione Sepharose FF column (GE Healthcare) pre-equilibrated with PBS containing 1 mM DTT. The GST-fused YwfE was eluted with 50 mM Tris–HCl pH 8.0, 50 mM NaCl, 1 mM DTT, 15 mM reduced glutathione. The GST tag was cleaved using PreScission protease (GE Healthcare) and reduced glutathione was removed by dialysis against 50 mM Tris–HCl pH 8.0, 50 mM NaCl and 1 mM DTT. The cleaved GST tag was then removed by chromatography on a Glutathione Sepharose FF column (GE Healthcare). The flowthrough fractions were concentrated by ultrafiltration using Vivaspin-20 10 000 MWCO (Sartorius) and loaded onto a HiLoad 16/60 Superdex 75 pg column (GE Healthcare) pre-equilibrated with PBS and 150 mM NaCl. Fractions containing pure YwfE were pooled, dialyzed extensively against 20 mM Tris–HCl pH 8.0 containing 1 mM DTT and concentrated to ∼20 mg ml−1 using Vivaspin-20 10 000 MWCO. The protein concentration was estimated by measuring the absorbance at 280 nm employing a molecular extinction coefficient of 38 850 M −1 cm−1. SeMet-substituted YwfE was overproduced in E. coli strain B834 (DE3) (Invitrogen) cultured in Core medium (Wako) with 50 mg l−1 l-selenomethionine and was purified using the same procedure as described above.
2.2. Analysis of the oligomeric state of YwfE
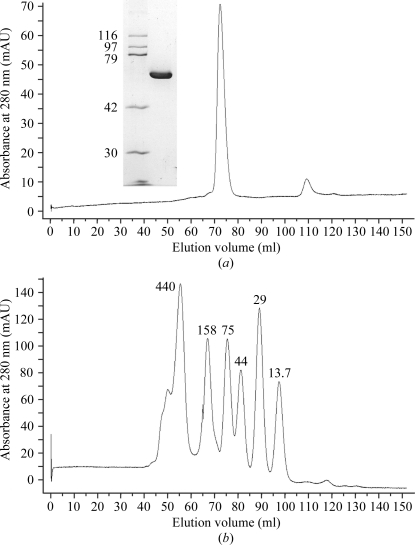
The molecular mass of full-length YwfE in solution was estimated by size-exclusion chromatography. About 500 µg purified YwfE (2.5 mg ml−1) was loaded onto a HiLoad 16/60 Superdex 200 pg column (GE Healthcare) pre-equilibrated with 50 mM Tris–HCl pH 8.0 and 100 mM NaCl at a flow rate of 1 ml min−1. The eluate was monitored by absorbance at 280 nm. The molecular mass was estimated by comparison with protein standards using LMW and HMW gel-filtration calibration kits (GE Healthcare): ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (44 kDa), carbonic anhydrase (29 kDa) and ribonuclease A (13.7 kDa). Blue dextran 2000 was used to identify the void volume of the column.
2.3. Crystallization
Crystallization screening was performed using the hanging-drop vapour-diffusion method at 293 K. Initial crystallization trials were carried out using the commercial crystallization reagent kits JCSG Core I, II, III and IV Suites from Qiagen. Drops were prepared by manually mixing 2 µl protein solution (8 mg ml−1) containing 10 mM ADP, 10 mM MgCl2, 100 mM l-Ala-l-Gln with 2 µl reservoir solution and were equilibrated against 500 µl reservoir solution. Both full-length and deletion-mutant YwfE (Lys4–Tyr468) crystals appeared in 3 d in screen conditions consisting of 20%(w/v) polyethylene glycol (PEG) in the presence of various salts at 0.2 M. After optimizing the conditions, deletion-mutant crystals were grown in reservoir solution consisting of 17%(w/v) PEG 3350, 0.3 M NaCl, 0.1 M MES–NaOH pH 6.75, 5%(v/v) ethylene glycol, 1 mM DTT. SeMet-YwfE (Lys4–Tyr468) crystals were obtained under similar conditions to those used for the native protein.
2.4. X-ray data collection and processing
Diffraction data were collected on the BL-5A beamline at the Photon Factory (PF; Tsukuba, Japan) using an ADSC Quantum 210 CCD detector. 20%(v/v) ethylene glycol was used as a cryoprotectant for data collection under cryogenic conditions. The crystals were soaked in cryoprotectant-containing mother liquor with ADP, MgCl2 and l-Ala-l-Gln and were flash-cooled in a nitrogen stream at 100 K. Totals of 230 and 360 diffraction images were collected with oscillation steps of 0.25 and 1.0°, respectively, for the native and SeMet data sets. The diffraction data sets were processed with HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
Recombinant full-length YwfE (Met1–Val472) from B. subtilus was expressed in E. coli and purified by two chromatographic steps, with a yield of ∼9 mg purified protein from 1 l culture. SDS–PAGE analysis of the purified protein showed a single band of molecular mass ∼55 kDa (Fig. 1 ▶), which is close to the calculated molecular mass of 52 578 Da with an extra N-terminal sequence (Gly-Pro-Leu-Gly-Ser) remaining after removal of the GST tag. To analyze the oligomeric state of the enzyme, purified protein was separated on a Superdex 200 gel-filtration column under low ionic strength conditions and the molecular mass was estimated with respect to the standard proteins (Fig. 1 ▶). The protein eluted at a position corresponding to a molecular mass of about 105 kDa, suggesting that YwfE forms a dimer in solution. Thus, the possibility arises that the quaternary structure of YwfE is the same as those of other amino-acid ligases which catalyze the formation of an α-peptide bond, such as Ddl and RizA.
Figure 1.
Elution profiles from a Superdex 200 gel-filtration column (GE Healthcare) of (a) YwfE and (b) molecular-mass standard proteins in 50 mM Tris–HCl pH 8.0 and 0.15 M NaCl. Inset in (a): SDS–PAGE (10% acrylamide gel) analysis of purified YwfE protein (3 µg) with Coomassie Blue staining. The molecular masses of the standard proteins are indicated in kDa.
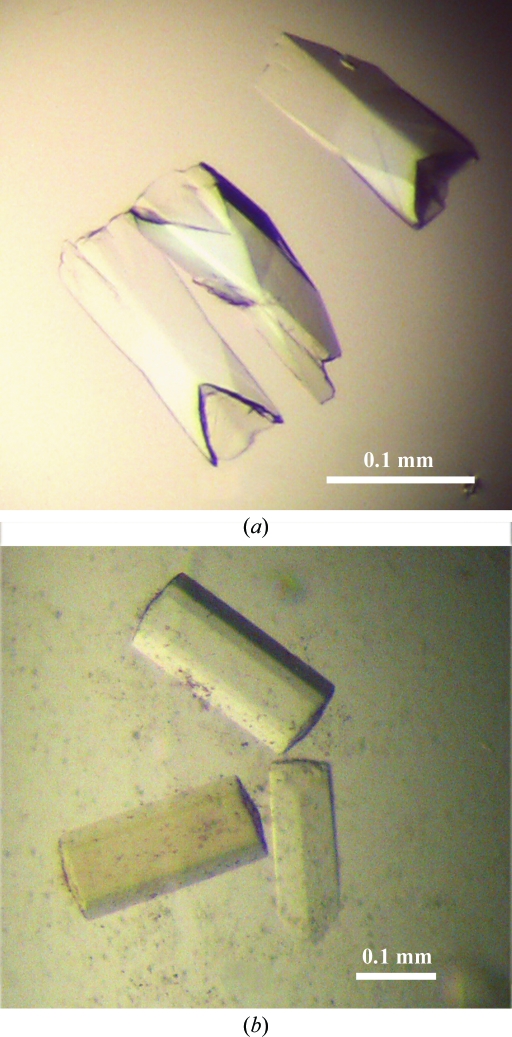
In the initial crystallization screen of full-length YwfE in the presence of ADP, MgCl2 and l-Ala-l-Gln, clusters of needle-shaped crystals appeared in 3 d. Most of the conditions contained 20%(w/v) PEG with 0.2 M salt. After optimization of the conditions, crystals with maximum dimensions of 150 × 50 × 50 µm were obtained in a week (Fig. 2 ▶). However, these crystals diffracted very weakly to 8.0 Å resolution using a synchrotron X-ray source. In order to obtain higher resolution data sets, we generated a deletion mutant of YwfE (Lys4–Tyr468). This construct was designed to remove an expected flexible sequence in the N- and C-termini based on the results of secondary-structure prediction. After optimization of the crystallization conditions using the mutant protein, hexagonal crystals were obtained with dimensions of 200 × 100 × 100 µm in a week (Fig. 2 ▶) using a reservoir solution consisting of 17%(w/v) PEG 3350, 0.3 M NaCl, 0.1 M MES–NaOH pH 6.75, 5%(v/v) ethylene glycol, 1 mM DTT. The crystallization conditions were almost the same as those for the full-length construct. The addition of ethylene glycol as an additive reagent improved the quality of the crystals, while the addition of glycerol or PEG 400 had little effect on crystallization. Diffraction data were collected on PF BL-5A at a wavelength of 1.0000 Å using an ADSC Quantum 210 CCD detector. The crystals belonged to the hexagonal system, with space group P6122 or P6522 and unit-cell parameters a = b = 90.85, c = 250.31 Å. Using the method of Matthews (1968 ▶), the solvent content of the crystal was calculated to be 57.3%, with a Matthews coefficient of 2.88 Å3 Da−1, assuming the presence of one molecule in the asymmetric unit. The data set was collected and processed to 1.9 Å resolution; the data-collection statistics are summarized in Table 1 ▶. We have not yet obtained a crystal of YwfE in the presence of adenosine 5′-(β,γ-methylene)triphosphate (AMP-PCP) with l-Ala.
Figure 2.
Native crystals of (a) full-length (Met1–Val472) and (b) deletion-mutant (Lys4–Tyr468) YwfE.
Table 1. Diffraction data statistics of YwfE crystals.
Values in parentheses are for the highest resolution shell.
| Native YwfE | SeMet YwfE | |
|---|---|---|
| Wavelength (Å) | 1.0000 | 0.9791 |
| Space group | P6522 | P6522 |
| Unit-cell parameters (Å) | a = b = 90.85, c = 250.31 | a = b = 90.50, c = 249.91 |
| Resolution (Å) | 30–1.9 (1.94–1.90) | 30–2.8 (2.85–2.80) |
| Total reflections | 206504 | 612297 |
| Unique reflections | 47336 (3201) | 28009 (1405) |
| Completeness (%) | 96.1 (100.0) | 99.5 (100.0) |
| Multiplicity | 4.4 (4.9) | 21.9 (23.0) |
| 〈I/σ(I)〉 | 35.8 (5.0) | 67.6 (20.3) |
| Rmerge† | 0.050 (0.346) | 0.078 (0.359) |
R
merge = 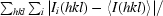
 , where Ii(hkl) is the ith measurement of the intensity of reflection hkl and 〈I(hkl)〉 is the mean value of reflection hkl.
, where Ii(hkl) is the ith measurement of the intensity of reflection hkl and 〈I(hkl)〉 is the mean value of reflection hkl.
For phasing by anomalous dispersion using the selenium edge, we also produced SeMet-labelled YwfE (Lys4–Tyr468) crystals in the same manner as for the native protein. A SAD data set was collected on PF BL-5A at a wavelength of 0.97910 Å using an ADSC Quantum 210 CCD detector. The crystals belonged to the hexagonal system, with space group P6122 or P6522 and unit-cell parameters a = b = 90.50, c = 249.91 Å. The obtained unit-cell parameters were almost the same as those of the native crystal. Structure determination was performed using the SAD method as implemented in automated experimental phasing with Crank (Ness et al., 2004 ▶) from the CCP4 suite (Winn et al., 2011 ▶). Preliminary phase calculations obtained in space group P6522 gave better statistics than those in P6122. Eight Se-atom positions were identified. The resulting electron-density map was interpretable. Further refinement and model building are now in progress and structural details will be described in a separate paper.
Acknowledgments
We thank the beamline scientists at Photon Factory for assistance with data collection. This work was supported by a JGC-S Scholarship Foundation grant to TT.
References
- Arai, T. & Kino, K. (2010). Biosci. Biotechnol. Biochem. 74, 1572–1577. [DOI] [PubMed]
- Eisenberg, D., Gill, H. S., Pfluegl, G. M. & Rotstein, S. H. (2000). Biochim. Biophys. Acta, 1477, 122–145. [DOI] [PubMed]
- Fan, C., Moews, P. C., Walsh, C. T. & Knox, J. R. (1994). Science, 266, 439–443. [DOI] [PubMed]
- Galperin, M. Y. & Koonin, E. V. (1997). Protein Sci. 6, 2639–2643. [DOI] [PMC free article] [PubMed]
- Holden, H. M., Thoden, J. B. & Raushel, F. M. (1999). Cell. Mol. Life Sci. 56, 507–522. [DOI] [PMC free article] [PubMed]
- Kino, K., Arai, T. & Tateiwa, D. (2010). Biosci. Biotechnol. Biochem. 74, 129–134. [DOI] [PubMed]
- Kino, K., Kotanaka, Y., Arai, T. & Yagasaki, M. (2009). Biosci. Biotechnol. Biochem. 73, 901–907. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Ness, S. R., de Graaff, R. A., Abrahams, J. P. & Pannu, N. S. (2004). Structure, 12, 1753–1761. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–325. [DOI] [PubMed]
- Polekhina, G., Board, P. G., Gali, R. R., Rossjohn, J. & Parker, M. W. (1999). EMBO J. 18, 3204–3213. [DOI] [PMC free article] [PubMed]
- Tabata, K., Ikeda, H. & Hashimoto, S. (2005). J. Bacteriol. 187, 5195–5202. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zhang, Y., Morar, M. & Ealick, S. E. (2008). Cell. Mol. Life Sci. 65, 3699–3724. [DOI] [PMC free article] [PubMed]