Abstract
Background
The lack of genomic data available for mites limits our understanding of their biology. Evolving high-throughput sequencing technologies promise to deliver rapid advances in this area, however, estimates of genome size are initially required to ensure sufficient coverage.
Methods
Quantitative real-time PCR was used to estimate the genome sizes of the burrowing ectoparasitic mite Sarcoptes scabiei, the non-burrowing ectoparasitic mite Psoroptes ovis, and the free-living house dust mite Dermatophagoides pteronyssinus. Additionally, the chromosome number of S. scabiei was determined by chromosomal spreads of embryonic cells derived from single eggs.
Results
S. scabiei cells were shown to contain 17 or 18 small (< 2 μM) chromosomes, suggesting an XO sex-determination mechanism. The average estimated genome sizes of S. scabiei and P. ovis were 96 (± 7) Mb and 86 (± 2) Mb respectively, among the smallest arthropod genomes reported to date. The D. pteronyssinus genome was estimated to be larger than its parasitic counterparts, at 151 Mb in female mites and 218 Mb in male mites.
Conclusions
This data provides a starting point for understanding the genetic organisation and evolution of these astigmatid mites, informing future sequencing projects. A comparitive genomic approach including these three closely related mites is likely to reveal key insights on mite biology, parasitic adaptations and immune evasion.
Keywords: Sarcoptes scabiei, Psoroptes ovis, Dermatophagoides pteronyssinus, genome size
Background
There is a distinct paucity of genomic data available for the class Acari (mites and ticks), and subsequently our understanding of their biology is limited. More information regarding mite genomics would greatly assist the development of novel control strategies underpinned by molecular approaches. Three astigmatid mites of particular medical and veterinary interest are the itch mite Sarcoptes scabiei (Sarcoptidae), the sheep scab mite Psoroptes ovis (Psoroptidae) and the house dust mite Dermatophagoides pteronyssinus (Pyroglyphidae).
Scabies remains a truly neglected disease, caused by the burrowing ectoparasitc "itch mite" S. scabiei. The scarcity of molecular data on S. scabiei has been due in part to very low parasite burden in most patients, and the historical lack of in vitro culture and animal models. A major advance was the creation of S. scabiei var. hominis cDNA libraries resulting in a database of ~43,000 Expressed Sequence Tags (ESTs), providing substantial molecular data for this parasite and securing a solid base for recombinant biology [1,2]. More recently, a tractable porcine model of scabies has been established [3], providing large amounts of mite material for molecular studies. P. ovis is a non-burrowing, ectoparastic mite causing the highly infectious disease 'sheep scab' in sheep and cattle, responsible for major economic losses and serious welfare concerns [4]. There is a similar dearth of understanding regarding parasite biology, but recently over 1,500 ESTs have been generated and deposited in public databases, representing the largest molecular data resource on P. ovis to date [5]. Finally, although allergies to house-dust mites are extremely common, remarkably little genetic information is available on the causative agents, the free-living mites D. pteronyssinus, D. farinae and Euroglyphus maynei. Research efforts have mainly focused on characterisation and generation of recombinant house dust mite allergens for diagnostics and immunotherapy [6]. However, a dataset of ~3000 ESTs is available [7], and the mitochondrial genome of D. pteronyssinus has recently been sequenced [8].
The Ixodes scapularis (black-legged tick) genome sequencing project marked the beginning of the genomics era for the field of acarology. As previously observed, there appears to be no patterns regarding genome size in the Acari. Flow cytometry based genome estimates indicated a large haploid genome for all Ixodida with a mean of 1281 Mbp (approx. 1.31 pg) for the Argasidae and 2671 Mbp (approx. 2.73 pg) for the Ixodidae [9]. An exciting development for mite genomics has been the publication of a preliminary genome survey for the honey-bee mite Varroa destructor [10]. At 565 Mbp, the Varroa genome is larger than many insects, including its host Apis melifera (262 Mbp). In contrast, the two-spotted spider mite Tetranychus urticae is predicted to possess a genome of only 75 Mb (0.08 pg) [11], however, this estimate was recently revised upward to 90.7 Mbp [12]. A similarly small nuclear genome of 88-90 Mbp was estimated in the phytoseiid mite Metaseiulus occidentalis [13], with the orbatid mite Archegozetes longisetosus genome estimated to be 150 Mbp [12].
Attempts to estimate the S. scabiei genome size accurately using flow cytometry have been impeded by the inability to obtain sufficient cell numbers from egg and whole-body preparations. The nuclei are very small in comparison to nuclei from mosquito cell lines, suggesting that the scabies genome may also be small (Fischer, unpublished observations). Similar issues were reported in M. occidentalis, where flow cytometry failed to resolve genome size, with estimates ranging from 35-160 Mbp, depending on egg age [13]. Due to the small size of mites, tissue dissections are difficult, hindering aquisition of homogenous cell preparations. An alternative approach utlilising quantitative-PCR (qPCR) was suggested to be particularly useful for organisms where the genome size is expected to be small and the availability of genetic material limited [13]. Based on on a pioneering study by Wilhelm [14], this method has proven reliable for a number of species, including Saccharomyces cerevisiae, Xiphorphours maculaus, Homo sapiens [14], Musca domestica, and Drosophila melanogaster [15]. Here, we use qPCR to estimate the genome sizes of S. scabiei, P. ovis and D. pteronyssinus, with the purpose of informing future sequencing projects. Additionally, the chromosome number of S. scabiei was determined, providing a starting point for understanding the genetic organisation and evolution for this species.
Methods
Source of samples
Scabies mites (Sarcoptes scabiei var. suis) were obtained from a colony maintained on pigs (Sus scrofa) at the Centre for Advanced Animal Studies (CAAS), University of Queensland, Gatton, QLD, Australia. Mites were isolated from heavily infested skin crusts as described previously [3]. Sheep scab mites (Psoroptes ovis) were harvested from infested donor lambs maintained at the Moredun Research Institute as previously described [16]. Ethical approvals for this work were obtained from the Queensland Institute of Medical Research and Queensland Department of Employment, Economic Development and Innovation, and the Moredun Research Institute Experiment Committees respectively. House dust mites (Dermatophagoides pteronyssinus), separated into the two sexes, were purchased from the Siriraj Dust Mite Centre for Services and Research, Department of Parasitology, Mahidol University, Bangkok, Thailand. Isolated mites were stored at -80°C until further processing. The methods to separate sexes and adults from larvae and nymphs of the individual mite species are well established in the three laboratories involved. Adult female scabies and psoroptes mites are easy to distinguish from males due to their much larger size. The adult female scabies mites are approximately 500 μm long compared to the smaller approximately 250 μm long males. In addition adult scabies mite males can be separated from larvae and nymphs based on size, a darker, and more sclerotized cuticle and leg number. Mature female Psoroptes are 550-750 μm long, with a striate cuticle and four long and 16 short dorsal somatic setae [17,18] while males are about one-fourth smaller, and they have an additional, larger posterodorsal cuticular plate and a pair of posteroventral adanal suckers. Adult female house dust mites measure approximately the same length as males but are 0.32 mm in width compared to 2.4 mm wide males. Males are more sclerotized with enlarged legs I and III [19]
DNA extraction
Genomic DNA (gDNA) preparations used for qPCR included: a) S. scabiei var. suis female mites (100) and mixed life-stage mites (~500); b) P. ovis female mites (75); c) D. pteronyssinus male (10 mg) and female (15 mg) mites; d) as a positive control for the method, Pichia pastoris strain GS115 gDNA was used.
Genomic DNA was extracted by homogenising mites using a motorised micropestle in a purpose-designed eppendorf tube (Kontes, Kindle Chase) over liquid-nitrogen. The homogenate was resuspended in 2 ml of digestion buffer (800 mM Guanidine HCl; 30 mM Tris-Cl, pH8.0; 30 mM EDTA, pH8.0; 5% Tween-20; 0.5%Triton X-100), supplemented with 2 μL RNase A (Qiagen, 20 mg/ml) and incubated at 37°C for 30 min. 100 μL proteinase K (Qiagen, 20 mg/ml) was added and incubated at 50°C for 1 hour with gentle agitation. The lysate was spun at 4000 g at 4°C for 10 min and the supernatant was loaded on to a Qiagen genomic tip 20/G of the Qiagen Blood & Cell Culture DNA Mini Kit (Qiagen, Doncaster, Australia). DNA extraction was performed according to manufacturer's instructions. Final DNA pellets were resuspended in 30-100 μL 15 mM Tris-EDTA Buffer and aliquots stored at -80°C. DNA concentration and purity was measured by a) measuring absorbance at 260 nm using the ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, USA), b) Qubit-IT pico green HS dsDNA reagent (Invitrogen, Mulgrave, Australia) in a fluorometer (Biotek Synergy-4) and c) agarose gel electrophoresis with ethidium bromide staining. Only DNA samples for which the three quantification methods concurred were used for subsequent qPCR.
PCR primer design and preparation of qPCR standards
Based on their successful use in similar studies [13], and availability of sequence information, two single copy nuclear genes were selected for this study--actin and elongation factor 1α (EF1α). Primers were designed from EST sequence available for S. scabiei actin [EU624346], EF1α [JQ236667]; Psoroptes ovis actin [BQ834989], EF1 α [BQ835070]; D. pteronyssinus EF1α [EU152830]. For positive control experiments PCR primers were also designed for P. pastoris actin [AF216956] and elongation factor 3 (EF3) [FN392322] (Table 1).
Table 1.
Primers used in this study
| Gene | Primer | Sequence (5'-3') | Size |
|---|---|---|---|
| Actin (S. scabiei) | SsAct F | CAACCATCCTTCTTGGGTATG | 311 bp |
| SsAct R | CCAGCTTCGTCGTATTCTTGT | ||
| Actin (P. ovis) | PoAct F | CATCAAGGTGTCATGGTTGG | 225 bp |
| PoAct R | GGCTTTTGGATTCAATGGAG | ||
| Actin (P. pastoris) | PpAct F | AGCGCGCCCATTTCTTACCGT | 203 bp |
| PpAct R | TCTCCGGCGTATCCGGCCTT | ||
| EF1a (S. scabiei) | SsEF1a F | TTGGCTTATACCTTGGGTGTG | 181 bp |
| SsEF1a R | CACCGTTCCATCCAGAGATT | ||
| EF1a (D. pteronyssius) | DpEF1a F | CCCGTGAACATGCTTTGCTTGCC | 175 bp |
| DpEF1a R | ACAAAAGCGACGGTAGCCGGA | ||
| EF1a (P. ovis) | PoEF1a F | CAAATATGCCTGGGTTTTGG | 205 bp |
| PoEF1a R | TTCACCAGTACCAGCAGCAA | ||
| EF3 (P. pastoris) | PpEF3 F | GTCGCCATTTTGGATGCTAT | 251 bp |
| PpEF3 R | AGGGTGTGGACAGTTTCTGG |
To ensure primer and PCR conditions were optimal, that the PCR was specific, and to prepare plasmid standards, a series of validation PCRs were run prior to commencing qPCR for each mite species and primer combination. Firstly, gradient PCRs were performed to determine optimum annealing temperatures. Reactions contained 0.2 mM dNTPs, 0.4 μM of each primer, 2.5 mM MgCl2 , 0.2 U Taq Polymerase (AmpliTaq Gold, Applied Biosystems), and 1 μL template gDNA. Cycling times were 95°C for 10 minutes, followed by 30 cycles of 94°C for 15 s, 55-65°C for 30 s, and 72°C for 30 s. Products were visualised on 1.5% agarose gels, purified (QiaQuick purification kit, Qiagen) and cloned into pGEM-T vectors (Promega, Alexandria, Australia). After sequencing to confirm product specificity, plasmids were linearised with NotI, purified and quantified as described previously.
Quantitative PCR
Quantitative real-time PCR was performed using Sensimix SYBR green (Bioline, Alexandria, Australia) in a Rotorgene 6000 cycler (Qiagen). Reactions contain 1× SYBR green mastermix, 0.4 μM of each primer, and 1 μL template DNA. Each run consisted of a series of linearised plasmid standards (seven, 10-fold dilutions), genomic DNA and a no-template control, with all reactions run in duplicate. Copy number of standards was calculated using a DNA weight to moles calculator (http://molbiol.edu.ru/eng/scripts/h01_07.html), which determined the number of copies based on the concentration and size of the plasmid. A standard curve was generated from the Ct values of the standards using Rotorgene software, which was used to calculate the copy number of the unknown gDNA. Only runs with a standard curve efficiency > 90% and < 0.5 Ct standard deviation of duplicates were utilised for calculation of genome estimates.
The weight of a single copy of the nuclear genome (C-value) in picograms was obtained by dividing the input template concentration by the qPCR derived copy number. The estimated genome size of the unknowns was calculated using the formula Genome size (bp) = (0.978 × 109) × C (pg). This is based on the formula of Dolezel [20], where the mean weight of one nucleotide base pair = 1.023 × 10-9 pg.
Determining the chromosome number in Sarcoptes scabiei
Female S. scabiei mites were isolated from infested pig skin and placed on glass petri dishes. Eggs that were deposited within a time frame of a few hours were processed, since they were most likely alive and provide dividing cells.
Superfrost slides (Manzel-Glaser, Braunschweig, Germany) that had been cleaned in 100% ethanol were dried and two eggs per slide were placed 2 cm apart on the slide surface. To dissolve the outer chorion, eggs were bathed in 10 μL 1% sodium hypochlorite in dH20 for 5 minutes. This solution was removed and replaced with 10 μL 1% colchicine (Sigma) in 0.7% sodium chloride/10 mM Tris pH 7.5. Eggs were incubated in the dark for 30 minutes in a humidity chamber, the colchicine solution was removed and replaced by 20 μL 0.075 M potassium chloride for 15 minutes. The hypotonic solution was removed and 20 μL fixative (3:1 methanol:acetic acid) added. A cover slip was placed on the slide and the eggs were squashed using the flat end of a pencil. The slide was placed on dry-ice for several minutes before the coverslip was removed. The slides and coverslip were air-dried before the addition of DAPI stain (ProLong Gold, Invitrogen). The cover slip was replaced, and the slides were viewed with the DeltaVision deconvolution microscope. The data generated through deconvolution (SoftWORx software) was used to generate a 360° rotational image of the chromosomal spread. Two people assessed the number of chromosomes in 18 eggs which had multiple cells in metaphase.
Results and Discussion
qPCR based genome estimation
qPCR on the positive control P. pastoris gDNA with actin and EF3 primers yielded a mean genome estimate of 8.7 ± 0.5 Mbp (Table 2), within 10% of the actual size of 9.43 Mbp [21], attesting to the validity of this technique.
Table 2.
qPCR based genome estimates of S. scabiei, P. ovis and D. pteronyssius
| Species | Sex | Gene | n | C (mean, ± SD, pg) | Genome size (mean, ± SD, Mbp) |
|---|---|---|---|---|---|
| S.scabiei | Female | Actin | 2 | 0.095 (± 0.042) | 93 (± 41) |
| EF1a | 3 | 0.091 (± 0.005) | 89 (± 14) | ||
| Mixed | Actin | 1 | 0.107 | 105 | |
| EF1a | 2 | 0.101 (± 0.014) | 98 (± 14) | ||
| Average | 0.099 (± 0.007) | 96 (± 7) | |||
| P. ovis | Female | Actin | 2 | 0.089 (± 0.001) | 87 ± (0.5) |
| EF1a | 3 | 0.086 (± 0.008) | 84 (± 8) | ||
| Average | 0.088 (± 0.002) | 86 (± 2) | |||
| D. pteronyssinus | Female | EF1a | 2 | 0.155 (± 0.0004) | 151 (± 0.4) |
| Male | EF1a | 2 | 0.223 (± 0.004) | 218 (± 4) | |
| P. pastoris | EF3 | 2 | 0.009 (0.0004) | 8.37 (± 0.4) | |
| Actin | 2 | 0.009 (± 0.001) | 9.05 (± 0.6) | ||
| Average | 0.009 | 8.71 (± 0.5) | |||
qPCR of two single-copy genes for two gDNA preparations of S. scabiei gave a mean genome estimate of 96 ± 7 Mbp (Table 2). There was no large difference overall between estimates using the different primer pairs, although the actin PCR gave a highly variable result with female mites, with variation in the EF1α PCRs lower. We were unable to sample male-only preparations due to insufficient numbers, but genome estimates of mixed life-stage gDNA were not significantly different to female only, an observation that agrees with previous suggestions that haplodiploidy or other forms of polyploidy is not a feature of Sarcoptes [22].
The genome size estimate for P.ovis female mites was smaller than S. scabiei, at 86 ± 2 Mbp. These estimates are very close to that of M. occidentalis (88-90 Mbp) and T. urticae (75-90 Mbp), collectively representing the smallest known arthropod genomes to date.
Interestingly, the genome of D. pteronyssinus is predicted to be larger than its parasitic relatives, at 151 Mbp for females and 218 Mbp for males (Table 2). The reason for the larger size estimate in males is not understood, and could be reflective of low sample numbers or different developmental stages. As the chromosome number, sex determination system and karyotype have not been determined for house dust mites, it is difficult to speculate without further investigation.
Although flow cytometry remains the "gold standard" for determination of genome size in the absence of sequencing, here we confirm the usefulness of the qPCR method, especially where genetic material is limited. While a limitation of the method is its sensitivity to inaccuracies in DNA concentration and PCR efficiency, it is a quick and useful way to establish general estimates of genome size where more accurate flow cytometry may not be possible.
Chromosome number in S. scabiei
Using 360° rotational images of chromosomal spreads from S. scabiei eggs, the chromosome number of embryonic cells from a single egg was either 17 or 18 very small chromosomes, of which the largest chromosome was < 2.5 μm (Figure 1). These findings accord with the qPCR results indicating a small scabies mite genome. While the cause for the disparate numbers is unknown, it may arise because of an XO sex determination mechanism, where males lack the sex chromosome and therefore have one less chromosome than the female. This occurs in a range of Ecdysozoan invertebrates including nematodes and arthropods, and has been demonstrated in the Acaridae [23]. These results again support the absence of haplodiploidy, where haploid males develop from unfertilised eggs and diploid females develop from fertilised eggs. Haplodiploidy in S. scabiei was also excluded in a previous study [22], as heterogeneity was demonstrated in microsatellite genotypes from male mites. Previous surveys of the Astigmata have demonstrated the prevailing sex determining mechanism to be diplo-diploidly, with chromosome numbers ranging from 10-18 [24,25].
Figure 1.
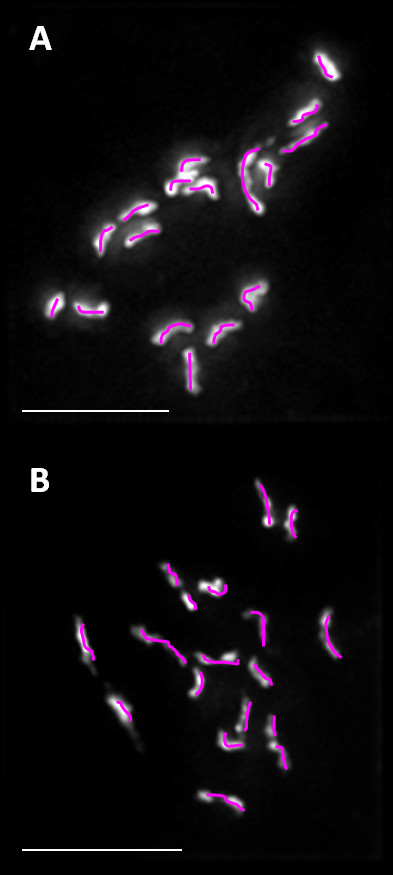
Chromosomal spreads of single Sarcoptes scabiei cells. A) 17 chromosomes can be counted while B) shows 18 chromosomes. Chromosomes are highlighted in pink for clarification. Scale bar = 5 μm.
Conclusions
There appears to be little correlation between genome size, chromosome number, organism complexity, or phylogenetic relatedness, commonly referred to as the "C-value enigma" [26]. However, key factors influencing small genome sizes may include small cell size (positively correlated), developmental complexity (negatively correlated), and maintenance or loss of non-coding DNA [27].
The smallest arthropod genome sequenced to date is that of the ectoparasitic body louse, Pediculus humanus humanus [28]. At 108 Mb, the body louse shows a reduced, but markedly functional genome. Relative to other sequenced arthropods, the most marked reductions were seen in gene families associated with environmental interaction [28], and thus it is has been postulated that parasitic species may have smaller genomes than their non-parasitic counterparts, as an adaptation to parasitism and ecological niche leads to the expansion and contraction of gene families [28,29]. Our current results accord with this hypothesis, as the free-living house dust mite genome was estimated to be larger than that of the parasitic Sarcoptes or Psoroptes mites. In contrast, the genome of V. destructor, also parasitic, is considerably larger at 565 Mb [10]. Interestingly S. scabiei has been shown to contain a multi-gene family of inactivated protease paralogues (SMIPPs), which to date have not been identified in either house dust mite or P. ovis [30,31]. It has been recently shown that these proteins can inhibit the human complement system, suggesting this gene family expansion is a specific adaptation to burrowing and immune evasion [32].
The degree of non-coding and repetitive DNA, intron size and transposable elements also appears to play a role in genome size. Ticks for example have up to 70% of their genome as non-coding/repetitive [9]. Conversely, the body louse genome has less than 1% of their genome devoted to transposable elements and introns, considerably less than Drosophila [28]. It has been suggested that transposable elements cannot be established in eukaryotic genomes < 100 Mb [33], so it will be very interesting to see whether these elements are present in the mites studied here. The majority of sequence data obtained to date from S. scabiei has been from cDNA, and only very few genome sequences are published. We have to date identified genomic sequences of 12 genes (7 complete, 5 incomplete) containing a total of 25 introns. The introns in all but one of the genes were all ≤ 100 bp with an average size of 69 bp for these 13 introns. Of the 11 introns in the remaining gene (encoding a large transmembrane protein), 7 were ≤ 100 bp with the remaining 4 introns being 547 bp , 248 bp , 560 bp and 381 bp . The average size of all the 25 different introns we have identified to date is 128 bp . Interestingly the average intron size in the V. destructor sodium channel gene is 2.7 kb [34], compared to only 203 bp in the partial S. scabiei gene sequence (domains II-IV)[35]. Altogether, this data supports the estimation of a very small genome in Sarcoptes.
Sequencing of the Sarcoptes scabiei genome is scheduled to commence in the near future, and we encourage the extension of efforts to also include D. pteronyssinus and P. ovis. A comparative genomic approach between these three skin-feeding mites is likely to highlight fascinating genetic similarities and differences between parasitic and free-living mites, and burrowing versus non-burrowing parasitic mites, and to provide essential information to inform control strategies for these important species. This would also be complemented by comparison with the recently sequenced P. humanus and V. destructor genomes. In this study, we have confirmed the small size of these mite genomes, which augurs well for the successful completion of future sequencing projects.
Competing interests
'The authors declare that they have no competing interests
Authors' contributions
KM conceived the study, carried out the cloning, qPCR and analysis, and drafted the manuscript. CW carried out the chromosomal studies. STGB provided P. ovis and participated in design of the study. DH carried out bioinformatic analysis, participated in study design and helped to draft the manuscript. JMc provided mites and helped to draft the manuscript. KF participated in design and coordination of the study, provided mites and carried out DNA extractions, all authors read and approved the final manuscript.
Contributor Information
Kate E Mounsey, Email: kate.mounsey@qimr.edu.au.
Charlene Willis, Email: charlene.willis@qimr.edu.au.
Stewart TG Burgess, Email: stewart.burgess@moredun.ac.uk.
Deborah C Holt, Email: deborah.holt@menzies.edu.au.
James McCarthy, Email: j.mccarthy@uq.edu.au.
Katja Fischer, Email: katja.fischer@qimr.edu.au.
Acknowledgements
We thank Andrew Kelly and Beverley Hutchinson from DEEDI for the maintence of our mange pig colony, Melanie Rampton and Simone Reynolds for assistance with mite collections. This work was supported by the Australian National Health and Medical Research Council (program grant 496600, project grant 496635, training fellowship 490345), the UK Department for Environment, Food and Rural Affairs (Project OD0553), and the Scottish Government.
References
- Fischer K, Holt DC, Harumal P, Currie BJ, Walton SF, Kemp DJ. Generation and characterization of cDNA clones from Sarcoptes scabiei var. hominis for an expressed sequence tag library: identification of homologues of house dust mite allergens. Am J Trop Med Hyg. 2003;68:61–64. [PubMed] [Google Scholar]
- Fischer K, Holt DC, Wilson P, Davis J, Hewitt V, Johnson M, McGrath A, Currie BJ, Walton SF, Kemp DJ. Normalization of a cDNA library cloned in lambda ZAP by a long PCR and cDNA reassociation procedure. BioTechniques. 2003;34:250–252. doi: 10.2144/03342bm03. 254. [DOI] [PubMed] [Google Scholar]
- Mounsey K, Ho MF, Kelly A, Willis C, Pasay C, Kemp DJ, McCarthy JS, Fischer K. A tractable experimental model for study of human and animal scabies. PLoS Negl Trop Dis. 2010;4:e756. doi: 10.1371/journal.pntd.0000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek AH, Huntley JF. Sheep scab: the disease, pathogenesis and control. J Comp Path. 2003;128:79–91. doi: 10.1053/jcpa.2002.0627. [DOI] [PubMed] [Google Scholar]
- Burgess ST, Nisbet AJ, Kenyon F, Huntley JF. Generation, analysis and functional annotation of expressed sequence tags from the ectoparasitic mite Psoroptes ovis. Parasit Vectors. 2011;4:145. doi: 10.1186/1756-3305-4-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WR, Hales BJ, Smith WA. House dust mite allergens in asthma and allergy. Trends Mol Med. 2010;16:321–328. doi: 10.1016/j.molmed.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Angus AC, Ong ST, Chew FT. Sequence tag catalogs of dust mite-expressed genomes: utility in allergen and acarologic studies. Am J Pharmacogenomics. 2004;4:357–369. doi: 10.2165/00129785-200404060-00003. [DOI] [PubMed] [Google Scholar]
- Dermauw W, VanLeeuwen TV, Vanholme B, Tirry L. The complete mitochondrial genome of the house dust mite Dermatophagoides pteronyssinus (Trouessart): a novel gene arrangement among arthropods. BMC Genomics. 2009;10:107. doi: 10.1186/1471-2164-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraci NS, Johnston SJ, Robinson JP, Wikel SK, Hill CA. Variation in genome size of agrasid and ixodid ticks. Insect Biochem Mol Biol. 2007;37:399–408. doi: 10.1016/j.ibmb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Cornman SR, Schatz MC, Johnston SJ, Chen YP, Pettis J, Hunt G, Bourgeois L, Elsik C, Anderson D, Grozinger CM, Evans JD. Genomic survey of the ectoparasitic mite Varroa destructor, a major pest of the honey bee Apis mellifera. BMC Genomics. 2010;11:602. doi: 10.1186/1471-2164-11-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic M, Khila A, Lee KW, Bjelica A, Grbic V, Whistlecraft J, Verdon L, Navajas M, Nagy L. Mity model: Tetranychus urticae, a candidate for chelicerate model organism. BioEssays. 2007;29:489–496. doi: 10.1002/bies.20564. [DOI] [PubMed] [Google Scholar]
- Hanrahan SJ, Johnston SJ. New genome size estimates of 134 species of arthropods. Chromosome Res. 2011;19:809–823. doi: 10.1007/s10577-011-9231-6. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A, Hoy M. The nuclear genome of the phytoseiid Metaseiulus occidentalis (Acari: Phytoseiidae) is amongst the smallest known in arthropods. Exp App Acarcol. 2009;47:263–273. doi: 10.1007/s10493-008-9227-x. [DOI] [PubMed] [Google Scholar]
- Wilhelm J, Pingoud A, Hahn M. Real-time PCR-based method for the estimation of genome sizes. Nucleic Acids Res. 2003;31:e56. doi: 10.1093/nar/gng056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Scott JG. Use of quantitative real-time polymerase chain reaction to estimate the genome size of the house-fly Musca domestica genome. Insect Mol Biol. 2006;15:835–837. doi: 10.1111/j.1365-2583.2006.00690.x. [DOI] [PubMed] [Google Scholar]
- Burgess ST, Frew D, Nunn F, Watkins CA, McNeilly TN, Nisbet AJ, Huntley JF. Transcriptomic analysis of the temporal host response to skin infestation with the ectoparasitic mite Psoroptes ovis. BMC Genomics. 2010;11:624. doi: 10.1186/1471-2164-11-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders A, Froggatt P, Wall R, Smith KE. Life-cycle stage morphology of Psoroptes mange mites. Med Vet Entomol. 2000;14:131–141. doi: 10.1046/j.1365-2915.2000.00223.x. [DOI] [PubMed] [Google Scholar]
- Sweatman GK. On the life history and validity of the species in Psoroptes, a genus of mange mites. Can J Zool. 1958;36:905–929. doi: 10.1139/z58-078. [DOI] [Google Scholar]
- Suggars AL. House Dust Mites: A Review. J Entomol Sci. 1987;S1:3–15. [Google Scholar]
- Dolezel J, Bartos J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry A. 2003;51:127–128. doi: 10.1002/cyto.a.10013. [DOI] [PubMed] [Google Scholar]
- De Schutter K, Lin YC, Tiels P, Van Hecke A, Glinka S, Weber-Lehmann J, Rouze P, Van de Peer Y, Callwaert N. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotech. 2009;27:561–566. doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- Walton S, Dougall A, Pizzutto S, Holt D, Taplin D, Arlian L, Morgan M, Currie B, Kemp D. Genetic epidemiology of Sarcoptes scabiei (Acari: Sarcoptidae) in northern Australia. Int J Parasitol. 2004;34:839–849. doi: 10.1016/j.ijpara.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Oliver JH. Cytogenetics of mites and ticks. Ann Rev Entomol. 1977;22:407–429. doi: 10.1146/annurev.en.22.010177.002203. [DOI] [PubMed] [Google Scholar]
- Izraylevich S, Gerson U, Wysoki M. Karyotype and sex determining mechanism of the mite Hemisarcoptes coccopagus Meyer (Acariformes: Astigmata: Hemisarcoptidae) Int J Acarol. 1995;21:229–232. doi: 10.1080/01647959508684064. [DOI] [Google Scholar]
- Norton RA, Kethley JB, Johnson EE, O'Connor BM. In: Evolution and diversity of sex ratio in insects and mites. Wrensch DL, Ebber MA, editor. New York: Chapman & Hall; 1993. Phylogenetic perspectives on genetic systems and reproductive modes of mites; pp. 8–99. [Google Scholar]
- Gregory TR. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol Rev. 2001;76:65–101. doi: 10.1017/s1464793100005595. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Genome size and developmental complexity. Genetica. 2002;115:131–146. doi: 10.1023/A:1016032400147. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, Clark JM, Lee SH, Robertson HM, Kennedy RC, Elhaik E, Gerlach D, Kriventseva EV, Elsik CG, Graur D, Hill CA, Veenstra JA, Walenz B, Tubio JM, Ribeiro JM, Rozas J, Johnston JS, Reese JT, Popadic A, Tojo M, Raoult D, Reed DL, Tomoyasu Y, Kraus E, Mittapalli O, Margam VM. et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci USA. 2010;107:12168–12173. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JS, Yoon KS, Strycharz JP, Pittendrigh BR, Clark JM. Body Lice and Head Lice (Anoplura: Pediculidae) have the smallest genomes of any hemimetabolous insect reported to date. J Med Ent. 2007;44:1009–1012. doi: 10.1603/0022-2585(2007)44[1009:BLAHLA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Holt DC, Fischer K, Allen GE, Wilson D, Wilson P, Slade R, Currie BJ, Walton SF, Kemp DJ. Mechanisms for a novel immune evasion strategy in the scabies mite Sarcoptes scabiei: a multigene family of inactivated serine proteases. J Invest Dermatol. 2003;121:1419–1424. doi: 10.1046/j.1523-1747.2003.12621.x. [DOI] [PubMed] [Google Scholar]
- Holt DC, Fischer K, Pizzutto SJ, Currie BJ, Walton SF, Kemp DJ. A multigene family of inactivated cysteine proteases in Sarcoptes scabiei. J Invest Dermatol. 2004;123:240–241. doi: 10.1111/j.0022-202X.2004.22716.x. [DOI] [PubMed] [Google Scholar]
- Bergström FC, Reynolds S, Johnstone M, Pike RN, Buckle AM, Kemp DJ, Fischer K, Blom AM. Scabies mite inactivated serine protease paralogs inhibit the human complement system. J Immunol. 2009;182:7809–7817. doi: 10.4049/jimmunol.0804205. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Wang R, Huang ZY, Dong KE. Molecular characterisation of an arachnid sodium channel gene from the varroa mite (Varroa destructor) Insect Biochem Mol Biol. 2003;33:733–739. doi: 10.1016/S0965-1748(03)00068-7. [DOI] [PubMed] [Google Scholar]
- Pasay C, Walton S, Fischer K, Holt D, McCarthy J. PCR-based assay to survey for knockdown resistance to pyrethroid acaricides in human scabies mites (Sarcoptes scabiei var. hominis) Am J Trop Med Hyg. 2006;74:649–657. [PubMed] [Google Scholar]


