Abstract
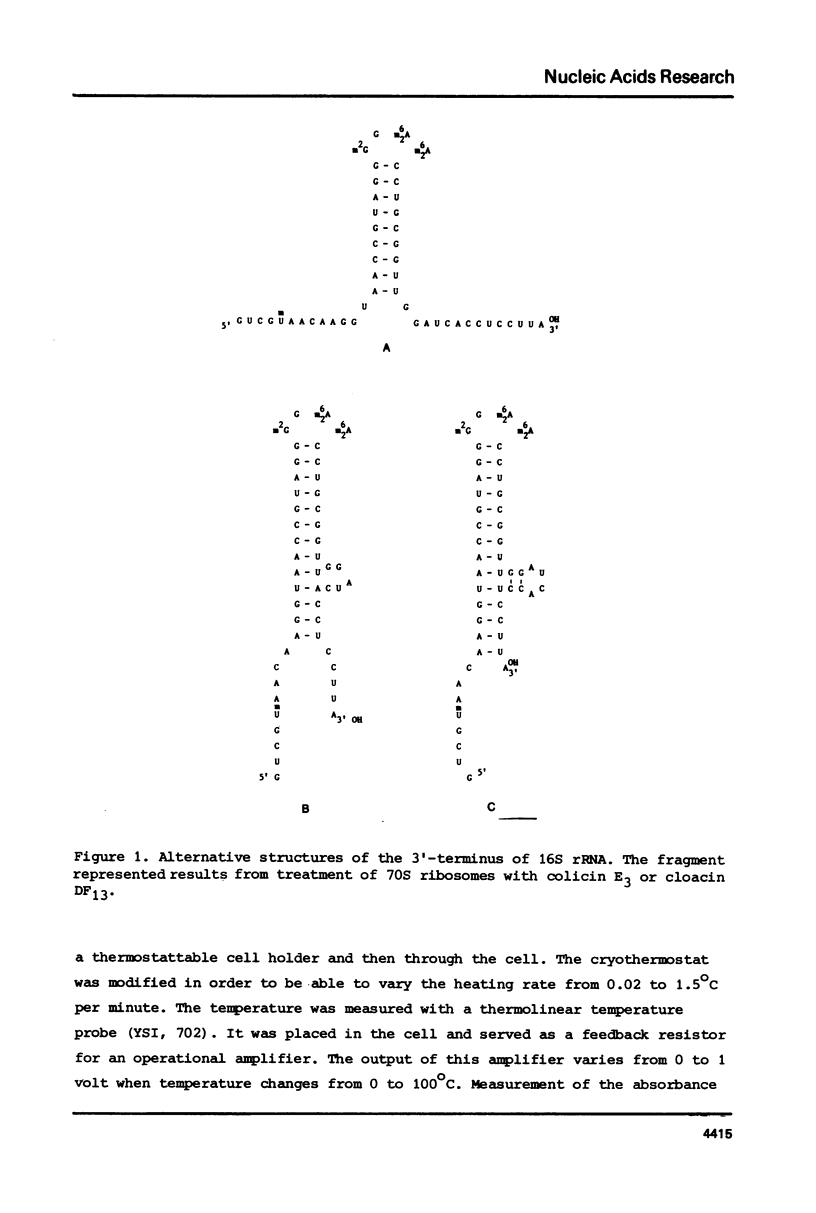
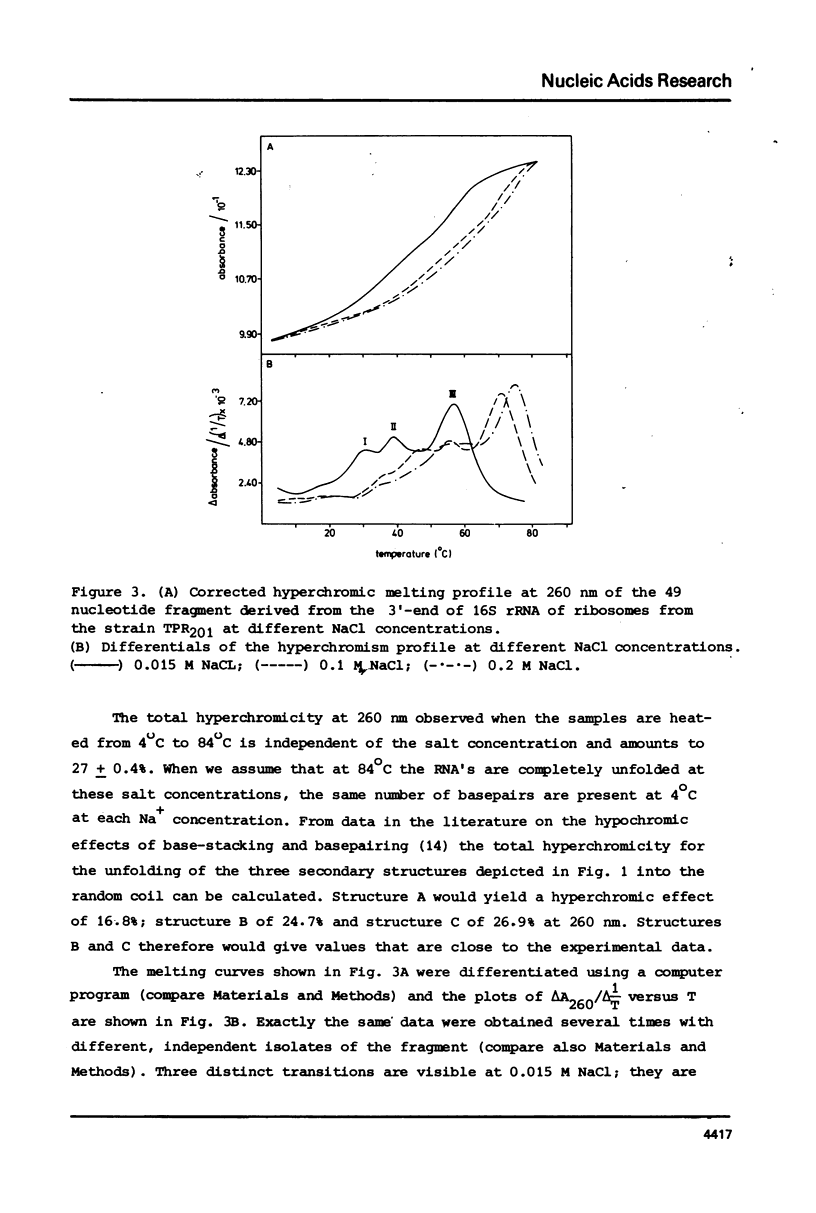
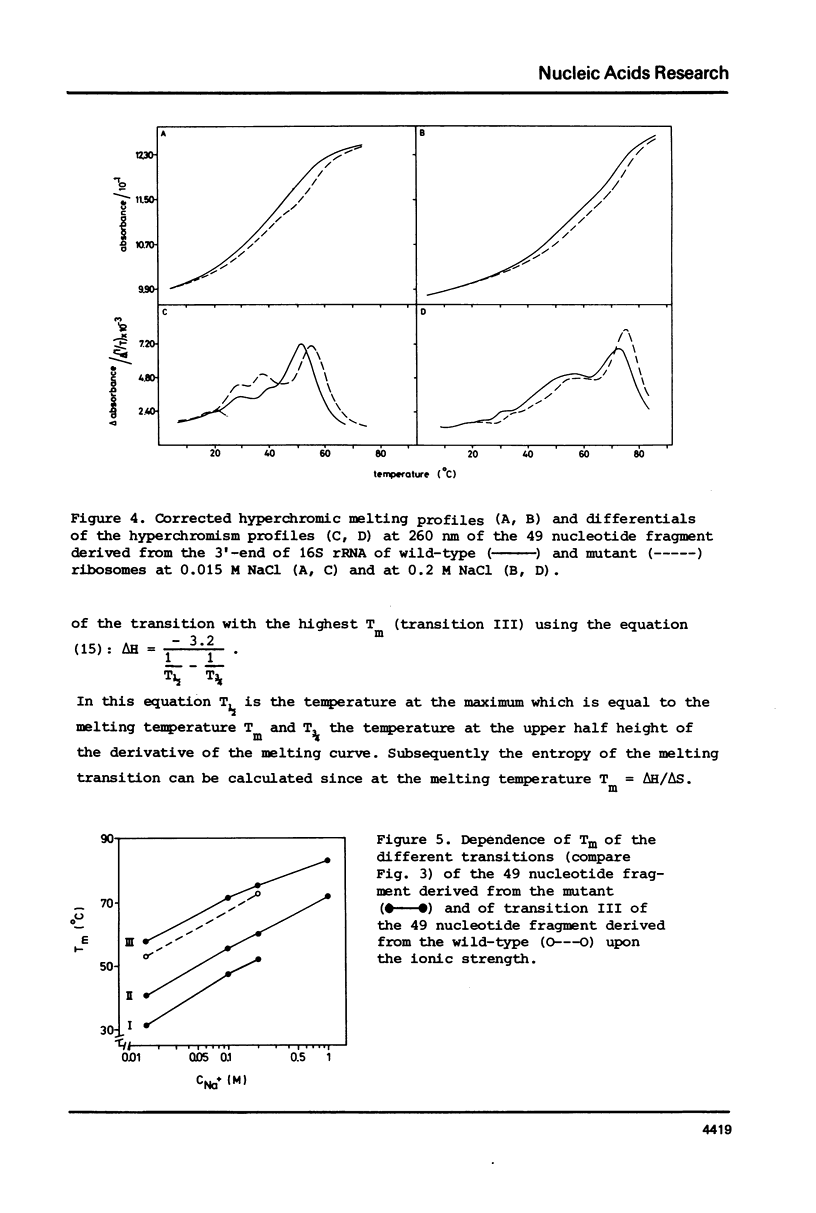
Fragments comprising the 49 nucleotides from the 3'-end have been purified from 16S ribosomal RNA of wild-type Escherichia coli and from a kasugamycin-resistant mutant that specifically lacks dimethylation of two adjacent adenines near the 3'-terminus. These fragments, obtained after treatment of ribosomes in vitro with the bacteriocin cloacin DF13, were used to study the effect of the methyl groups on the temperature dependent unfolding of double-stranded regions. Both fragments contain at least 3 independent melting transitions, of which the one with the highest Tm corresponds with the unfolding of a nine-basepair long central hairpin. Dimethylation of the adenines in the loop of this hairpin lowers the melting temperature (Tm) by approximately 2 degrees C at 0.2 M NaCl and by about 5 degrees C at 0.15 M NaCl. It is suggested that m6(2)Am6(2)A is more antagonistic to loop formation that ApA and that the function of the methyl groups is to help to destabilize the 3'-terminal hairpin in 16S rRNA in order to facilitate intermolecular interactions.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baan R. A., Hilbers C. W., Van Charldorp R., Van Leerdam E., Van Knippenberg P. H., Bosch L. High-resolution proton magnetic resonance study of the secondary structure of the 3'-terminal 49-nucleotide fragment of 16S rRNA from Escherichia coli. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1028–1031. doi: 10.1073/pnas.74.3.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baan R. A., van Charldorp R., van Leerdam E., van Knippenberg P. H., Bosch L., de Rooij J. F., van Boom J. H. The 3'-terminus of 16 S ribosomal RNA of Escherichia coli. Isolation and purification of the terminal 49-nucleotide fragment at a milligram scale. FEBS Lett. 1976 Dec 1;71(2):351–355. doi: 10.1016/0014-5793(76)80968-4. [DOI] [PubMed] [Google Scholar]
- Cole P. E., Yang S. K., Crothers D. M. Conformational changes of transfer ribonucleic acid. Equilibrium phase diagrams. Biochemistry. 1972 Nov 7;11(23):4358–4368. doi: 10.1021/bi00773a024. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Electrophoretic analysis of the unfolding of proteins by urea. J Mol Biol. 1979 Apr 5;129(2):235–264. doi: 10.1016/0022-2836(79)90279-1. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E., Dahlberg J. E. Binding of ribosomal protein S1 of Escherichia coli to the 3' end of 16S rRNA. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2940–2944. doi: 10.1073/pnas.72.8.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotz C., Brimacombe R. An experimentally-derived model for the secondary structure of the 16S ribosomal RNA from Escherichia coli. Nucleic Acids Res. 1980 Jun 11;8(11):2377–2395. doi: 10.1093/nar/8.11.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. 3. Small internal loops resulting from mismatches. J Mol Biol. 1973 Aug 5;78(2):301–319. doi: 10.1016/0022-2836(73)90118-6. [DOI] [PubMed] [Google Scholar]
- Helser T. L., Davies J. E., Dahlberg J. E. Change in methylation of 16S ribosomal RNA associated with mutation to kasugamycin resistance in Escherichia coli. Nat New Biol. 1971 Sep 1;233(35):12–14. doi: 10.1038/newbio233012a0. [DOI] [PubMed] [Google Scholar]
- Olsthoorn C. S., Haasnoot C. A., Altona C. Circular dichroism studies of 6-N-methylated adenylyladenosine and adenylyluridine and their parent compounds. Thermodynamics of stacking. Eur J Biochem. 1980 May;106(1):85–95. doi: 10.1111/j.1432-1033.1980.tb05999.x. [DOI] [PubMed] [Google Scholar]
- Poldermans B., Bakker H., Van Knippenberg P. H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16S ribosomal RNA of Escherichia coli. IV. The effect of the methylgroups on ribosomal subunit interaction. Nucleic Acids Res. 1980 Jan 11;8(1):143–151. doi: 10.1093/nar/8.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldermans B., Van Buul C. P., Van Knippenberg P. H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16 S ribosomal RNA of Escherichia coli. II. The effect of the absence of the methyl groups on initiation of protein biosynthesis. J Biol Chem. 1979 Sep 25;254(18):9090–9093. [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A., Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler P., Carbon P., Zuker M., Ebel J. P., Ehresmann C. Structural organization of the 16S ribosomal RNA from E. coli. Topography and secondary structure. Nucleic Acids Res. 1981 May 11;9(9):2153–2172. doi: 10.1093/nar/9.9.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazawa I., Koike T., Inoue Y. Stacking properties of a highly hydrophobic dinucleotide sequence, N6, N6-dimethyladenylyl(3' leads to 5')N6, N6-dimethyladenosine, occurring in 16--18-S ribosomal RNA. Eur J Biochem. 1980 Aug;109(1):33–38. doi: 10.1111/j.1432-1033.1980.tb04764.x. [DOI] [PubMed] [Google Scholar]
- Van Charldorp R., Heus H. A., Van Knippenberg P. H. Adenosine dimethylation of 16S ribosomal RNA: effect of the methylgroups on local conformational stability as deduced from electrophoretic mobility of RNA fragments in denaturing polyacrylamide gels. Nucleic Acids Res. 1981 Jan 24;9(2):267–275. doi: 10.1093/nar/9.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duin J., Kurland C. G., Dondon J., Grunberg-Mangago M., Branlant C., Ebel J. P. New aspects of the IF3-ribosome interaction. FEBS Lett. 1976 Feb 15;62(2):111–114. doi: 10.1016/0014-5793(76)80030-0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Gupta R., Siegel R. B., Stahl D. A., Kop J., Crawford N., Brosius J., Gutell R., Hogan J. J. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980 May 24;8(10):2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R. C., Steitz J. A., Moore P. B., Crothers D. M. The 3' terminus of 16S rRNA: secondary structure and interaction with ribosomal protein S1. Nucleic Acids Res. 1979 Dec 20;7(8):2399–2418. doi: 10.1093/nar/7.8.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]


