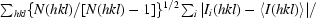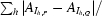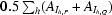CPIP1 from N. tabacum (NtCPIP1) is a plant heat-shock 40-type protein that participates in effector–host interactions during potyviral infections. Whereas crystals of full-length wild-type NtCPIP1 only diffracted to 8.0 Å resolution, diffraction from crystals of a shortened variant lacking the first 127 amino acids extended to 2.4 Å resolution.
Keywords: plant heat-shock protein 40, viral infections, potyviruses
Abstract
Chaperones promote many different molecular processes, including the folding, targeting and degradation of proteins. The best-studied chaperone system consists of the Hsp70s and their co-chaperones the Hsp40s. Chaperone function can be hijacked by viruses in plants. Potato virus Y interacts via its coat protein with an Hsp40 from Nicotiana tabacum, referred to as NtCPIP1, in order to regulate replication. To understand the molecular determinants of this mechanism, different variants of NtCPIP1 were expressed, purified and crystallized. While crystals of wild-type NtCPIP1 diffracted to 8.0 Å resolution, the deletion mutant NtCPIP1-Δ(1:127) crystallized in space group P21212 and diffracted to 2.4 Å resolution.
1. Introduction
Chaperones play a pivotal role in protein quality control and protein targeting (Bukau et al., 2006 ▶; Hartl et al., 2011 ▶). Chaperones not only segregate misfolded proteins from properly folded proteins, but their ability to interconvert folded and unfolded proteins places them at the centre of important regulatory crossroads such as the rescuing of mistrapped proteins, the assembly and disassembly of protein complexes and the translocation and trafficking of proteins to various compartments, as well as the targeting of proteins for degradation. Chaperones can be grouped into several major families, small heat-shock proteins (Hsps), Hsp40, Hsp70, Hsp90 and Hsp110, which are traditionally distinguished by their migration behaviour during polyacrylamide gel electrophoresis (Hartl et al., 2011 ▶). The structural insights available into the mechanism of function of the various chaperone-family members have been summarized in a recent review (Saibil, 2008 ▶).
In the Hsp70/Hsp40 system, Hsp70 toggles between a high-affinity and a low-affinity substrate-binding state, thereby helping to untangle misfolded polypeptide chains (Hartl et al., 2011 ▶). Misfolded substrates are delivered to Hsp70 by Hsp40. Hsp40 proteins contain a highly conserved J-domain of about 75 amino acids and a substrate-binding domain to which the cargo protein is attached before it is delivered to Hsp70. Depending on the location of these two domains in the Hsp40 polypeptide chain, as well as the presence of additional sequence features in the linker between these domains, Hsp40 proteins have been divided into different subfamilies (Qiu et al., 2006 ▶).
Arabidopsis thaliana encodes as many as 116 J-domain-containing proteins (Rajan & D’Silva, 2009 ▶).The number of J-domain proteins in Nicotiana tabacum is not known, but a similar number is expected. One of these, namely coat protein interacting protein 1 (NtCPIP1), caught our attention since it participates in the mechanism by which potyviruses infect plant cells (Hafrén et al., 2010 ▶; Hofius et al., 2007 ▶). Yeast two-hybrid experiments identified a specific Hsp40 protein from N. tabacum that interacts with the capsid protein of potato virus Y via its substrate-binding domain. Tobacco plants display increased tolerance towards potyviral infections when a deletion variant of NtCPIP1 that lacks the J-domain is overproduced in planta. This shows that fully functional Hsp40s are required for the infectivity and replication of potyviruses (Hofius et al., 2007 ▶). Subsequent studies have shown that NtCPIP1 enables the disassembly and subsequent degradation of the capsid protein by the Hsp70/Hsp40 system. The current hypothesis is that in the early phase of viral replication the number of coat proteins must be controlled so that the viral ssRNA can be readily replicated and transcribed without being blocked by the interaction with capsid proteins (Hafrén et al., 2010 ▶).
As a step towards understanding the role of NtCPIP1 in potyviral infections, we successfully expressed, purified and crystallized two different variants of NtCPIP1.
2. Materials and methods
2.1. Protein production and purification
The gene for NtCPIP1 from N. tabacum (UniProtKB Q6EIX9) was originally provided in a pQE9 vector (U. Sonnewald, personal communication). The gene encoding full-length wild-type NtCPIP1 (NtCPIP1-wt) was amplified using the primers 1for090309 (5′-GTCGTACCATGGGCGTTGATTAC-3′) and 1rev090309 (5′-TGATTCCTCGAGTTAGTCAACAGTCCT-3′). The gene fragment encoding amino acids from Val128 to the C-terminal residue Asp306 [NtCPIP1-Δ(1:127)] was amplified using the forward primer Nt1.4 (5′-ATATAGGCCATGGTGGAGAACAAGTTG-3′) and the reverse primer 1rev090309 via polymerase chain reaction. Both genes were cloned into a pBADM-11 vector (EMBL, Heidelberg, Germany) using the restriction enzymes NcoI and XhoI and fused to an N-terminal hexahistidine tag followed by a cleavage site for tobacco etch virus (TEV) protease. Successful cloning was confirmed by DNA-sequence analysis.
Protein production was very similar for the NtCPIP1-wt and the NtCPIP1-Δ(1:127) constructs. The vectors encoding the two variants were transformed into Escherichia coli BL21 (DE3) cells (Merck, Darmstadt, Germany). The cells were grown to an OD600 of 0.5 at 310 K in LB medium containing 100 µg ml−1 ampicillin. Protein overproduction was induced with 0.01% l-(+)-arabinose. Cells were harvested after 4 h via centrifugation and the pellet was stored at 253 K. For cell disruption by sonication, 11 g of cells was thawed and resuspended in 50 mM sodium phosphate buffer pH 8.0, 300 mM NaCl, 10 mM imidazole, 0.5 mM EDTA pH 8.0. Subsequently, one Complete protease-inhibitor cocktail tablet (Roche, Basel, Switzerland) was added. After centrifugation for 1 h at 100 000g and 277 K, the supernatant was loaded onto a 5 ml HisTrap FF column (GE Healthcare, Munich, Germany) and the target protein was eluted with a linear gradient of 30–500 mM imidazole in sodium phosphate buffer pH 8.0 containing 300 mM NaCl and 1 mM dithiothreitol (DTT). To remove the hexahistidine tag, 1 mg TEV protease was added per 100 mg NtCPIP1-wt, resulting in an NtCPIP1 protein that starts with an additional N-terminal Gly-Ala sequence. In the case of NtCPIP1-Δ(1:127) three additional amino acids remained at the N-terminus after cleavage, namely glycine, alanine and methionine. Subsequently, uncleaved protein and TEV protease were removed after passage through a HiPrep 26/10 desalting column (GE Healthcare) by a second 5 ml HisTrap FF column. After a second buffer exchange on a HiPrep 26/10 Desalting column, the proteins were additionally purified on a MonoS 5/50 GL column (GE Healthcare) using a linear gradient of 50 mM–1 M NaCl in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 8.0, 1 mM DTT. The final purification step was performed in 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM DTT using a HiLoad Superdex 200 16/60 column (GE Healthcare). Peak fractions containing pure NtCPIP1-wt or NtCPIP1-Δ(1:127) were pooled and concentrated to a suitable concentration for crystallization using Vivaspin concentrators (Vivascience, Hanover, Germany). The protein was stored at 253 K after cooling in liquid nitrogen.
2.2. Circular-dichroism spectroscopy
The spectra of both variants of NtCPIP1 were recorded using a J-815 CD spectrometer (Jasco, Tokyo, Japan). Both proteins were dialysed against 20 mM sodium phosphate buffer pH 8.0. NtCPIP1-wt was used at a concentration of 0.31 mg ml−1, whereas NtCPIP1-Δ(1:127) was concentrated to 0.13 mg ml−1. The spectra were scanned from 260 to 190 nm in 0.1 nm steps at 293 K. The secondary-structure content was analyzed using Spectra Manager v.2 (Jasco).
2.3. Crystallization
NtCPIP1-Δ(1:127) was concentrated to a final concentration of 8.5 mg ml−1 in 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM DTT using Vivaspin concentrators. The protein concentration was determined photometrically at a wavelength of 280 nm using a molecular weight of 20 152 Da and a molar extinction coefficient of 6990 M −1 cm−1. Crystals of NtCPIP1-Δ(1:127) were grown directly from PEG/Ion Screen (Hampton Research, Aliso Viejo, California, USA) using the sitting-drop vapour-diffusion method at 292 K. 0.2 µl protein solution was mixed with 0.1 µl reservoir solution [0.2 M ammonium citrate tribasic pH 7.0, 20%(w/v) polyethylene glycol (PEG) 3350] and equilibrated against 70 µl reservoir solution.
NtCPIP1-wt was concentrated to 10.0 mg ml−1 in the same buffer as NtCPIP1-Δ(1:127). Determination of the protein concentration was performed assuming an extinction coefficient of 22 920 M −1 cm−1 and a molecular weight of 34 363 Da. In the case of NtCPIP1-wt, initial crystals could be grown in 1.5 M ammonium sulfate, 0.1 M Tris–HCl pH 8.5, 12% glycerol from the Crystallization Extension Kit for Proteins (Sigma–Aldrich, St Louis, Missouri, USA) at 292 K. The best crystals were obtained using Additive Screen (Hampton Research) by mixing 0.2 µl protein solution with 0.1 µl reservoir solution consisting of 1.5 M ammonium sulfate, 0.1 M Tris–HCl pH 8.0, 12% glycerol, 0.015 mM 7-cyclohexyl-1-heptyl-β-d-maltoside (CYMAL-7); the drop was equilibrated against 70 µl reservoir solution at 292 K.
2.4. Data collection
Crystals of NtCPIP1-Δ(1:127) were soaked in cryoprotection solution consisting of reservoir solution diluted with 25%(v/v) PEG 400. Crystals of NtCPIP1-wt were flash-cooled in reservoir solution diluted with 25%(v/v) glycerol. Diffraction data sets were collected from crystals of both proteins at 100 K on beamline BL14.1 at the BESSY-II synchrotron (Berlin, Germany) on an MX-225 3 × 3 CCD detector (Rayonics, Evanston, Illinois, USA). Diffraction images for NtCPIP1-wt were recorded using 0.5° oscillation steps with a crystal-to-detector distance of 600.8 mm, whereas the diffraction data for the truncated protein were collected in 1° oscillation steps with a crystal-to-detector distance of 276.6 mm. Subsequently, data were indexed, integrated and scaled using XDS and XSCALE (Kabsch, 2010 ▶). Data-collection statistics are given in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| NtCPIP1-Δ(1:127) | NtCPIP1-wt | |
|---|---|---|
| Beamline | BESSY-MX BL14.1 | |
| Detector | Rayonics MX-225 3 × 3 CCD detector | |
| Temperature (K) | 100 | |
| Space group | P21212 | C222 |
| Unit-cell parameters (Å, °) | a = 65.6, b = 141.5, c = 50.9, α = β = γ = 90 | a = 136.8, b = 260.8, c = 378.2, α = β = γ = 90 |
| Wavelength (Å) | 0.9180 | 0.9180 |
| Resolution range (Å) | 35.0–2.38 (2.44–2.38) | 20.0–8.00 (8.21–8.00) |
| Unique reflections | 19599 (1409) | 6569 (538) |
| Multiplicity | 4.0 (3.8) | 3.8 (3.8) |
| Completeness (%) | 99.6 (98.1) | 87.2 (95.6) |
| Mean I/σ(I) | 15.8 (2.5) | 8.7 (1.6) |
| Rmeas† (%) | 7.6 (61.1) | 16.8 (85.3) |
| Rmrgd-F‡ (%) | 13.9 (83.2) | 26.6 (110.2) |
3. Results and discussion
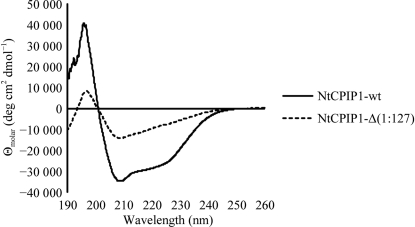
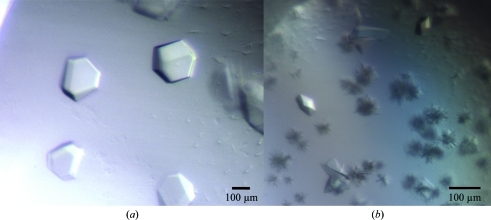
Recombinant NtCPIP1-wt was solubly expressed from pBADM-11 in E. coli BL21 (DE3) cells. A three-step purification procedure consisting of HisTrap, ion-exchange and size-exclusion chromatography yielded 3.8 mg pure protein per litre of cell culture. Circular-dichroism (CD) spectroscopy demonstrated that the purified protein was properly folded and displays α-helical and β-stranded secondary-structure elements (Fig. 1 ▶). Secondary-structure estimation predicted 24% α-helical, 35% β-strand, 14% turn and 25% random-coil content. Initial crystallization hits were obtained using the vapour-diffusion method (Fig. 2 ▶ a). However, crystals did not diffract to high resolution and subsequent rounds of optimization failed to improve the crystal quality. Diffraction data were collected on beamline BL14.1 at the BESSY-II synchrotron. The diffraction data for NtCPIP1-wt were indexed using XDS in the orthorhombic space group C222 to a resolution of 8.0 Å, with unit-cell parameters a = 136.8, b = 260.8, c = 378.2 Å (Table 1 ▶). The program POINTLESS confirmed the presence of three perpendicular twofold symmetry axes (Evans, 2006 ▶; Winn et al., 2011 ▶). At present, we cannot rule out space group C2221 as the correct space group. Based on the measured unit-cell parameters and the molecular mass of 34 363 Da, the calculated Matthews coefficient of between 2.01 and 3.44 Å3 Da−1 (with corresponding solvent contents of 38.8 and 64.3%) indicates that 14–24 monomers might be present in the asymmetric unit.
Figure 1.
Circular-dichroism spectra of recombinantly produced NtCPIP1-wt and NtCPIP1-Δ(1:127).
Figure 2.
(a) Crystals of NtCPIP1-wt in space group C222. (b) Crystals of NtCPIP1-Δ(1:127) in space group P21212 with dimensions of 0.05 × 0.05 × 0.07 mm.
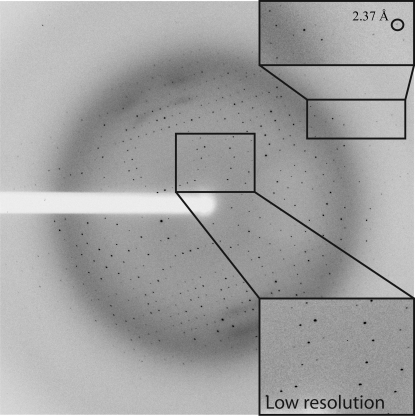
Since the crystals of NtCPIP1-wt did not diffract to high resolution, a shortened variant lacking the N-terminal J-domain and a flexible linker between the domains was cloned. NtCPIP1-Δ(1:127) was also produced in high amounts in E. coli BL21 (DE3). 1 l cell culture yielded 2.3 mg highly pure protein after several purification steps. We expected the CD spectrum of NtCPIP1-Δ(1:127) to be shifted towards higher β-strand content since this variant lacks the predicted α-helical J-domain. Indeed, the predicted α-helical, β-strand, turn and random-coil contents were 17, 37, 20 and 25%, respectively (Fig. 1 ▶). High-throughput crystallization screening yielded crystals with dimensions of 0.05 × 0.05 × 0.07 mm within a few days (Fig. 2 ▶ b). Crystals were directly flash-cooled from the initial screens and diffraction data were collected at the BESSY-II synchrotron (Fig. 3 ▶). Indexing and integration of the diffraction data using XDS indicated that the crystal belonged to space group P21212, with unit-cell parameters a = 65.6, b = 141.5, c = 50.9 Å (Table 1 ▶). The Matthews coefficient is 2.94 Å3 Da−1 assuming the presence of two monomers in the asymmetric unit. In this case, the crystal solvent content is 58.2%.
Figure 3.
Diffraction pattern extending to a maximum resolution of 2.4 Å recorded from an NtCPIP1-Δ(1:127) crystal.
We are confident that we will be able to solve the structure of NtCPIP1-Δ(1:127) by molecular replacement as the crystal structure of the substrate-binding domain of Hsp40 from Cryptosporidium parvum (PDB entry 2q2g; Structural Genomics Consortium, unpublished work) displays 40% sequence identity to NtCPIP1. We expect to encounter some difficulties in solving the structure of NtCPIP1-wt by molecular replacement. The expected large number of molecules in the asymmetric unit as well as the limited resolution of 8.0 Å renders molecular replacement highly unlikely. Nevertheless, the structure of a full-length Hsp40 would be welcome since to our knowledge no crystal structure of a full-length Hsp40 protein has yet been determined. If we succeed in improving the resolution limit of NtCPIP1-wt, we might possibly be able to locate the J-domain and the substrate-binding domain and show for the first time how these domains are oriented with respect to each other and thereby influence each other’s function. We expect that the crystal structure of the substrate-binding domain of NtCPIP1, namely of NtCPIP1-Δ(1:127), in conjunction with the modelling of ligand binding to NtCPIP1 will ultimately help us to understand the susceptibility of plants to potyvirus infections.
Acknowledgments
We would like to thank Uwe Müller and Manfred Weiss at the BESSY-II synchrotron (Berlin, Germany) for help with data collection and Madhumati Sevvana for help with data reduction and discussion. This work was supported by the Deutsche Forschungsgemeinschaft within the framework of the collaborative research project SFB 796 (Teilprojekt A3).
References
- Bukau, B., Weissman, J. & Horwich, A. (2006). Cell, 125, 443–451. [DOI] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol. 4, 269–275. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Hafrén, A., Hofius, D., Rönnholm, G., Sonnewald, U. & Mäkinen, K. (2010). Plant Cell, 22, 523–535. [DOI] [PMC free article] [PubMed]
- Hartl, F. U., Bracher, A. & Hayer-Hartl, M. (2011). Nature (London), 475, 324–332. [DOI] [PubMed]
- Hofius, D., Maier, A. T., Dietrich, C., Jungkunz, I., Börnke, F., Maiss, E. & Sonnewald, U. (2007). J. Virol. 81, 11870–11880. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Qiu, X.-B., Shao, Y.-M., Miao, S. & Wang, L. (2006). Cell. Mol. Life Sci. 63, 2560–2570. [DOI] [PMC free article] [PubMed]
- Rajan, V. B. & D’Silva, P. (2009). Funct. Integr. Genomics, 9, 433–446. [DOI] [PubMed]
- Saibil, H. R. (2008). Curr. Opin. Struct. Biol. 18, 35–42. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.