Abstract
Background
The envelope (env) protein of the human endogenous retrovirus type K (HERV-K) family is commonly expressed on the surface of breast cancer cells. We assessed whether HERV-K env is a potential target for antibody-based immunotherapy of breast cancer.
Methods
We examined the expression of HERV-K env protein in various malignant (MDA-MB-231, MCF-7, SKBR3, MDA-MB-453, T47D, and ZR-75-1) and nonmalignant (MCF-10A and MCF-10AT) human breast cell lines by immunoblot, enzyme-linked immunosorbent assay, immunofluorescence staining, and flow cytometry. Anti-HERV-K env monoclonal antibodies (mAbs; 6H5, 4D1, 4E11, 6E11, and 4E6) were used to target expression of HERV-K, and antitumor effects were assessed by quantifying growth and apoptosis of breast cancer cells in vitro, and tumor growth in vivo in mice (n = 5 per group) bearing xenograft tumors. The mechanisms responsible for 6H5 mAb–mediated effects were investigated by microarray assays, flow cytometry, immunoblot, and immunofluorescence staining. The expression of HERV-K env protein was assessed in primary breast tumors (n = 223) by immunohistochemistry. All statistical tests were two-sided.
Results
The expression of HERV-K env protein in malignant breast cancer cell lines was substantially higher than nonmalignant breast cells. Anti–HERV-K-specific mAbs inhibited growth and induced apoptosis of breast cancer cells in vitro. Mice treated with 6H5 mAb showed statistically significantly reduced growth of xenograft tumors compared with mice treated with control immunoglobulin (control [mIgG] vs 6H5 mAb, for tumors originating from MDA-MB-231 cells, mean size = 1448.33 vs 475.44 mm3; difference = 972.89 mm3, 95% CI = 470.17 to 1475.61 mm3; P < .001). Several proteins involved in the apoptotic signaling pathways were overexpressed in vitro in 6H5 mAb–treated malignant breast cells compared with mIgG-treated control. HERV-K expression was detected in 148 (66%) of 223 primary breast tumors, and a higher rate of lymph node metastasis was associated with HERV-K-positive compared with HERV-K-negative tumors (43% vs 23%, P = .003).
Conclusion
Monoclonal antibodies against HERV-K env protein show potential as novel immunotherapeutic agents for breast cancer therapy.
CONTEXT AND CAVEATS
Prior knowledge
Human endogenous retroviruses (HERVs) are overexpressed in several types of tumors. The envelope protein of HERV-K (HERV-K env) is suggested to trigger an antigen-specific immune response in breast cancer and influence the disease progression.
Study design
Expression of HERV-K env protein was examined in various malignant and nonmalignant human breast cell lines. Anti-HERV-K env monoclonal antibodies were used to target expression of HERV-K, and antitumor effects were assessed in vitro as well as in mice bearing xenograft tumors. Association between HERV-K env protein expression in primary breast tumors and rate of lymph node metastasis was also assessed.
Contribution
Expression of HERV-K env protein was higher in malignant breast cancer cells compared with nonmalignant breast cells. Anti–HERV-K-specific monoclonal antibodies inhibited growth and induced apoptosis of breast cancer cells in vitro. Mice treated with 6H5 monoclonal antibody showed statistically significantly reduced tumor growth compared with control mice. HERV-K expression was associated with a higher rate of lymph node metastasis compared with no expression.
Implications
HERV-K env is a potential target for antibody-based immunotherapy of breast cancer, and monoclonal antibodies against the antigen show potential as novel immunotherapeutic agents.
Limitations
HERV-K may not be the only member of the HERV family that is involved in breast cancer etiology. This study was done in mice, and the efficacy of the antibody is not known in breast cancer patients.
From the Editors
The germline human endogenous retroviruses (HERVs) and other retroviral elements containing long terminal repeat-like sequences constitute up to 8% of the human genome (1). It is thought that none of these germline viral sequences encodes an infectious virus, but hormonal stimuli and stress factors can induce transcription of retroviral proteins and viable viral particles from several genomic loci that can be detected as cellular antigens and/or viral particles in tumor tissues and blood samples from cancer patients (2–4). Members of the HERV type K family (HERV-K) appear to have the full complement of open reading frames typical of replication-competent mammalian retroviruses (5,6). HERV-K-encoding loci are thought to be transcriptionally silent in normal cells but become active after malignant transformation, as found in germ cell tumors (7). As a consequence, HERV-K genes are found to be overexpressed in several types of cancer cell lines and tumors including germ cell tumors (8), melanoma (9), and human breast and ovarian tumors (10–15). The envelope protein of HERV-K (HERV-K env) consists of a 55-kDa surface subunit (SU) and a 39-kDa transmembrane subunit (16). Our group has previously reported that the expression of HERV-K env transcripts in breast cancer triggers an antigen-specific immune response and observed along with others that HERV-K expression may influence disease pathophysiology or outcome (2,3,9,17,18). A T cell response against HERV-K was detected in peripheral blood mononuclear cells from breast cancer patients stimulated with autologous dendritic cells pulsed with HERV-K env surface antigens, but a response was not detected in matched normal control subjects (18).
Although published data suggest that HERV-K expression may be associated with disease progression in breast cancer (19), almost nothing is known about the functional role of HERV-K proteins in tumor biology. In this study, we used the anti-HERV-K monoclonal antibodies (mAbs) developed previously in our laboratory (18) to investigate whether endogenous retroviral protein-specific mAbs can induce apoptosis and regression of breast tumors in a mouse xenograft model. Antitumor activity of the mAbs, if detected, may have important clinical implications.
Patients, Materials, and Methods
Cell lines, Cell Culture, and Reagents
Malignant human breast cancer cell lines (MDA-MB-231, SKBR3, MDA-MB-453, T47D, and ZR-75-1), a nonmalignant human breast epithelial cell line (MCF-10A), and human embryonic kidney 293T (HEK293T) cells were obtained from the American Type Culture Collection (Rockville, MD). The MDA-MB-435EB1 human breast cancer cell line, which was established by transfecting c-erbB-2 cDNA into MDA-MB-435 and which overexpresses v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) (ERBB2), was a gift from Dr Michael Rosenblum at The University of Texas MD Anderson Cancer Center (Houston, TX). The IDC51N, IDC51T, and IDCm73T cells were cultured from breast cancer patient tissues. The breast cancer cells and HEK293T cells were maintained in Dulbecco’s modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with penicillin (100 U/mL) (Invitrogen), streptomycin (100 μg/mL) (Invitrogen), and fetal bovine serum (FBS; 10%) (Atlanta Biologicals, Lawrenceville, GA), which is the complete DMEM. The v-Ha-ras Harvey rat sarcoma viral oncogene homolog (HRAS)–expressing, weakly tumorigenic MCF-10AT cells were purchased from The Barbara Ann Karmanos Cancer Institute (Detroit, MI). MCF-10A and MCF-10AT cells were maintained in DMEM: Nutrient Mixture F-12 (DMEM/F-12) (Invitrogen) supplemented with penicillin (100 U/mL) (Invitrogen), streptomycin (100 μg/mL) (Invitrogen), horse serum (5%) (Invitrogen), insulin (10 μg/mL) (Sigma-Aldrich, St Louis, MO), epidermal growth factor (20 ng/mL) (Invitrogen), and hydrocortisone (0.5 μg/mL) (Sigma-Aldrich). MCF-7, a malignant human breast cancer cell line, was a gift from Dr Melinda Hollingshead (Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis [DCTD], National Cancer Institute, Bethesda, MD). These cells were maintained in DMEM media supplemented with penicillin (100 U/mL) (Invitrogen), streptomycin (100 μg/mL) (Invitrogen), and FBS (10%) (Atlanta Biologicals). The identity of cell lines was confirmed by short tandem repeat genetic profiling.
Patient Samples
Primary tumor and uninvolved normal human breast cells were isolated from surgery specimens obtained from breast cancer patients at The University of Texas MD Anderson Cancer Center according to an approved Institutional Review Board (IRB) protocol (LAB04-0083). Breast cancer cells (T) and matched uninvolved normal epithelial cells (N) were successfully cultured from two patients, Acc 51 and Acc 71, and used for this study. Patient Acc 51 was a 47-year-old woman of white race with invasive mammary carcinoma (nuclear grade 2, estrogen receptor positive [ER+], progesterone receptor positive [PR+], without metastasis), and patient Acc 73 was a 70-year-old woman of white race with invasive ductal carcinoma (IDC) (nuclear grade 2, ER+, PR+, HER2 negative with lymph node metastasis). Paraffin-embedded tumor specimens (n = 223) were obtained from breast cancer patients who resided in the greater Baltimore area, as described previously (20). Patients were recruited at the University of Maryland Medical Center (UMD), the Baltimore Veterans Affairs Medical Center, Union Memorial Hospital, Mercy Medical Center, and the Sinai Hospital in Baltimore between February 15, 1993, and August 27, 2003. All patients signed a consent form. Clinical and pathological information was obtained from medical records and pathology reports. Disease staging was performed according to the TNM staging system of the American Joint Committee on Cancer (AJCC) (21) and the Union for International Cancer Control (UICC) (22). Low stage was defined as stage I or II, and high stage was defined as stage III or IV. The Nottingham grading system (23) was used to determine the tumor grade using the cumulative score of glandular differentiation, nuclear pleomorphism, and mitotic count. Low grade was defined as grade 1 or 2, and high grade was defined as grade 3.The collection of the tumor specimens and clinical and pathological information was reviewed and approved by the University of Maryland IRB for the participating institutions (UMD protocol number 0298229). IRB approval was then obtained at all institutions (Veterans Affairs Medical Center, Union Memorial Hospital, Mercy Medical Center, and Sinai Hospital). The research was also reviewed and approved by the Office of Human Subjects Research (OHSR) at the National Institutes of Health (OHSR number 2248).
Production and Affinity Purification of Antigens and Antibodies
HERV-K SU gene was cloned into a pGEX-6P1-glutathione S-transferase (GST) expression vector (GE Healthcare Life Sciences, Piscataway, NJ) to obtain HERV-K env SU-GST recombinant fusion protein (K-GST). Protein expression was induced with isopropylthiogalactoside (IPTG) in Escherichia coli (E coli) BL-21 (DE3), and fusion proteins were affinity purified using an ÄKTA Fast Protein Liquid Chromatography (FPLC) system (GE Healthcare Life Sciences) equipped with a GSTrap column as described previously (18). HERV-K SU was also cloned into a 6xHis-tagged pQE30 expression vector (Qiagen, Chatsworth, CA). The resultant construct (K-Q18), a recombinant fusion protein consisting of HERV-K env SU tagged with 6xHis, was expressed by induction with IPTG in M15 E coli and affinity purified using the ÄKTA system. Purified K-Q18 fusion protein was used to immunize 6- to 8-week-old female BALB/c mice for production of hybridoma using standard technology. Previously, we developed mAbs against HERV-K, member 7 (18). Mouse anti-HERV-K mAbs 6H5, 4D1, 4E11, 6E11, and 4E6 were selected for specific binding to K-GST antigen by enzyme-linked immunosorbent assay (ELISA) and immunoblot. These mAbs were produced from their respective hybridomas by the ascites method in female BALB/c mice and purified using Protein G HP resin (GE Healthcare Life Sciences) as described previously (18). Briefly, hybridoma cells were produced by immunizing 6- to 8-week-old female BALB/c mice with K-Q18 fusion protein. The splenocytes of the immunized mice were fused with a myeloma cell line to derive hybridoma cells. Hybridoma cells that generated anti-HERV-K antibodies were selected by ELISA using K-GST protein. Sample endotoxin content was determined by limulus amebocyte lysate (LAL) assay (Lonza, Basel, Switzerland). Any endotoxin was removed from antibody preparations by affinity purification using Detoxi-Gel resin columns (Pierce, Rockford, IL) as per manufacturer's instructions.
A single-chain variable fragment (scFv) antibody was generated from 6H5 mAb as described previously (24) using the Recombinant Phage Antibody System (GE Healthcare Life Sciences) to investigate possible advantages of using this antibody compared with HERV-K mAbs with respect to tumor uptake and immunogenicity. Briefly, the scFv clone G11D10 was selected using a colony lift assay followed by ELISA using K-GST protein as antigen. G11D10 scFv antibody was induced by IPTG in E. coli HB2151, and the periplasmic extract was clarified using a 0.45-μm cellulose acetate filter (Corning Life Sciences, Corning, NY) and purified by ÄKTA FPLC using a HiTrap anti-Etag column (GE Healthcare Life Sciences). The purity of the antibodies and fusion proteins was evaluated by Coomassie blue staining or immunoblot.
Immunoblot Analysis
Total protein lysates of cells (MCF-10A, MCF-10AT, SKBR3, T47D, MDA-MB-231, and MCF-7) were used for immunoblot analysis (50 μg protein per lane) as described previously (18,24). The following primary antibodies were used: 6H5 mAb (1 μg/mL final concentration) and mouse anti-chicken β-actin (ACTB) mAb (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) (1:1000 dilution), followed by treatment with anti-mouse immunoglobulin (mIgG) HRP (1:5000 dilution) (Sigma-Aldrich). Rabbit anti-human caspase 3, mouse anti-human caspase 8, and mouse anti-human caspase 9 antibodies (Sigma-Aldrich; 1:1000 dilution) were used to detect cleavage of caspase proteins by immunoblot. Rabbit anti-human polyclonal antibody to active caspase 7 (Abcam, Cambridge, MA; 1 μg/mL final concentration) was used for detection of active caspase 7. Mouse anti-human cell death–inducing DFFA-like effector A (CIDEA) antibody (Abnova, Taipei, Taiwan; 1:1000 dilution) was used for detection of CIDEA expression in cells treated with 6H5 mAb or mIgG. Other primary antibodies used for immunoblot were mouse anti-human tumor protein p53 (TP53, 1 μg/mL final concentration), rabbit anti-human tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain (TNFRSF10D, 0.5 μg/mL final concentration), mouse anti-human cyclin-dependent kinase inhibitor 1A (CDKN1A, 1:500 dilution), mouse anti-human cyclin-dependent kinase 5 (CDK5) (all from Sigma-Aldrich); mouse anti-human myogenic differentiation 1 (MYOD1, 1 μg/mL final concentration), mouse anti-human glycosylphosphatidylinositol anchored molecule like protein (GML, 1 μg/mL final concentration), rabbit anti-human TP53-regulated apoptosis–inducing protein-1 (TP53AIP1, 1 μg/mL final concentration) (all from Abcam); mouse anti-human FAS ligand (FASLG; TNF superfamily, member 6, 1:200 dilution), goat anti-human tumor necrosis factor receptor superfamily, member 8 (TNFSF8, 1:200 dilution) (both FASLG and TNFSF8 antibodies from Santa Cruz Biotechnology, Santa Cruz, CA), and antitumor necrosis factor receptor superfamily, member 25 (TNFRSF25, also known as DR3) (rabbit anti-human DR3, 1:1000 dilution) (Sigma-Aldrich). Samples treated with 1000 U of peptide-N-glycosidase F (PNGase F; New England Biolabs, Ipswich, MA) to remove all N-linked carbohydrates for glycoprotein analysis were incubated at 37°C overnight in reaction buffer provided by the manufacturer. These PNGase F-treated samples were resolved on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed using a glycoprotein detection kit (GLYCOPRO; Sigma) and an immunoblot assay with 6H5 mAb (1 μg/mL final concentration). All immunoblots were performed using 0.2 μm polyvinylidene fluoride (PVDF) membranes (Immun-blot; Bio-Rad, Hercules, CA) and blocked with 3% bovine serum albumin (BSA) in Tris-buffered saline (20 mM Tris [pH 7.5], 150 mM NaCl) with 0.2% Tween-20.This experiment was repeated at least two independent times.
Metabolic Turnover Assay
We examined the metabolic turnover of the target antigen HERV-K env protein in the absence of 6H5 mAb using a biotinylation pulse-chase assay described previously (25). MDA-MB-231 and MCF-7 cell surface proteins were pulsed with biotin using a Sulfo-NHS-SS-biotin reagent (Pierce), as per manufacturer’s instructions, and incubated at 37°C for 0, 5, 15, 45, 90, and 180 minutes. At each time point the cells were lysed, biotinylated proteins were pulled down with streptavidin resin, and the pull-down proteins were analyzed by immunoblot for detection of HERV-K env expression (surface and full-length envelope protein) using 6H5 mAb (21). This experiment was repeated at least three independent times.
Enzyme-Linked Immunosorbent Assay
Sensitivity and specificity of antibody–antigen interaction were analyzed by ELISA, as described previously (15,24,26). For cell ELISA, breast cells (MDA-MB-231, MCF-7, SKBR3, MCF-10A) (2500 cells per well) were seeded in a 96-well plate and incubated at 37°C overnight. After washing the plate one time with 1× phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), the liquid was removed, and the plate was incubated at 37°C overnight to dry. Cells were fixed with 3.7% formaldehyde (Sigma-Aldrich) for 8 minutes, washed 5 times with 1× PBS, and then blocked for 2 hours at room temperature with 3% BSA in 1× PBS containing 0.05% Tween-20. Serial dilutions of 6H5 mAb (1:1000, 1:3000, 1:9000, 1:27 000, 1:81 000, 1:243 000, 1:729 000, and 1:2 187 000) or mIgG (using the same dilutions used for 6H5 mAb) were added, and the plate was incubated overnight at 4°C. After washing with 1× PBS, anti-mIgG–HRP (1:4000) was added, and the plate was incubated for 1 hour at room temperature. After washing with 1× PBS, the substrate 2,2’-Azino-bis(3-Ethylbenzthiazoline-6-Sulfonic Acid) (ABTS) (Sigma-Aldrich) was added for 10–30 minutes, and the plate was read on a Wallac Victor 2 V Microplate Reader (PerkinElmer, Waltham, MA). This experiment was repeated at least three independent times.
Immunofluorescence, Fluorescence-Activated Cell Sorting (FACS), and Antibody Conjugation Assays
For immunofluorescence assays, breast cells (MCF-10A, MCF-10AT, MCF-7, MDA-MB-231, MDA-MB-453) were cultured in six-well plates (2 × 105 cells per well) containing glass coverslips for 24 hours. Cells were fixed with 3.7% paraformaldehyde at room temperature for 8 minutes followed by washing six times with 1× PBS. Cells were incubated with 6H5 mAb (10 μg/mL) for 60 minutes followed by six washes with 1× PBS. Cells were incubated with goat anti-mIgG conjugated with AlexaFluor 488 (anti-mIgG-AF488) (Invitrogen) for 30 minutes followed by six washes with 1× PBS. Coverslips were mounted onto slides using 10% glycerol. Slides were examined with the use of a Zeiss LSM 510 confocal imaging system (Zeiss, Heidelberg, Germany). Cells were subjected to immunofluorescence staining with antibodies against caspases 3, 8, and 9, CDKN1A, and CDK5 (1 μg/100 μL per slide). To detect cytoplasmic expression of HERV-K env, cells were treated with 0.1% Triton X-100 as described previously (15). This experiment was repeated at least three independent times.
For FACS assays, breast cells (MCF-10A, MCF-10AT, MCF-7, MDA-MB-231, SKBR3, T47D, IDC51N and IDC51T, and MDA-MB-453) were removed from culture flasks using 1× trypsin and added to a 96-well plate (1 × 106 cells per well). Cells were incubated with 6H5 mAb (1 μg/106 cells) for 1 hour at room temperature followed by six washes with 1× PBS. Cells were incubated with goat anti-mIgG-Alexa Fluor 647 (Invitrogen) (mIgG conjugated with AlexaFluor 647, 1 μg/106 cells) for 30 minutes, washed six times with 1× PBS, resuspended in 250 μL 1× PBS, and analyzed on a FACS Array (BD Biosciences, Franklin Lakes, NJ). For blocking experiments, 6H5 mAb (1 μg) was preincubated with K-GST (1 μg) for 30 minutes at room temperature before adding to cells. This experiment was repeated at least three independent times.
A quantitative indirect immunofluorescence (QIFI) assay was used for the quantitation of surface envelope levels. QIFI beads, which are microbeads coated with different known amounts of mIgG of IgG1 subclass to mimic cells coated with mAbs at saturating concentration, were stained with goat anti-mIgG-Alexa Fluor 647 (1 μg per 1 × 106 cells). The peak fluorescence position of each bead population vs the number of antibody molecules per calibration bead (1700, 11 000, 54 000, 194 000, and 561 000 molecules per bead) (determined by Dako, Glostrup, Denmark) was plotted, as described previously (27). The number of surface molecules on the cells was determined with a calibration curve, so that arbitrary units of mean fluorescence intensity (MFI) can be transformed to molecules of equivalent fluorochrome values that can be compared from one assay to another.
For antibody conjugation to AlexaFluor dye, 20 μg of antibody in PBS (pH 8.0) were incubated with 1 μg of carboxylic acid succinimidyl ester–activated AlexaFluor dye (Invitrogen) at room temperature in the dark for 1 hour. The conjugate was purified from free dye using size exclusion chromatography (Bio-Rad P30 gel) in spin column format. Propidium iodide (Sigma-Aldrich) or 4’-6-diamidino-2-phenylindole (DAPI) (Invitrogen) was used for nuclear staining. To detect cytoplasmic expression of HERV-K env, cells were treated with 0.1% Triton X-100, as described previously (15).
Kinetic Binding Assays
Surface plasmon resonance assays are used to evaluate kinetics of binding, primarily the rate of association (Ka) and dissociation (Kd). We used these assays to assess affinity or strength of binding of mAb or scFv antibody to its antigen by determining the affinity constant (KD) values of 6H5 mAb and G11D10 scFv antibody for the HERV-K env protein. Assays were performed on a Biacore X100 instrument at 10 μL/min flow rate. Two sensor surfaces (flow cell 1 and flow cell 2) of a Biacore CM5 sensor chip (BIACORE, Piscataway, NJ) were activated with 0.4M N-ethyl-N’-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC)/0.1 M N-hydroxysuccinimide (NHS). Two hundred resonance units (RU) of recombinant HERV-K SU env protein, that is, approximately 200 pg/mm2, were immobilized onto flow cell 2 in 20 mM sodium acetate (pH 4.5). One RU corresponds to 1 pg/mm2 of mass bound to the sensor surface. Flow cell 1 contained no ligand. Both flow cells were then quenched using 1 M ethanolamine. In separate experiments, a series of concentrations (3, 6, 12, 24, and 48 nM) of 6H5 mAb or G11D10 scFv antibody were prepared in HEPES-buffered saline (10 mM HEPES, 150 mM sodium chloride), with 3 mM EDTA and 0.005% Tween-20 (HBS-EP) running buffer (pH 7.4) and injected onto both the ligand (K-GST: flow cell 2) and control flow cells. Kinetic constants were determined from blank normalized curves using Biacore X100 evaluation software (BIACORE) by fitting to the 1:1 Langmuir binding model. The affinity of the antibody (6H5 mAb or G11D10 scFv antibody) for its ligand (K-GST) is determined by measuring the binding kinetics of the interaction and expressed as KD for the interaction of a single antibody combining site with a single epitope. All concentrations were injected in duplicate.
Cycling of HERV-K env Protein and Net Antibody Internalization Assays
For qualitative and quantitative analysis of cycling of HERV-K env protein between the cell surface and intracellular stores, a cycling assay was used. MCF-7, MDA-MB-231, and T47D cells were incubated in 96-well U-bottom plates with unlabeled 6H5 mAb (10 μg/mL) at 4°C for 20 minutes, washed one time with 1× PBS, and incubated with unlabeled 6H5 mAb at 37°C for different time intervals (0, 1, 5, 15, and 45 minutes) as described previously (28). At each time point, cells in one-half of the wells were labeled with 5 μg/mL of goat anti-mIgG-Alexa Fluor 647 at 4°C for 30 minutes to detect HERV-K env protein that remained on the cell surface. The percentage of internalization at each time point was 100 minus the percentage of label at time 0. Cells in the other half of the wells were treated with citrate phosphate buffer (0.131 M citric acid, 0.066 M sodium monohydrogen phosphate, pH 3) to strip 6H5 mAb from the surface and then reincubated with 4D1 mAb conjugated to AlexaFluor 555 (Invitrogen), to determine whether 6H5 mAb influenced the cell surface binding of HERV-K env protein.
Cell Growth and Cell Proliferation Assays
The viability of cell lines (MCF-10AT, MCF-7, MDA-MB-231, SKBR3, IDCm73T, and T47D) as a function of antibody dosage was tested in the presence or absence of 6H5 mAb (10 μg/mL per dose). Untreated and mIgG (10 μg/mL)-treated cells were used as controls in all assays. Cells were seeded in a 12-well plate (2 × 104 cells per well) and incubated overnight for attachment. One group of cells was treated with 6H5 mAb (10 μg/mL per dose) or control mIgG (10 μg/mL per dose) administered at 24, 48, and 72 hours after seeding and harvested at 96 hours after seeding (designated as dose 3). In a second protocol, two doses were administered at 48 and 72 hours after seeding and harvested at 96 hours after seeding (designated as dose 2). Finally, one dose was administered at 72 hours after seeding and harvested at 96 hours after seeding (designated as dose 1). Cells in each well treated with 6H5 mAb or mIgG were counted after trypan blue staining using a hemocytometer. Triplicate fields containing at least 100 cells per field were counted, and because the volume counted was known, the counts were converted to total cell numbers per well. The counts of cells treated with 6H5 mAb or mIgG under dose 1, dose 2, or dose 3 conditions were plotted.
To evaluate the effect of antibody concentration on cell proliferation, breast cell lines were plated in a 96-well plate (3000 cells per well) and incubated overnight for attachment. Cells were treated with a serial dilutions of 6H5 mAb (2.13 × 10−12, 1.07 × 10−11, 5.33 × 10−11, 2.67 × 10−10, 1.33 × 10−9, 6.67 × 10−9, 3.33 × 10−8 M) or mIgG (3.33 × 10−8 final concentration) for 72 hours, and cell proliferation was determined using the CellTiter 96 AQ Non-Radioactive Cell Proliferation Assay (MTS assay; Promega, Madison, WI). The absorbance produced by chromophore in all positively stained cells in each well was measured at 490 nm using a Wallac Victor 2 V Microplate Reader (Perkin Elmer, Waltham, MA). Cytotoxicity was determined by staining with crystal violet (26). The absorbance was measured at 600 nm using a Wallac Victor 2 V Microplate Reader.
Bromodeoxyuridine (BrdU) Incorporation and Cell Cycle Assay
Cell cycle analysis was performed using BrdU, an analog of the DNA precursor thymidine, which incorporates into newly synthesized DNA. MCF-10A, MCF-10AT, MDA-MB-453, T47D, MCF-7, MDA-MB-231, and SKBR-3 cells (2.5 × 105 per well) were plated in a six-well plate (BD Biosciences) in their respective media. Culture media containing FBS was replaced with serum-free media 24 hours before analysis. Cells were pulsed with BrdU (30 μmol/mL) (APC BrdU Flow Kit; BD Biosciences) and cultured with 10 μg/mL 6H5 mAb or mIgG at 37°C for 24, 48, and 72 hours. Cells were resuspended in cytofix/cytoperm buffer (BD Biosciences) and placed on ice for 30 minutes. After washing with 1× PBS, cells were resuspended in cytoperm plus buffer (BD Biosciences) and incubated on ice for 10 minutes. After washing with 1× PBS, cells were refixed with cytofix/cytoperm buffer on ice for 5 minutes. After washing with 1× PBS, cells were resuspended in DNase (300 μg/mL, APC BrdU Flow Kit; BD Biosciences) in Dulbecco's phosphate-buffered saline (DPBS; Sigma-Aldrich) and incubated at 37°C for 1 hour. Cells were washed, stained with anti-BrdU conjugated with allophycocyanin (APC) at room temperature for 20 minutes, and then analyzed on a BD FACSArray. For total cellular DNA staining, 7-amino-actinomycin D (7-AAD) was used. This two-color flow cytometry analysis quantitates cells that are actively synthesizing DNA (BrdU incorporation) in terms of their cell cycle position, which is defined by 7-AAD staining intensities.
Ki-67 assay
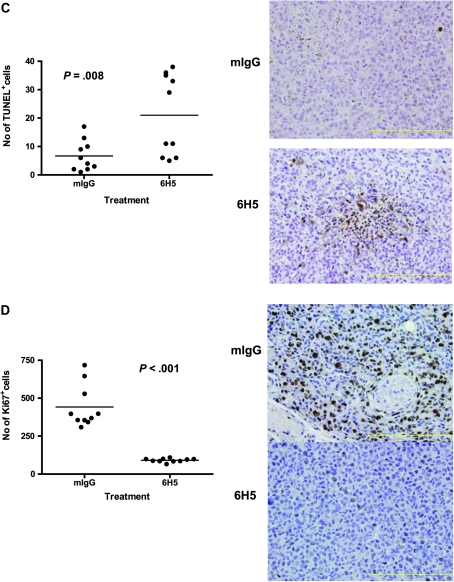
Ki-67 is a nuclear antigen and proliferation marker expressed only in cycling cells. Consequently, assessment of Ki-67 staining on paraffin-embedded tumor sections is an estimate of the proliferation index of individual tumors. Immunohistochemistry (IHC) was performed on paraffin-embedded tumor sections using mouse anti-human Ki-67 mAb (BD Bioscience) and a VECTASTAIN Elite ABC Kit (Vector Laboratories, Burlingame, CA). After deparaffinization, tumor sections were covered in 1 μg of anti-Ki-67 antibody diluted in 100 μL blocking buffer (1% BSA in 1× PBS) and incubated at room temperature for 60 minutes. Ten random fields were used for counting the Ki-67-positive cells under an Olympus 1X51 microscope (Olympus, Center Valley, PA) using the 20× objective.
Apoptosis Assays
Annexin V staining.
Annexin V staining precedes the loss of membrane integrity that characterizes the later stages of cell death, and therefore the vital dye 7-AAD, in conjunction with annexin V, was used to allow discrimination of early apoptotic cells. To measure the effects of 6H5 mAb on cellular apoptosis in vitro, breast cells MCF-10A, MCF-10AT, MCF-7, MDA-MB-231, MDA-MB-453, and T47D (5 × 105 cells/mL) were grown in their respective media and treated with 6H5 mAb at various concentrations (0, 1.25, 2.5, 5, and 10 μg/mL) at 37°C for 16 hours, or were treated with various mAbs (4D1, 4E11, 6H5, and 6E11) at a constant concentration of 10 μg/mL, or with an mIgG control antibody. Treated cells were harvested and resuspended in 200 μL 1× binding buffer (BD Pharmingen) at 1 × 106 cells per well. Cells were stained with annexin V-APC (red; BD Pharmingen) at room temperature for 15 minutes followed by one wash with 1× annexin V binding buffer. Cells were then incubated with 7-AAD-phycoerythrin-cyanide 7 (PE-Cy7) at room temperature for 10 minutes, and samples were analyzed on a BD FACSArray Bioanalyzer. The percentage of annexin V–positive cells for each cell line treated with 6H5 mAb was determined using FlowJo software (version 7.2.5).
Deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay.
The ApopTag Peroxidase In Situ Apoptosis Detection kit (Chemicon International, Temecula, CA) was used for measuring apoptosis in paraffin-embedded tumor sections by labeling and detecting DNA strand breaks by the TUNEL method. Tumor sections (5 μM) were labeled at the free 3’OH DNA termini in situ with digoxigenin-labeled and unlabeled nucleotides added to the DNA by terminal deoxynucleotidyl transferase (TdT). The labeled DNA fragments then bind an anti-digoxigenin antibody conjugated to a peroxidase reporter molecule, which generates a strong stain from chromogenic substrates. Numbers of apoptotic cells were counted in 10 randomly selected high-power fields. Each slide was examined on at least two separate occasions by two individuals.
Analysis of Apoptosis and TP53 Signaling Pathways
Breast cancer cells (MCF-7) were treated with 6H5 mAb or mIgG (10 μg/mL) at 37°C for 24 hours, and total RNA was extracted using TRI Reagent Solution (Applied Biosystems, Foster, CA) and subjected to quantitative polymerase chain reaction (qPCR) analyses using Human Apoptosis and Human p53 Signaling Pathway PCR cDNA arrays, both from SA Biosciences (Frederick, MD). RNA extracts of cells treated with 6H5 mAb or mIgG were used for cDNA synthesis by reverse transcription (RT) using a RT2 First Strand Kit (SA Biosciences), per manufacturer's instructions. Briefly, RNA extracts (1 μL containing 0.5 μg) were mixed with 10 μL of the RT cocktail and incubated at 42°C for exactly 15 minutes. The reaction was immediately stopped by heating at 95°C for 5 minutes. Water (91 μL) was added, and the mixture was kept on ice until the PCR reaction. cDNA (102 μL reaction volume) obtained from cells treated with 6H5 mAb or mIgG was added to RT2 qPCR master mix (1350 μL), and 1248 μL water was added, for a total volume of 2700 μL; 25 μL of the mixture was aliquoted per well into the wells of a 96-well plate that contained 84 predispensed pathway gene–specific primer sets plus wells containing five housekeeping genes and three RNA and PCR quality controls (SA Biosciences). A model 7900HT real-time PCR system (Applied Biosystems) was used with a two-step cycling program for qPCR reactions: 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute. The relative gene expression was determined by the difference in the Ct values (ΔΔCt) method; Ct is the PCR cycle at which the sample reaches the threshold, and ΔΔCt is ΔCt,sample − ΔCt,reference, where ΔCt,sample is the Ct value for any 6H5 mAb–treated sample normalized to the endogenous housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ΔCt,reference is the Ct value for the mIgG-treated sample also normalized to the endogenous housekeeping gene. Each array contained a panel of 96 primer sets for 84 relevant pathway- or disease-focused genes plus five housekeeping genes and three RNA and PCR quality controls. The relative expression levels for genes expressed after 6H5 mAb or mIgG treatment were determined.
In Vivo Studies
Mice were randomized into treatment groups, and researchers were blinded to the assignment of treatment groups in all in vivo studies. Anti-HERV-K sera were obtained from BALB/c mice immunized with K-GST as described previously(18). Briefly, K-GST fusion proteins (100 μg in 100 μL 1× PBS with 100 μL Freund's complete adjuvant; Sigma-Aldrich) were used for subcutaneous immunization of 6- to 8-week-old female BALB/c mice (n = 3 per group), weighing between 21 and 25 g (NCI, Frederick, MD). Three weeks later, K-GST fusion proteins (100 μg in 100 μL 1× PBS with 100 μL Freund's incomplete adjuvant) were administered subcutaneously in weekly boosters for 3 weeks. Ten days after the final boost, the mice (n = 3 per group) were killed, and sera were collected by heart puncture. Anti-HERV-K serum antibody titers were determined by ELISA and immunoblot using K-GST fusion protein antigen.
Female immunodeficient athymic NCr-nu/nu mice (n = 15 per cell line), 6 to 8 weeks old and weighing between 21 and 25 g (NCI), were inoculated subcutaneously in the fourth mammary fat pad with 5 × 106 cells of breast cancer cell lines MCF-7 or MDA-MB-231 on day 1. The mice were randomly divided (n = 5 per group) and treated with 6H5mAb, or G11D10 scFvantibody, or mIgG, on days 4, 6, and 8. The 6H5 mAb or G11D10 scFv antibodies were administered intravenously at a dosage of 0.33 mg per injection, for a total dose of 1 mg per mouse over the 3-day dosing period. Immunodeficient mice were also treated with 6H5-r-Gel (0.33 mg per injection) or sera (200 μL per mouse) by intravenous injection. Mice (n = 5 per group) were injected intravenously with 0.33 mg of mIgG per injection as controls (n = 5 mice in all control groups). Additional control sera were obtained from unimmunized mice (n = 5). Tumor volumes (L × W × D; L = length; W = width; D = depth) for each group were compared. At least two independent experiments were done using the MCF-7 xenograft model, and at least three independent experiments were done using MDA-MB-231 xenografts. Mice were housed in a temperature-, humidity-, and light-controlled room and fed a stock regular diet. All mouse experiments were approved by the Institutional Animal Care and Use Committees (IACUC, protocol number 01-04-00733) of the University of Texas MD Anderson Cancer Center. The ventilated caging housing system consists of a polycarbonate or polysulfone cage, a polycarbonate or polysulfone filter lid with woven-fiber filter media, and a stainless steel wire-bar lid. The individual cages are maintained on a HEPA-filtered, ventilated, suspended cage-type rack. Per Keeling Center for Comparative Medicine and Research practice, the maximum mouse population per mouse cage is five adult mice of any size or age. Mice were placed in a chamber filled with CO2 and after breathing stopped and the mice were unconscious, euthanasia was completed by cervical dislocation. Tumors were excised on the final day of the study—day 31, day 39, or day 40. The tumor tissues were dissected, transferred to 10% neutral buffered formalin (Fisher Scientific, Pittsburgh, PA), and embedded into paraffin blocks. Five-micrometer-thick formalin-fixed and paraffin-embedded biopsy sections were used for histological diagnosis, TUNEL, and Ki-67 assays.
All animal facilities of The University of Texas MD Anderson Cancer Center are under the direction of full-time veterinarians and are fully accredited by the American Association of Accreditation of Laboratory Animal Care. The University of Texas MD Anderson Cancer Center complies with the National Institutes of Health's policy on animal welfare, the Animal Welfare Act and all applicable federal, state, and local laws.
Analysis of HERV-K env Expression by IHC
IHC was performed on 5-μm formalin-fixed paraffin-embedded tissue sections using standard protocols and the VECTASTAIN Elite ABC Kit as described previously (18). The expression of antigens was evaluated with the following primary antibodies: 6H5 mAb (1:200 dilution) for HERV-K env and mouse anti-mouse inducible nitric oxide synthase (NOS2; amino acids 961-1144) mAb (1:250 dilution) (DakoCytomation Envision System). Briefly, the slides were baked in an oven at 60°C for 1 hour, then deparaffinized with 100% xylene at room temperature for 1 minute, then hydrated in a graded alcohol series consisting of two 30-second dips each in 100% and 95% ethyl alcohol diluted in water (total volume is 5 mL) at room temperature, and then hydrated in water. Sections were incubated in 3% hydrogen peroxidase in water at room temperature for 10 minutes to block endogenous peroxidase activity. After washing the slides for 5 minutes in water, blocking solution (four drops of stock horse serum in 10 mL of 1× PBS; horse serum was provided in the VECTASTAIN Elite ABC Kit) was added, and the slides were incubated at room temperature for 30 minutes. Slides were then incubated with 6H5 mAb (1 μg in 150 μL normal serum buffer per slide) at 4°C for 16 hours. After three washes with 1× PBS, slides were incubated for 30 minutes with anti-mIgG-HRP secondary antibody (1:600 dilution) in blocking buffer. After three washes with 1× PBS, slides were incubated in 3,3-diaminobenzidine for 5 minutes and counterstained with hematoxylin. Envelope expression was categorized by intensity (0 = absent; 1 = weak; 2 = moderate; 3 = strong) and distribution (percent tumor positive for envelope) (18). Intensity and distribution scores were multiplied to obtain the final score (0–300) for envelope expression in a tumor. The expression level of NOS2 was categorized as low and high based on a combined score of intensity and distribution, as described previously (20).
Statistical Analysis
For cell culture studies, differences between treatment groups were analyzed by Student's t test, and means, standard deviations, and 95% confidence intervals (CIs) were calculated using GraphPad Prism 5 (GraphPad Software Inc, San Diego, CA). The χ2 and Fisher exact tests were used to assess associations between HERV-K env expression and clinicopathologic features of breast cancer, such as grade (grade 1 or 2 vs grade 3), disease stage (TNM stage I or II vs III or IV), node status (negative vs positive), or tumor hormone receptor status (ER α negative vs ER α positive), as described previously (29). Adjusted logistic regression was used to analyze dichotomized data for independent association with clinicopathologic features of breast cancer and to calculate odds ratios (ORs), 95% confidence intervals, and P values.
A repeated measures linear mixed model was applied to analyze the difference in tumor growth in vivo, allowing for one level of variation between mice and another level of variation within mice (repeated tumor size measures over time). Observations were recorded longitudinally for individual mice. Using a human breast cancer xenograft model, we observed previously in our laboratory (F. Wang-Johanning, unpublished data) that the control group had a mean tumor volume of 2000 mm3 (SD = 856 mm3) by day 36, and the group treated with an anti-HERV-K mAb had a mean tumor volume of 170 mm3 (SD = 24.5 mm3). Using these numbers as a guide, five mice per group were needed to ensure a power of 80% to detect a difference at alpha level of 0.05. All statistical tests were two-sided, and all P values less than .05 were considered statistically significant. Data analysis was performed using R statistical free software (http://www.r-project.org/).
Results
Characterization of Anti-HERV-K env Antibodies
We checked the purity of 6H5 mAb, G11D10 scFv antibody, and K-GST fusion protein (Supplementary Figure 1, A–C, available online). We observed in an immunoblot assay that G11D10 scFv subclones (A6, A10, E8, and F10) were able to bind to K-GST protein (Supplementary Figure 1, D, available online). Several assays were used to determine the specificity and sensitivity of 6H5 mAb and G11D10 scFv antibody binding to K-GST (Supplementary Figure 1, E, available online). The sensitivity of several mAbs was determined by ELISA assays, which showed that all mAbs and G11D10 scFv antibody had similar binding sensitivity (Supplementary Figure 1, E, left panel, available online). Specificity of binding of 6H5 mAb to K-GST protein and not to other HERV-K fusion proteins, such as Np9, Rec, or the HERV-K transmembrane env fusion protein was demonstrated (Supplementary Figure 1, E, right panel, available online). Biacore analysis revealed that 6H5 mAb had a greater affinity for its target antigen (association rate constant [Ka] = 2.43 × 105 M−1s−1; dissociation rate constant [Kd] = 3.54 × 10−4 s−1; and affinity constant [KD] = 1.46 × 10−9 M) compared with G11D10 scFv antibody (Ka = 3.33 × 105 M−1s−1; Kd = 1.73 × 10−3 s−1; and KD = 5.21 × 10−9 M) (Supplementary Figure 1, F, available online).
The kinetics of HERV-K env internalization after antibody binding was quantified by a flow cytometry-based cycling assay. After binding with 6H5 mAb, the MFI decreased rapidly on the MCF-7, MDA-MB-231, and T47D breast cancer cell surface as incubation time increased from 0 to 45 minutes (Supplementary Figure 1, G, available online), indicating rapid internalization of the envelope–antibody complex. The percentage of internalization at 15 and 45 minutes was 47.96% and 57.92% for MCF-7 cells, 41.94% and 64.52% for MDA-MB-231 cells, and 36.6% and 39% for T47D cells, respectively. Another anti-HERV-K mAb, 4D1, was able to bind to HERV-K env on the cell surface after the 6H5 mAb molecules bound to the surface were stripped by acid buffer treatment (Supplementary Figure 1, G, available online), and the intensity of fluorescence decreased rapidly on the breast cancer cell surface as incubation time increased. No substantial change in cell surface binding level of the conformation-dependent 4D1 antibody was detected, indicating that 6H5mAb did not alter the expression level of HERV-K on the cell surface and did not substantially affect the localization or the function of HERV-K.
Surface Expression of HERV-K env Protein on Breast Cancer Cell Lines
To determine the expression of HERV-K env protein on the cell surface of human breast epithelial cells, surface proteins were biotinylated and pulled down with streptavidin in the absence of 6H5 mAb. HERV-K env protein expression on the surface of breast cancer cells was confirmed by metabolic turnover assay using 6H5 mAb (Figure 1, A). The full-length HERV-K env (66-kDa) and the HERV-K SU env (40-kDa) proteins from MDA-MB-231 and MCF-7 cells were detected by immunoblot. These results provide clear evidence for the expression of HERV-K env proteins on the surface of these two breast cancer cell lines.
Figure 1.
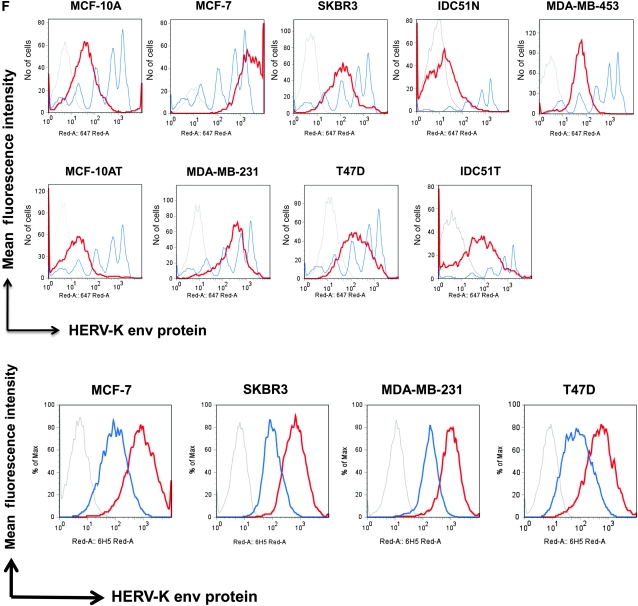
Analysis of envelope protein of human endogenous retrovirus type K (HERV-K env) expression in breast cancer cells. A)Metabolic turnover assay of biotin-pulsed HERV-K env in MDA-MB-231 and MCF-7 cells. The full-length and surface subunit env proteins of HERV-K were detected by immunoblot analysis using 6H5 monoclonal antibody (mAb). One representative blot of two independent experiments is shown. B) Immunoblot analysis of HERV-K env expression in breast cell lines. Protein lysates (50 μg per lane) from each cell line were loaded on each lane, and 6H5 mAb was used for detection. ACTB was used as the protein loading control. One representative blot of two independent experiments is shown. C) Deglycosylation analysis using peptide-N-glycosidase F (PNGase F). Cell lysates were treated with or without PNGase F. Glycosylated and deglycosylated HERV-K env proteins were detected using glycoprotein stained using periodic acid Schiff (top panel) or in an immunoblot analysis using 6H5 mAb (bottom panel). One representative blot of two independent experiments is shown. D) Surface expression of HERV-K env protein assessed on a panel of breast cell lines by cell enzyme-linked immunosorbent assay using 6H5 mAb as the primary antibody and mIgG as the negative control. OD = optical density. Results are representative of two independent experiments done. E) Immunofluorescent staining using 6H5 mAb showing the level of surface expression of HERV-K env protein (green fluorescence) on malignant (MCF-7 and MDA-MB-231) and nonmalignant (MCF-10A and MCF-10AT) breast cells (left panel). Scale bar = 20 μm. Surface (membrane) and cytoplasmic expression of HERV-K env protein (green fluorescence) in MCF-10A, MCF-7, MDA-MB-231, and MDA-MB-453 cells (right panel). Results are representative of two independent experiments. Scale bar = 20 μm. F) Flow cytometry assay of surface molecules of HERV-K env protein on breast cell lines. Cells were first treated with 6H5 mAb (red), followed by goat anti-mIgG-Alexa Fluor 647. Cells treated with isotoype control (IgG2a) followed by goat anti-mIgG-Alexa Fluor 647 treatment were used as control (gray). The mean fluorescence intensity of HERV-K for each cell line was calculated according to a calibration equation using quantitative indirect immunofluorescence beads (blue). Each of the five blue peaks from left to right represents a standard number of 1700, 11 000, 54 000, 194 000, and 561 000 beads, respectively. The mean fluorescence intensity of HERV-K was also compared between tumor cells (IDC51T) and matched uninvolved breast cells (IDC51N) obtained from a patient diagnosed with invasive ductal carcinoma (top panel). The number of surface molecules of HERV-K env protein in MCF-7, SKBR3, MDA-MB-231, and T47D cells treated with 6H5 mAb preincubated with K-GST (1 μg K-GST per 10 μg of 6H5 mAb) (blue) was compared with non-preincubated 6H5 mAb–treated cells (red) (bottom panel). Results are representative of at least three independent flow cytometry assays.
It is important for a potential immunotherapeutic antibody to be able to specifically bind to the target protein that is uniquely expressed by cancer cells and not be able to nonspecifically bind to nonmalignant cells that do not express the target. We found that HERV-K env protein was expressed in all tested breast cancer cell lines to a greater extent than in the weakly tumorigenic MCF-10AT breast cell line (Figure 1, B). In addition, no HERV-K env expression was observed in the non-neoplastic breast epithelial cell line MCF-10A. HERV-K env protein in the breast cancer cell lines was sensitive to PNGase F cleavage (Figure 1, C, top panel), indicating that it is a glycoprotein. HERV-K env protein was detected in breast cancer cells treated with or without PNGase F by immunoblot using 6H5 mAb (Figure 1, C, bottom panel), indicating that the antibody recognized both the glycosylated and non-glycosylated forms of the envelope proteins.
To analyze if there was a difference in surface expression of HERV-K env in different breast cell lines, we conducted a cell ELISA using 6H5 mAb or mIgG. Increased optical density was observed with increasing concentrations of 6H5 mAb, indicating increased level of HERV-K env expression in MDA-MB-231, MCF-7, and SKBR3 breast cancer cells, compared with mIgG as the primary antibody. In contrast, 6H5 mAb vs mIgG antibody showed no substantial change in optical density in MCF-10A cells (Figure 1, D).
A successful antibody therapy also requires target protein localization on the cell surface. Next, we tested the surface (membrane) expression of HERV-K env protein in malignant and nonmalignant breast cell lines by immunofluorescence using 6H5 mAb. Surface expression of HERV-K env protein was substantially higher in malignant than in nonmalignant breast cell lines (Figure 1, E, left panel). Both membrane and cytoplasmic expression of HERV-K env protein was detectable by immunofluorescence in breast cancer cell lines (Figure1, E, right panel).
We also wished to determine whether an anti-HERV-K mAb conjugated to a toxin could specifically target breast cancer cells expressing HERV-K. We found that 6H5 mAb was able to deliver r-Gel (a plant toxin) into the target cancer cells by internalization, which was detected using anti-r-Gel polyclonal antibodies after treating cells with r-Gel-conjugated 6H5 mAb (6H5-r-Gel antibody) (Supplementary Figure 2, A, available online). The intensity of the r-Gel signal was much weaker in MCF-10A than in breast cancer cells.
The expression of HERV-K env protein on the surface of breast epithelial cells was also quantified by flow cytometry using 6H5 mAb (Figure1, F, top panel). A statistically significantly higher MFI was detected on the malignant cells compared with nonmalignant breast cell lines, which reflects the number of HERV-K SU molecules expressed on the cell surface (mean number of SU molecules on cell surface, MDA-MB-231 vs MCF-10A: 173 078 vs 39 759 molecules, difference = 133 319 molecules, 95% CI = 71 900 to 194 739 molecules, P = .001; MDA-MB-231 vs MCF-10AT: 173 078 vs 20 965 molecules, difference = 152 123 molecules, 95% CI = 96 506 to 207 720 molecules, P < .001) (see Supplementary Table 1, available online, for other cell lines). When we analyzed HERV-K env expression in primary cells cultured from patient tumors (n = 26), the number of cells expressing HERV-K env in IDC51T cells, a primary tumor cell line originating from IDC patient Acc 51, was statistically significantly higher than IDC51N cells, an epithelial cell line originating from uninvolved tissue adjacent to breast tumor tissue from IDC patient Acc 51 (mean percentage of HERV-K surface molecules, Acc 51T vs Acc 51N: 80.40% vs 54.20%; difference = 26.20%, 95% CI = 21.23% to 31.17%, P < .001) (Figure 1, F, top panel). Increased surface expression of HERV-K env protein on MCF-7, SKBR3, MDA-MB-231, and MDA-MB-435EB1 cell lines was noted with increasing amounts of 6H5 mAb (Supplementary Figure 2, B, available online). Binding of 6H5 mAb to HERV-K SU env protein on breast cancer cells was greatly reduced if 6H5 mAb was preincubated with K-GST fusion protein (Figure1, F, bottom panel), which further confirms the specificity of 6H5 mAb for HERV-K env protein.
In summary, HERV-K env proteins were highly expressed on the surface of breast cancer cells and rapidly internalized by the majority of breast cancer cells in a time-dependent manner. The results were consistent with the metabolic turnover rate of this protein in the absence of its antibody.
Effect of 6H5 mAb on Growth, Proliferation, Cell Viability, and Cell Cycle of Breast Cancer Cells
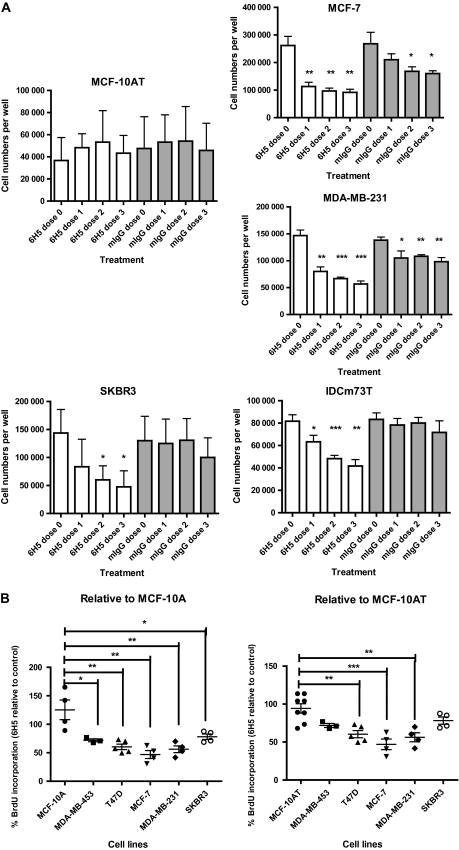
We hypothesized that the HERV-K env antigen can be targeted by antibody therapy because of the abundance of HERV-K env on the surface of breast cancer cells. We tested whether 6H5 mAb–treated breast cancer cells showed reduced cell growth by conducting an assay of cell counts. A statistically significant decrease in cell number was observed in several breast cancer cell lines (MCF-7, MDA-MB-231, SKBR3) as well as primary cells cultured from an IDC biopsy with metastasis to lymph nodes (IDCm73T) when cells were treated with one, two, and three doses of 6H5 mAb (10 μg/mL per dose; designated as doses 1, 2, and 3, respectively) compared with either untreated (dose 0) cells or cells treated with control mIgG (mean number of MDA-MB-231cells, control vs dose 3: 146 667 vs 56 667 cells, difference = 90 000 cells, 95% CI = 70 924 to 109 076 cells, P < .001; mean number of IDCm73T cells, control vs dose 3: 81 667 vs 41 667 cells, difference = 40 000 cells, 95% CI = 26 914 to 53 086 cells, P = .001) (Table 1 and Figure 2, A). No statistically significant change in growth was found in transformed but non-tumorigenic MCF-10AT cells treated with 6H5 mAb compared with mIgG.
Table 1.
Effect of 6H5 mAb on growth of breast cancer cells*
| Cell line | Mean No. of cells |
|||||||||||||
| Dose 0 vs 1 |
Dose 0 vs 2 |
Dose 0 vs 3 |
||||||||||||
| Dose 0 | Dose 1 | Difference | 95% CI | P† | Dose 2 | Difference | 95% CI | P† | Dose 3 | Difference | 95% CI | P† | N | |
| MCF-10AT | 36 667 | 48 333 | −11 667 | −50 652 to 27 318 | .45 | 53 333 | −16 667 | −73 142 to 39 809 | .46 | 43 333 | −6667 | −48 818 to 35 484 | .68 | 3 |
| MCF-7 | 261 667 | 113 333 | 148 333 | 89 628 to 207 039 | .002 | 96 667 | 165 000 | 109 096 to 220 904 | .001 | 91 667 | 170 000 | 113 524 to 226 476 | .001 | 3 |
| MDA-MB-231 | 146 667 | 80 000 | 66 667 | 44 966 to 88 368 | .001 | 66 667 | 80 000 | 62 689 to 97 311 | <.001 | 56 667 | 90 000 | 70 924 to 109 076 | <.001 | 3 |
| SKBR3 | 143 833 | 83 333 | 60 500 | −43 213 to 164 213 | .18 | 60 000 | 83 833 | 5659 to 162 008 | .041 | 47 500 | 96 333 | 14 841 to 177 826 | .031 | 3 |
| IDCm73 | 81 667 | 63 333 | 18 333 | 5247 to 31 420 | .018 | 48 333 | 33 333 | 22988 to 43679 | <.001 | 41 667 | 40 000 | 26 914 to 53 086 | .001 | 3 |
The nonmalignant breast cell line MCF-10AT; breast cancer cell lines MCF-7, MDA-MB-231, and SKBR3; and a cell line cultured from the primary tumor of a breast cancer patient (IDCm73T) were left untreated (dose 0), were dosed with 6H5 mAb (10 μg/mL/dose), and continuously exposed for to mAb for 24 hours (dose 1), were treated at 24 and 48 hours and continuously exposed to mAb (dose 2) or were treated at 24, 48, and 72 hours and continuously exposed to mAb (dose 3). Cells were stained with trypan blue and living cells were counted from four fields under low power microscopy. Mean values from at least three independent experiments are presented. CI = confidence interval; mAb = monoclonal antibodies; N = number of independent experiments.
P values were calculated using a two-sided Student's test.
Figure 2.
Effect of 6H5 monoclonal antibody (mAb) on growth of breast cells. A) Growth of MCF-10AT, MCF-7 MDA-MB-231, SKBR3, and IDCm73T breast cells after treatment with 10 μg/mL of 6H5 mAb. Cells were treated with 6H5 mAb or control mIgG for 24, 48, or 72 hours and counted after staining with trypan blue to determine the effect of antibody treatment on cell growth. Experiment was performed three times. Means and 95% confidence intervals (error bars) are presented. *P ≤ .05, **P ≤ .005, and ***P ≤ .001, compared with dose 0, calculated using two-sided Student's t test. B) DNA replication and cell cycle analysis by bromodeoxyuridine (BrdU) incorporation assay. Cells were treated with 10 μg/mL of either 6H5 mAb or mIgG (control) for 3 days and stained with 7-amino-actinomycin D coupled with immunofluorescent BrdU to determine the total cellular DNA in cells treated with mAbs. Each point represents a single measurement of BrdU incorporation. Experiment was performed three independent times. Means (horizontal bars) and 95% confidence intervals (error bars) are presented. *P ≤ .05, **P ≤ .005, and ***P ≤ .001, compared with MCF-10A or MCF-10AT, calculated using two-sided Student's t test.
Next, we analyzed cell proliferation by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay (Supplementary Figure 3, A, available online) and cell viability by crystal violet assay (Supplementary Figure 3, B, available online). The MTS assay showed that proliferation of 6H5 mAb–treated MDA-MB-231 and T47D cells was inhibited to a greater extent than proliferation of MCF-10AT cells, and inhibition occurred in a dose-dependent manner. The crystal violet assay confirmed the MTS assay and showed reduced viability of 6H5 mAb–treated (three-dose regimen as above) breast cancer cells (MDA-MB-231 and T47D cells), but little or no effect on the viability of MCF-10AT, indicating that 6H5 mAb was cytotoxic specifically toward breast cancer cells. These assays provided evidence that an anti-HERV-K antibody can inhibit proliferation of breast cancer cells, but not nonmalignant breast cells, via specific targeting of the endogenous HERV-K env on the surface of the breast cancer cells.
Because 6H5 mAb was able to inhibit cancer cell growth and proliferation, we performed a BrdU assay to label the newly synthesized DNA of replicating cells during the S phase of the cell cycle. Statistically significantly reduced BrdU incorporation was observed in breast cancer cells (BrdU incorporation in 6H5 mAb–treated cells as a percentage of mIgG-treated controls, MDA-MB-231 vs MCF-10A: 56.22% vs 125.30%, difference = 69.08%, 95% CI = 24.42% to 113.7%, P = .009; MDA-MB-231 vs MCF-10AT: 56.22% vs 94.30%, 95% CI = 16.27% to 59.89%, P = .003) (Figure 2, B and Table 2 for other breast cancer cell lines).
Table 2.
BrdU incorporation in breast cancer and nonmalignant breast cells*
| Cell line | Breast cancer cells vs control MCF-10A |
Breast cancer cells vs control MCF-10AT |
||||||
| Mean percent of BrdU-positive cells relative to control | Difference | 95% CI | P† | Difference | 95% CI | P† | N | |
| MCF-10A | 125.30 | — | — | — | — | — | — | 4 |
| MCF-10AT | 94.30 | 30.99 | −1.60 to 63.58 | .06 | — | — | — | 8 |
| MCF-7 | 46.96 | 78.33 | 32.80 to 123.9 | .006 | 47.34 | 24.86 to 69.82 | <.001 | 4 |
| MDA-MB-231 | 56.22 | 69.07 | 24.42 to 113.7 | .009 | 30.08 | 16.27 to 59.89 | .003 | 4 |
| SKBR3 | 78.16 | 47.13 | 3.33 to 90.93 | .039 | 16.14 | −5.02 to 37.30 | .12 | 4 |
| T47D | 60.36 | 64.63 | 26.90 to 103.0 | .005 | 33.94 | 14.71 to 53.17 | .003 | 5 |
| MDA-MB-453 | 71.84 | 53.45 | .65 to 106.2 | .048 | 22.46 | −1.35 to 46.26 | .062 | 3 |
Nonmalignant breast cell lines (MCF-10A or MCF-10AT) and breast cancer cell lines (MCF-7, MDA-MB-231, SKBR3, T47D, and MDA-MB-453) were treated with 6H5 mAb or mIgG (10 μg/mL per dose of each antibody) for 24 hours. The BrdU incorporation percentage of 6H5 mAb–treated vs mIgG-treated cells (control) was compared. A total of 5000 BrdU-positive cells were counted for each cell line. Mean values from at least three independent experiments are presented. BrdU = Bromodeoxyuridine; CI = confidence interval; mAb = monoclonal antibodies; N = number of independent experiments. — = not applicable.
P values reflect comparisons between MCF-10A cells and each breast cancer cell line, or between MCF-10AT cells and each breast cancer cell line, and were calculated using a two-sided Student's t test.
S-phase arrest was observed in MCF-7, MDA-MB-231, and ZR-75-1 cells, and G0/G1 arrest in T47D cells treated with 6H5 mAb (Supplementary Figure 3, C, available online). These data suggest that the observed decreased growth rates of cancer cells treated with 6H5 mAb is because of their slower progression through the DNA synthesis phase for most breast cancer cell lines.
Effect of 6H5 mAb on Apoptosis of Breast Cancer Cells and Signaling Pathways
Apoptosis.
After we found that treatment of cancer cells with 6H5 mAb leads to inhibition of growth and proliferation, we tested whether this inhibition was the result of increased apoptosis. Annexin V assay showed induction of apoptosis in breast cancer cells treated with 6H5 mAb compared with mIgG treatment (percentage of apoptosis in 6H5 mAb–treated cancer and noncancer cells, MCF-7 vs MCF-10A: 48.11% vs 9.04%, difference = 39.07%, 95% CI = 30.75% to 47.39%, P < .001; MCF-7 vs MCF-10AT: 48.11% vs 12.63%, difference = 35.48%, 95% CI = 27.14% to 43.84%, P < .001; MDA-MB-231 vs MCF-10A: 51.74%vs 9.04%, difference = 42.70%, 95% CI = 33.16% to 52.24%, P < .001; MDA-MB-231 vs MCF-10AT: 51.74% vs 12.63%, difference = 39.11%, 95% CI = 29.49% to 48.74%, P < .001 (see Supplementary Table 2, available online, for summary and number of independent experiments for each cell line). Examples of independent experiments are shown in Figure 3, A and Supplementary Figure 4, A, available online. Dose-dependent induction of apoptosis was observed in MCF-7 and T47D cells but not in MCF-10A and MCF-10AT cells (Supplementary Figure 4, A, available online). Thus, an increased dose of 6H5 mAb–induced greater percentages of apoptosis in the cancer cells than in noncancer cells. When using several other anti-HERV-K mAbs (4E11, 6E11, and 4D1), all exhibited cytotoxicity toward malignant breast cell lines, but not toward nonmalignant breast cells, similar to 6H5 mAb (Supplementary Figure 4, A, available online).
Figure 3.
Effect of 6H5 monoclonal antibody (mAb) on apoptosis of breast cancer cells. A) Cells were treated with 6H5 mAb (red line) or control mIgG (gray line) (10 μg/mL of each antibody) for 16 hours, stained with annexin V–allophycocyanin and 7-AAD-phycoerythrin-cyanide 7, and analyzed by flow cytometry. B) Effect of 6H5 mAb treatment on cell death–inducing DFFA-like effector A (CIDEA) protein expression. Breast cancer cell lines were treated with 6H5 mAb or mIgG (10 μg/mL of each antibody) for 24 hours and analyzed for changes in protein expression by immunoblot using a mouse anti-human CIDEA antibody. ACTB was used as the protein loading control. Results are representative of two independent assays. C) The effect of 6H5 mAb treatment on expression of TP53 and TP53AIP1 proteins. MCF-7 and MDA-MB-231 breast cancer cell lines were treated with 6H5 mAb or mIgG (10 μg/mL of each antibody) for 24 hours, and an immunoblot assay was done using mouse anti-human TP53 and rabbit anti-human TP53AIP1 antibodies. ACTB was used as the protein loading control. Results are representative of two independent assays. D) Expression of active caspases 3 and 9 was assessed by immunoblot assay in ZR-75-1 and MDA-MB-231 breast cancer cells treated with 6H5 mAb or 6E11 mAb, or with mIgG (10 μg/mL of each antibody) for 24 hours using rabbit anti-human caspase 3 and mouse anti-human caspase 9 antibodies. ACTB was used as the protein loading control (top panel). Expression of active caspase 8 was assessed by immunoblot assay in MDA-MB-453 and MCF-7 breast cancer cells treated with 6H5 mAb (10 , 25, or 50 μg/mL), or with mIgG (10 μg/mL) for 24 hours using mouse anti-human caspase 8 antibody (bottom panel). Results are representative of at least two independent assays. E) Immunofluorescence assay to assess the expression of caspase proteins in MDA-MB-231 cells treated with 6H5 mAb or mIgG (10 μg/mL of each antibody) for 24 hours using rabbit anti-human caspase 3, mouse anti-human caspase 8, and mouse anti-human caspase 9 antibodies. Results are representative of two independent assays. Scale bar = 10 μm. F) Immunofluorescence assay to assess the expression of CDK5 and CDKN1A proteins. MDA-MB-231 cells were treated with 6H5 mAb or mIgG (10 μg/mL of each antibody) using mouse anti-human CDK5 and mouse anti-human CDKN1A antibodies. Results are representative of at least two independent assays. Scale bar = 10 μm.
Expression of Apoptosis Genes.
Because anti-HERV-K mAbs not only inhibited breast cancer cell growth but also induced apoptosis, we investigated the mechanism of apoptosis induction by evaluating expression of key genes (at transcriptional and translational levels) involved in apoptosis in 6H5 mAb-treated MCF-7 cells. Using a cDNA microarray–specific for apoptosis genes, an initial screening showed that TNFRSF25, TNFSF8, and CIDEA genes were overexpressed in 6H5-treated MCF-7 cells compared with mIgG-treated cells (data not shown). Furthermore, immunoblot assays showed increased protein levels of these genes (Figure 3, B and Supplementary Figure 4, B, available online). Increased expression of TNFSF8, FASLG, GML, and MYOD1 proteins was detected in MDA-MB-453 and MCF-7 breast cancer cells treated with 6H5 mAb compared with mIgG, and enhanced expression of CIDEA protein was detected in MDA-MB-231 and MDA-MB-453 breast cancer cell lines treated with 6H5 mAb, compared with mIgG. Our immunoblot data showed that CIDEA, a cell death activator, is present in normal breast cells but absent in the breast cancer cells analyzed. Treatment with 6H5 mAb greatly increased CIDEA expression in breast cancer cells, which suggests that it may be responsible in part for the increased apoptosis we observed after 6H5 mAb treatment. Normal breast cells also showed increased expression of CIDEA after antibody treatment, but this was against a background of already high CIDEA expression. Flow cytometry analysis (Table 3 and Supplementary Figure 4, C and D, available online) showed that expression of apoptotic genes increased less than twofold in nonmalignant breast cells (MCF-10A and MCF-10AT) but increased by at least twofold or more in the breast cancer cell lines, except TNFSF8 and TNFRSF10D.
Table 3.
Effect of anti-HERV-K antibody treatment on protein expression in breast cell lines, analyzed by flow cytometry*
| Fold change in expression relative to HEK293 |
Fold change in expression relative to MCF-10AT |
|||||||
| Protein | Cell line | Mean fold change | Difference | 95% CI | P† | Difference | 95% CI | P† |
| CIDEA | HEK293T | 1.04 | — | — | — | — | — | — |
| MCF-10AT | 2.44 | 1.40 | 1.05 to 1.77 | <.001 | — | — | — | |
| MCF-7 | 6.86 | 5.82 | 5.51 to 6.14 | <.001 | 4.42 | 4.24 to 4.60 | <.001 | |
| MDA-MB-231 | 15.12 | 14.08 | 12.78 to 15.38 | <.001 | 12.67 | 11.40 to 13.94 | <.001 | |
| SKBR3 | 15.95 | 14.91 | 14.60 to 15.23 | <.001 | 13.5 | 13.32 to 13.68 | <.001 | |
| T47D | 8.14 | 7.10 | 6.79 to 7.42 | <.001 | 5.69 | 5.51 to 5.87 | <.001 | |
| Caspase 3 | HEK293T | 1.03 | — | — | — | — | — | — |
| MCF-10AT | 1.83 | 0.800 | 1.62 to 4.58 | .004 | — | — | — | |
| MCF-7 | 3.51 | 2.47 | 1.53 to 3.42 | .013 | 1.67 | 0.99 to 2.36 | .043 | |
| MDA-MB-231 | 16.75 | 15.71 | 12.65 to 18.78 | .001 | 14.91 | 11.92 to 17.91 | <.001 | |
| SKBR3 | 3.65 | 1.82 | 1.51 to 2.12 | <.001 | 2.62 | 1.89 to 3.34 | <.001 | |
| T47D | 4.71 | 3.68 | 2.96 to 4.40 | <.001 | 2.88 | 2.58 to 3.17 | <.001 | |
| Caspase 7 | HEK293T | 1.63 | — | — | — | — | — | — |
| MCF-10AT | 4.06 | 2.43 | 2.13 to 2.74 | <.001 | — | — | — | |
| MCF-7 | 4.87 | 3.24 | 2.94 to 3.54 | <.001 | .800 | 0.71 to 0.90 | <.001 | |
| MDA-MB-231 | 32.32 | — | — | — | — | — | — | |
| SKBR3 | 18.68 | 17.05 | 16.76 to 17.35 | <.001 | 14.62 | 14.54 to 14.70 | <.001 | |
| T47D | 13.67 | 12.04 | 11.75 to 12.34 | <.001 | 9.61 | 9.53 to 9.69 | <.001 | |
| Caspase 8 | HEK293T | 1.01 | — | — | — | — | — | — |
| MCF-10AT | 2.47 | 1.46 | 1.20 to 1.73 | <.001 | — | — | — | |
| MCF-7 | 7.63 | 6.62 | 6.36 to 6.88 | <.001 | 5.16 | 5.06 to 5.26 | <.001 | |
| MDA-MB-231 | 14.05 | 13.04 | 5.96 to 20.12 | .007 | 11.58 | 4.51 to 18.65 | .011 | |
| SKBR3 | 23.7 | 22.7 | 22.44 to 22.95 | <.001 | 21.23 | 21.15 to 21.32 | <.001 | |
| T47D | 6.49 | 5.49 | 5.23 to 5.74 | <.001 | 4.02 | 3.94 to 4.11 | <.001 | |
| Caspase 9 | HEK293T | 1.04 | — | — | — | — | — | — |
| MCF-10AT | 3.69 | 2.65 | 2.52 to 2.79 | <.001 | — | — | — | |
| MCF-7 | 4.34 | 3.3 | 3.13 to 3.46 2 | <.001 | .64 | 0.52 to 0.66 | <.001 | |
| MDA-MB-231 | 24.28 | 23.24 | 10.31 to 36.18 | .008 | 2.59 | 7.65 to 33.52 | .012 | |
| SKBR3 | 13.99 | 12.95 | 12.83 to 13.08 | <.001 | 10.3 | 10.24 to 10.36 | <.001 | |
| T47D | 5.83 | 4.79 | 4.65 to 4.92 | <.001 | 2.13 | 2.06 to 2.21 | <.001 | |
| TP53 | HEK293T | 1.24 | — | — | — | — | — | — |
| MCF-10AT | 1.93 | .698 | 0.376 to 1.02 | .004 | — | — | — | |
| MCF-7 | 4.24 | 3.01 | 2.67 to 3.34 | <.001 | 2.31 | 2.21 to 2.40 | <.001 | |
| MDA-MB-231 | 22.59 | — | — | — | — | — | — | |
| SKBR3 | 18.94 | 17.71 | 17.39 to 18.03 | <.001 | 17.01 | 16.99 to 17.03 | <.001 | |
| T47D | 6.19 | 4.95 | 4.63 to 5.27 | <.001 | 4.25 | 4.22 to 4.28 | <.001 | |
| FASLG | HEK293T | 1.15 | — | — | — | — | — | — |
| MCF-10AT | 7.03 | 5.88 | — | <.001 | — | — | — | |
| MCF-7 | 6.06 | 4.91 | 2.62 to 7.19 | .004 | .973 | −3.37 to 1.42 | .32 | |
| MDA-MB-231 | — | — | — | — | — | — | — | |
| SKBR3 | 20.84 | 19.69 | 13.83 to 25.56 | <.001 | 15.57 | 7.91 to 19.72 | .003 | |
| T47D | 10.33 | 9.18 | 5.57 to 12.80 | .002 | 3.22 | 0.386 to 6.99 | .068 | |
| CDKN1A | HEK293T | 1.13 | — | — | — | — | — | — |
| MCF-10AT | 3.37 | 2.24 | 2.00 to 2.47 | <.001 | — | — | — | |
| MCF-7 | 5.75 | 4.62 | 4.382 to 4.85 | <.001 | 2.38 | 2.32 to 2.44 | <.001 | |
| MDA-MB-231 | 33.11 | 21.97 | 28.00 to 35.94 | <.001 | 25.77 to 33.70 | <.001 | ||
| SKBR3 | 18.95 | 17.81 | 17.58 to 18.04 | <.001 | 15.57 | 15.53 to 15.62 | <.001 | |
| T47D | 6.59 | 5.46 | 5.22 to 5.69 | <.001 | 3.22 | 3.16 to 3.28 | <.001 | |
| CDK5 | HEK293T | 1.6 | — | — | — | — | — | — |
| MCF-10AT | 2.56 | .959 | 0.329 to 1.59 | .013 | — | — | — | |
| MCF-7 | 1.95 | .35 | 0.170 to 0.870 | .135 | −.609 | −1.26 to 0.039 | .060 | |
| MDA-MB-231 | 14.99 | 13.38 | 12.64 to 14.12 | <.001 | 12.42 | 11.58 to 13.26 | <.001 | |
| SKBR3 | 7.45 | 5.84 | 5.34 to 6.34 | <.001 | 4.88 | 4.25 to 5.52 | <.001 | |
| T47D | 7.56 | 5.95 | 4.51 to 16.42 | .226 | 5 | −5.47 to 15.46 | .30 | |
HEK293Tcells, the nonmalignant breast cell line MCF-10AT, and the breast cancer cell lines MCF-7, MDA-MB-231, SKBR3, and T47D, were treated with 6H5 mAb or mIgG (10 μg/mL per dose of each antibody) for 24 hours and analyzed for protein expression by flow cytometry. The fold changes in protein levels of CIDEA, caspase 3, 7, 8, and 9, TP53, CDKN1A, and CDK5of 6H5 mAb–treated or mIgG-treated cells were compared. The fold changes in mAb-treated breast cancer cells were compared with fold changes in mAb-treated HEK293T or MCF-10AT cells. The mean values from at least three independent experiments are presented. CI = confidence interval; CIDEA = cell death–inducing DFFA-like effector A; mAb = monoclonal antibody; — = not applicable.
P values reflect comparisons between HEK293T cells and each breast cancer cell line, or between MCF-10AT cells and each breast cancer cell line, and were calculated using a two-sided Student‘s t test.
TP53 Pathway.
Viral infection activates TP53 signaling as part of the innate immune response, which leads to inhibition of apoptotic signaling to allow the efficient manufacture and export of viruses before cell death (30). A possible effect of 6H5 mAb on TP53 pathway genes was determined by examining the expression of proteins involved in TP53-mediated signal transduction. Increased expression of FASLG, GML, MYOD1, TP53, and TP53AIP1 was observed in cells treated with 6H5 mAb compared with cells treated with mIgG, in immunoblot assays (Figure 3, C and Supplementary Figure 4, B, available online), as well as flow cytometry analysis (Table 3 and Supplementary Figure 4, C and D, available online). This suggests that blocking of HERV-K with 6H5 is not part of an innate immune response but rather that antibody treatment itself is inducing a TP53 signaling response.
Caspase Pathway.
Caspase enzymes play a central role in most types of apoptotic cell death (31). Induction of caspase 3 and caspase 9 expression was detected by immunoblot assay in MDA-MB-231 and ZR-75-1 cells treated with 6H5 or 6E11 mAb compared with cells treated with mIgG (Figure 3, D, top panel). Increased cleavage of caspase 8 was detected in MDA-MB-453 and MCF-7 cells treated with increasing amounts of 6H5 mAb (50 > 25 > 10 μg/mL) for 24 hours, indicating a dose–response effect (Figure 3, D, bottom panel). Immunofluorescence staining showed increased expression of caspases 3, 8, and 9 (Figure 3, E), and flow cytometry (Supplementary Figure 4, C–E, available online) showed the fold change in expression levels of these proteins in MDA-MB-231 cells treated with 6H5 mAb (10 μg/mL) for 24 hours relative to mIgG (10 μg/mL) treatment. The fold change in expression levels of caspases 3, 7, 8, and 9 in different breast cancer cells treated with different concentrations of 6H5 mAb relative to MCF-10AT cells was assessed in a flow cytometry analysis (Supplementary Figure 4, C–E, available online). Changes in expression of apoptotic proteins as assessed by flow cytometry are summarized in Table 3; HEK293T was used as a negative control. Expression of proteins was compared in each 6H5 mAb–treated or mIgG-treated cell line.
Expression of CDKN1A and CDK5.
The tumor suppressor gene TP53 and its downstream effector CDKN1A are thought to play major roles in the development of human malignancies. Binding sites for p53 in the promoter region of CDKN1A contribute to CDKN1A transcriptional induction after a p53 stress response (32). CDK5 activates p53 expression in breast cancer cells following carboplatin treatment, and the involvement of CDK5 in phosphorylating p53 has been documented in other cancer types (33). We investigated CDKN1A and CDK5 in the cancer cells treated with anti-HERV-K mAb. Increased expression of CDKN1A and CDK5 was observed in MDA-MB-231 cells treated with 6H5 mAb, compared with cells treated with mIgG, both by immunofluorescence (Figure 3, F) and flow cytometry (Table 3, and Supplementary Figure 4, D, available online). The fold increase in expression levels of CDKN1A and CDK5 was statistically significantly higher in 6H5-treated MDA-MB-231 cells compared with 6H5-treated MCF-10AT cells (MDA-MB-231 vs MCF-10AT: CDKN1A, 33.11- vs 3.37-fold, difference = 29.74-fold, 95% CI = 28.00- to 35.94-fold, P < .001; CDK5, 14.99- vs 2.56-fold, difference = 12.43-fold, 95% CI = 11.58- to 13.26-fold, P < .001). Other cell lines are shown in Table 3.
Antitumor Effect of mAbs
In Vivo Studies in Mice.
Our previous experiments showed that 6H5 mAb inhibits breast cancer growth and induces breast cancer cells to undergo apoptosis in vitro. Therefore, we next investigated the anti tumor effect of 6H5 mAb in vivo using a linear mixed-effects model for data analysis. Female immunodeficient athymic NCr-nu/nu mice were injected with MDA-MB-231 cells and intravenously treated with 6H5 mAb (n = 5 mice), tumor sizes were statistically significantly reduced, in 6H5 mAb–treated mice compared with mIgG-treated mice (n = 5 mice) (mean tumor volume on day 39, mIgG vs 6H5 mAb: 1448.33 vs 475.44 mm3, difference = 972.89 mm3, 95% CI = 470.17 to 1475.61 mm3, P < .001; mean tumor volume on day 40, mIgG vs 6H5 mAb: 2343.75 vs 617.00 mm3, difference = 1726.75 mm3, 95% CI = 555.53 to 2897.97 mm3, P = .005) (Figure 4, A). Tumor sizes were also statistically significantly decreased, and growth was delayed in immunodeficient mice inoculated with MDA-MB-231 cells and treated with 6H5-r-Gel, compared with mice treated with mIgG (mean tumor volume on day 40, mIgG vs 6H5-r-Gel mAb, 2343.75 vs 366.90 mm3, difference = 1976.85 mm3, 95% CI = 874.54 to 3079.16 mm3, P < .001) (Supplementary Figure 5, A, available online). Tumor sizes were also reduced in immunodeficient mice bearing MCF-7 xenografts when treated with 6H5 mAb (n = 5 mice) compared with mice treated with mIgG (n = 5 mice) (mean tumor volume on day 31, mIgG vs 6H5 mAb: 1278.8 vs 456.84 mm3, difference = 822.00 mm3, 95% CI = 110.82 to 1533.18 mm3, P = .034, Figure 4, B). Treatment of MCF-7 xenografts with G11D10 scFv antibody also showed reduced tumor volume (mean tumor volume on day 31, mIgG vs G11D10 scFv antibody: 1278.80 vs 460.80 mm3, difference = 818.00 mm3, 95% CI = 225.02 to 1410.98 mm3, P < .001) (Supplementary Figure 5, B, available online). Treatment of immunodeficient mice bearing MCF-7 xenografts (n = 5 mice) when treated with anti-HERV-K sera obtained from BALB/c mice immunized with HERV-K fusion proteins (200 μL per dose) showed reduced tumor sizes compared with mice (n = 5) treated with mIgG (mean tumor volume on day 31, mIgG vs anti-HERV-K sera: 1278.80 vs 667.67 mm3, difference = 611.13 mm3, 95% CI = −226.10 to 1448.36 mm3, P = .015) (Supplementary Figure 5, B, available online).
Figure 4.
Antitumor effects of 6H5 monoclonal antibody (mAb) in immunodeficient mice carrying MDA-MB-231 xenograft tumors. A) Tumor sizes in MDA-MB-231 xenografts treated with 6H5 mAb (right panel) were compared with xenografts treated with mIgG (left panel) using a linear mixed-effects model. The solid arrows indicate the days of antibody injection. The tumor sizes of five mice on a particular day are shown, using a box and whiskers plot format. The horizontal line in the “box” indicates the median tumor size, the box represents interquartile range (25th and 75th percentiles), and the ends of the vertical lines or “whiskers” indicate the minimum and maximum data values. Open circles are outliers or suspected outliers. Error bars represent 95% confidence intervals from experiments performed two independent times with similar results. B) Tumor sizes were also assessed in MCF-7 xenografts treated with 6H5 mAb and compared with mIgG. Error bars represent with 95% confidence intervals from experiments performed two independent times with similar results. C) The number of TUNEL-positive (TUNEL+) cells in tumors from mice bearing MDA-MB-231 xenograft tumors treated with 6H5 mAb relative to mIgG-treated control tumors is shown. The horizontal bar represents the mean number of TUNEL+ cells in 6H5 mAb–treated and mIgG-treated tumors, and the solid circles represent positive cell counts in a single field (P = .008; Student's t test, two-sided) (left panel). Representative immunohistochemically stained sections of tumors from mIgG-treated and 6H5 mAb–treated mice are shown (right panel). Brown stain (TUNEL+) indicates apoptotic cells. These experiments were performed two independent times. Magnification = 400×. Scale bar = 200 μm. D) Ki-67-positive (Ki-67+) cell numbers were determined in xenograft tumors from mice injected with MDA-MB-231 cells and treated with 6H5 mAb compared with mIgG control. Representative immunohistochemical stains of tumors from 6H5 mAb–treated mice are shown: mIgG (right, top panel) and 6H5 mAb (right, bottom panel). The horizontal bar represents the mean number of Ki-67+cells in 6H5 mAb–treated and mIgG-treated tumors, and the solid circles represent positive cell counts in a single field (P < .001; calculated using a two-sided Student's t test) (left panel). These experiments were performed two independent times. Magnification = 400×. Scale bar = 200 μm.
It should be noted that tumor sizes still increased with time in mice receiving antibody treatment. It is possible that if the experiment continued for another month, the tumor sizes in the treatment group would have approached those of the controls, given the experimental design. The increase in tumor size with time in the treatment group is likely because of clearance of the antibody in the animals. We found that the antibody titers reached their highest values 1–6 hours after injection, and the antibody was cleared 1 week after injection (data not shown). To maintain antibody titers at high levels, more frequent injections would likely be necessary.
Next, we analyzed the tumors for apoptosis using TUNEL assay and for proliferation using Ki67 assay in tumor biopsies obtained from each group of mice (n = 5). Tumors were removed 14 days after initiation of mAb treatment. The number of TUNEL-positive cells in tumors of mice treated with 6H5 mAb was statistically significantly higher compared with tumors from mice treated with mIgG (mean number of TUNEL-positive cells in tumors, 6H5 mAb–treated vs mIgG-treated mice: 21.00 vs 6.7 cells, difference = 14.30 cells, 95% CI = 4.190 to 24.41 cells, P = .008) (Figure 4, C, left panel; right panel showing the TUNEL staining). A decrease in Ki-67-positive cells (mean number of Ki-67-positive cells in tumors, 6H5 mAb–treated vs mIgG-treated mice: 91.20 vs 442.00 cells, difference = 350.80 cells, 95% CI = 257.3 to 444.3 cells, P < .001) (Figure 4, D, left panel; right panel showing the Ki-67 staining), indicating that treatment with 6H5 mAb leads to both increased apoptosis and decreased proliferation in the xenografts. A subgroup of cells likely did not adequately respond to the antibody treatment, as evidenced by the lack of increase in TUNEL-positive cells in certain regions of the biopsy sections. This lack of response by all cells could be because of poor antibody delivery into this region, as the large size of 6H5 mAb (150 KDa) may prevent its complete penetration into xenografted tissues and may promote its degradation, thus limiting its complete exposure to all cells in the tissue.
Association Between HERV-K env Expression in Human Breast Tumors and Clinicopathologic Characteristics of Patients.
We examined HERV-K env expression in 223 human breast tumors by IHC. Four invasive breast tumors were analyzed for expression of HERV-K env (Figure 5). Expression HERV-K env, detected using 6H5 mAb, ranged from absent (Figure 5, top left image) to a rather strong expression in the tumor epithelium, as shown by the brown chromogen deposits in the other images in this figure. Of the 148 HERV-K-positive tumors, 64 had lymph node metastasis and 84 did not. Of the 75 HERV-K-negative tumors, 17 had lymph node metastasis and 58 did not. The clinicopathologic characteristics of patients including node status, TNM stage, ER status, and tumor grade are shown in Table 4. We did not find a statistically significant association between envelope expression and patient age at diagnosis, race or ethnicity, disease stage and grade, or tumor ER status (data not shown). However, envelope expression was statistically significantly associated with a node-positive status (P = .005, two-sided χ2 test), suggesting that envelope expression may enhance metastatic spread. Similar observations have been made by us previously (17), using biopsies from a different geographic location. In this study, 43% of the patients with an env-positive tumor had metastases to the lymph nodes, whereas only 23% of patients with an env-negative tumor were node positive. Thus, patients with HERV-K env–positive tumor had a statistically significantly increased odds of developing a node-positive disease when compared with patients with env-negative tumor (OR = 3.82, 95% CI = 1.78 to 8.2; adjusted for age, race or ethnicity, and tumor ER status). Additional analyses revealed that envelope expression was also associated with the expression of inducible NOS2, a marker of tissue inflammation and a response gene to infections (34). Tumors that were env positive were more likely to express high NOS2 than env-negative tumors (OR = 2.32, 95% CI = 1.13 to 4.80; adjusted for age, race or ethnicity, and tumor ER status).
Figure 5.
Expression of envelope protein of human endogenous retrovirus type K (HERV-K env) in invasive breast tumors from patients. Immunohistochemically stained sections were assessed in invasive breast tumors from four patients (representative of two independent experiments). HERV-K env was detected using 6H5 monoclonal antibody, shown as brown chromogen deposits. Top left tumor is negative for HERV-K env expression. Magnification = 100× for top right image and scale bar = 500 μm; magnification = 200× for all other images and scale bars = 200 μm.
Table 4.
Demographic and clinicopathologic features of breast cancer patients*
| Patient characteristic | No. (%) |
| Case patients, Total No. | 223 |
| Mean (±SD) age at diagnosis, y | 55.0 (±13.7) |
| Race | |
| AA | 136 (61) |
| EA | 87 (39) |
| TNM stage | |
| ≤II | 167 (81) |
| ≥III | 40 (19) |
| Node status | |
| Negative | 132 (63) |
| Positive | 76 (37) |
| Grade | |
| 1 and 2 | 96 (49) |
| 3 | 101 (51) |
| ER status | |
| Negative | 91 (41) |
| Positive | 131 (59) |
| HERV-K env | |
| Negative (score 0) | 75 (34) |
| Positive (score>0) | 148 (66) |
| NOS2 | |
| Low | 68 (30) |
| High | 155 (70) |
Patients with missing information were not included. Race was determined by self-identification. Disease staging was performed according to the TNM staging system of the American Joint Committee on Cancer (AJCC) (21) and the Union for International Cancer Control (UICC) (22). Grading was performed according to the Nottingham grading system (23) for breast cancer; overall grades 1, 2, and 3 were based on the cumulative score of glandular differentiation, nuclear pleomorphism, and mitotic count. ER status was determined by immunohistochemistry following guidelines for clinical laboratories to evaluate semiquantitatively ER expression in formalin-fixed paraffin-embedded tissues. A combined score (sum score) of intensity and distribution was used to categorize the immunohistochemical staining for inducible NOS2). Intensity received a score of 0, 1, 2, and 3 if the staining was negative, weak, moderate, or strong, respectively. The distribution received a score of 0, 1, 2, 3, and 4 (for staining distribution <10%, 10% to 30%, >30% to 50%, >50% to 80%, and >80% positive cells, respectively). A sum score was then divided into four groups as follows: negative = sum scores 0–1; weak = sum scores 2–3; moderate = sum scores 4–5; and strong = sum scores 6–7. For statistical analysis, NOS2 immunohistochemistry was dichotomized in low (negative and weak) and high (moderate and strong). The cut point for the statistical analysis of the HERV-K immunohistochemistry score in breast tumors was set at the median. Envelope expression was categorized by intensity (0 = absent; 1 = weak; 2 = moderate; 3 = strong) and distribution (percent tumor positive for envelope) (18). Intensity and distribution scores were multiplied to obtain the final score (0–300) for envelope expression in a tumor. AA = African American; EA = European American (ie, white race of European ancestry in America); ER = estrogen receptor; HERV-K env = envelope protein of human endogenous retrovirus type K; NOS2 = nitric oxide synthase 2.
Discussion
In this study, we have demonstrated that monoclonal and single-chain antibodies against the HERV-K env are capable of blocking growth and proliferation of human breast cancer cells in vitro and inhibiting tumor growth in mice bearing xenograft tumors. Results showed that treatment of breast cancer cells with anti-HERV-K env mAb induced apoptosis and activated the TP53 signaling pathway. Expression of HERV-K env was associated with a positive lymph node status in breast cancer patients.
Monoclonal antibodies are one of the most promising agents for therapy of human malignancies. However, numerous approaches with humanized mAbs failed as therapeutics in cancer therapy. The major problems associated with current antibody therapies are lack of antibody selectivity/specificity, low target expression in cells, and no involvement of target proteins in cancer proliferation, apoptosis, and metastasis (35). In this regard, an antibody against HERV-K env protein would meet the therapeutic criteria to address all of these limitations and could be efficacious in the therapy of breast cancers that failed other approaches.
We previously reported that HERV-K env protein can trigger a T cell immune response in breast cancer patients (18). Here, we show that breast cancer cell lines and human primary breast cancer cells express high numbers of surface molecules of this protein, which makes it an ideal candidate for B-cell targeting and development of antibody therapies. Consistent with this hypothesis, this study provided the first proof of principle experiment in showing for the first time that HERV-K env can be successfully targeted by a monoclonal antibody therapy, leading to antitumor effects of anti-HERV-K env antibodies in vitro and in vivo.
Our results from cell growth, MTS, and viability assays suggest that HERV-K env protein may play a role in cancer proliferation. Targeting env by anti-HERV-K mAb therapy suppressed the growth of breast cancer cells but not of nonmalignant breast cells. BrdU incorporation and cell cycle assays revealed that 6H5 mAb treatment induces delayed S-phase progression in the tested breast cancer cell lines. Treatment with 6H5 mAb of breast cancer cells also induced apoptosis, and the apoptotic cell numbers correlated with numbers of surface molecules of HERV-K env protein. These findings suggest that HERV-K expression has a previously unrecognized function in breast cancer biology and may serve as a prognostic marker for the disease. Indeed, HERV-K reverse transcriptase has been suggested as a breast cancer prognostic marker by others. In that study (19), HERV-K expression was found to correlate with poor prognosis for disease-free patients and with their overall survival. The presence of anti-HERV-K serum antibodies in melanoma patients was also found to be associated with a decreased disease-specific overall survival (36). This latter group proposed that humoral anti-HERV-K immune response may provide prognostic information additional to that of established melanoma markers.
Signaling pathways altered by expression of HERV-K env protein are involved in processes that include apoptosis, cell cycle, cell growth and proliferation, and DNA repair (summarized in Supplementary Figure 6, available online). Our results indicate that binding of an anti-HERV-K antibody to the cell surface env signals through TP53 pathway activation, as altered expression of genes in the TP53 pathway, was observed. TP53 is a tumor suppressor protein that plays a key role in apoptosis and senescence (including initiation of apoptosis and programmed cell death), activation of DNA repair proteins, and cell cycle arrest at the G1/S regulation point during DNA damage recognition. We speculate that either the internalization of the complex formed between env and the antibody or signaling through surface env after binding of the antibody leads to TP53 pathway activation. Future research will have to determine how the TP53 status of cancer cells influences the response to anti-HERV-K therapy, but our preliminary data indicate that both wild-type and mutant TP53 cells are targeted by anti-HERV-K antibodies. Treatment with 6H5 mAb also increased expression of CDK5 and CDKN1A (Figure 5 and Supplementary Figure 4, D, available online). Activation of TP53 is dependent on the functional level of CDK5, and its activation increases CDKN1A expression (33).
Infusion of anti-HERV-K env mAbs into immunodeficient mice injected with human breast cancer cells markedly reduced tumor development, and an analysis of tumor biopsies from mice revealed that the anti-HERV-K antibodies increased apoptosis, as judged by TUNEL-positive cells, and reduced proliferation, as judged by Ki-67+ cells. This observation of an antibody-induced tumor growth suppression both in vitro and in vivo suggests that HERV-K env protein may contribute to the aggressiveness of breast tumors. However, it should be noted that tumor size has proven to be a poor surrogate for a true regression of a cancer (37).
Although these mechanistic assumptions are preliminary and need evaluation by future research, these conclusions are corroborated by the analyses of breast cancer patient biopsies using IHC assays. Expression of HERV-K was found to be present in two-thirds of the 223 analyzed tumors in this study, and the expression of HERV-K env statistically significantly correlated with lymph node metastases. The findings are consistent with other studies, which also observed that HERV-K is reactivated in the majority of human breast tumors and is associated with prognosis (19). Other reports have suggested an increased expression of HERVs in human tumors, mainly at the transcriptional level (38). Thus, there is increasing evidence of HERV expression and activation in human cancer. It has been proposed that hormones and stress signals lead to HERV-K activation in human cancer cells. For example, the expression of HERV-K was statistically significantly elevated in MCF-7, T47D, and other breast cancer cells treated with estrogen, as reported by others and us (13,17). Despite the transcriptional activity of HERVs in a variety of human cancers, their potential roles in tumorigenicity and cancer progression are still poorly understood and urgently need further evaluation.
This study has a few limitations. First, a definitive mechanism by which anti-HERV-K mAbs and scFvs suppress cell growth and proliferation has not been determined. Although our results suggest involvement of the TP53 signaling pathway, other pathways may also be involved. Additional studies will be required to determine whether mechanisms usually associated with expression of retroviral genes, such as signaling molecules downstream of pattern recognition receptors that detect viral nucleic acid, are involved. Another limitation of this study is the possible involvement of other HERV family members in breast cancer etiology. HERV-K is one of a number of HERV family members who may modify the risk of breast cancer, and the efficacy of antibodies against these other HERV proteins has not been evaluated. A third limitation is that the efficacy of anti-HERV-K mAb immunotherapy has not been evaluated in breast cancer patients. Although our study using HERV-K env mAbs and our preclinical breast cancer vaccine study using HERV-K antigens (18) show promise in cell culture and mice bearing xenograft tumors, it will be necessary to test the antibodies and vaccines in clinical trials involving breast cancer patients.
In summary, we explored in a pilot study the feasibility of using anti-HERV-K antibodies for treatment of human breast cancer. To our knowledge, this is the first study to determine whether endogenous retroviruses are causative agents in breast cancer, and characterization of the effectiveness and mechanism of action of the neutralizing anti-HERV-K mAbs, has set the stage for producing new clinical reagents that can effectively target breast cancer. These antibodies induced selective growth suppression of cancer cells in vitro and in vivo, thus providing the first evidence to date that HERV-K expression could be linked to disease development and progression. Our novel observations suggest that HERV-K is a candidate therapeutic target for monoclonal antibody therapy for breast cancer.
Funding
This work was supported in part by the Department of Defense (BC052782 to F.W.-J), Susan G. Komen for the Cure (BCTR0402892 to F.W.-J), the National Institute of Environmental Health Sciences (ES007784 to G.L.J.), the Avon Foundation (07-2007-070 01 to G.L.J.), seed money provided by the Cattleman for Cancer Research Foundation (to F.W.-J and G.L.J.), and MD Anderson Cancer Center startup funds (to F.W.-J).
Supplementary Material
Footnotes
We thank Dr Kaoru Kiguchi for assistance with confocal microscopy. We thank Dr Michael G. Rosenblum for providing us with r-Gel conjugates and anti-rGel antibody, Dr George Georgiou for providing access to a Biacore 3000 instrument, and Dr Dean Tang for his helpful comments. We also thank personnel at the University of Maryland and the Baltimore Veterans Administration and the Surgery and Pathology Departments at the University of Maryland Medical Center, Baltimore Veterans Affairs Medical Center, Union Memorial Hospital, Mercy Medical Center, and Sinai Hospital for their contributions.
The authors are solely responsible for the study design, data collection, analysis and interpretation of the data, writing the article, and decision to submit the article for publication.
References
- 1.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Cho K, Lee YK, Greenhalgh DG. Endogenous retroviruses in systemic response to stress signals. Shock. Aug;30(2):105–116. doi: 10.1097/SHK.0b013e31816a363f. [DOI] [PubMed] [Google Scholar]
- 3.Contreras-Galindo R, Kaplan MH, Leissner P, et al. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J Virol. 2008;82(19):9329–9336. doi: 10.1128/JVI.00646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knossl M, Lower R, Lower J. Expression of the human endogenous retrovirus HTDV/HERV-K is enhanced by cellular transcription factor YY1. J Virol. 1999;73(2):1254–1261. doi: 10.1128/jvi.73.2.1254-1261.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ono M. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J Virol. 1986;58(3):937–944. doi: 10.1128/jvi.58.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer J, Sauter M, Racz A, Scherer D, Mueller-Lantzsch N, Meese E. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat Genet. 1999;21(3):257–258. doi: 10.1038/6766. [DOI] [PubMed] [Google Scholar]
- 7.Flockerzi A, Ruggieri A, Frank O, et al. Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV Transcriptome Project. BMC Genomics. 2008;9(Jul 29):354. doi: 10.1186/1471-2164-9-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleiman A, Senyuta N, Tryakin A, et al. HERV-K(HML-2) GAG/ENV antibodies as indicator for therapy effect in patients with germ cell tumors. Int J Cancer. 2004;110(3):459–461. doi: 10.1002/ijc.11649. [DOI] [PubMed] [Google Scholar]
- 9.Buscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005;65(10):4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 10.Seifarth W, Skladny H, Krieg-Schneider F, Reichert A, Hehlmann R, Leib-Mosch C. Retrovirus-like particles released from the human breast cancer cell line T47-D display type B- and C-related endogenous retroviral sequences. J Virol. 1995;69(10):6408–6416. doi: 10.1128/jvi.69.10.6408-6416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etkind PR, Lumb K, Du J, Racevskis J. Type 1 HERV-K genome is spliced into subgenomic transcripts in the human breast tumor cell line T47D. Virology. 1997;234(2):304–308. doi: 10.1006/viro.1997.8670. [DOI] [PubMed] [Google Scholar]
- 12.Ono M, Kawakami M, Ushikubo H. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J Virol. 1987;61(6):2059–2062. doi: 10.1128/jvi.61.6.2059-2062.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ejthadi HD, Martin JH, Junying J, et al. A novel multiplex RT-PCR system detects human endogenous retrovirus-K in breast cancer. Arch Virol. 2005;150(1):177–184. doi: 10.1007/s00705-004-0378-8. [DOI] [PubMed] [Google Scholar]
- 14.Wang-Johanning F, Frost AR, Johanning GL, et al. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin Cancer Res. 2001;7(6):1553–1560. [PubMed] [Google Scholar]
- 15.Wang-Johanning F, Liu J, Rycaj K, et al. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 2007;120(1):81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- 16.Ruggieri A, Maldener E, Sauter M, et al. Human endogenous retrovirus HERV-K(HML-2) encodes a stable signal peptide with biological properties distinct from Rec. Retrovirology. 2009;6(Feb 16):17. doi: 10.1186/1742-4690-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang-Johanning F, Frost AR, Jian B, Epp L, Lu DW, Johanning GL. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22(10):1528–1535. doi: 10.1038/sj.onc.1206241. [DOI] [PubMed] [Google Scholar]
- 18.Wang-Johanning F, Radvanyi L, Rycaj K, et al. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008;68(14):5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golan M, Hizi A, Resau JH, et al. Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia. 2008;10(6):521–533. doi: 10.1593/neo.07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prueitt RL, Boersma BJ, Howe TM, et al. Inflammation and IGF-I activate the Akt pathway in breast cancer. Int J Cancer. 2007;120(4):796–805. doi: 10.1002/ijc.22336. [DOI] [PubMed] [Google Scholar]
- 21.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 22.Sobin LH, Fleming ID. TNM Classification of Malignant Tumors, fifth edition (1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer. 1997;80(9):1803–1804. doi: 10.1002/(sici)1097-0142(19971101)80:9<1803::aid-cncr16>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 23.Galea MH, Blamey RW, Elston CE, Ellis IO. The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat. 1992;22(3):207–219. doi: 10.1007/BF01840834. [DOI] [PubMed] [Google Scholar]
- 24.Wang-Johanning F, Gillespie GY, Grim J, et al. Intracellular expression of a single-chain antibody directed against human papillomavirus type 16 E7 oncoprotein achieves targeted antineoplastic effects. Cancer Res. 1998;58(9):1893–1900. [PubMed] [Google Scholar]
- 25.Schmidt MM, Thurber GM, Wittrup KD. Kinetics of anti-carcinoembryonic antigen antibody internalization: effects of affinity, bivalency, and stability. Cancer Immunol Immunother. 2008;57(12):1879–1890. doi: 10.1007/s00262-008-0518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenblum MG, Cheung LH, Liu Y, Marks JW., III Design, expression, purification, and characterization, in vitro and in vivo, of an antimelanoma single-chain Fv antibody fused to the toxin gelonin. Cancer Res. 2003;63(14):3995–4002. [PubMed] [Google Scholar]
- 27.Bikoue A, Janossy G, Barnett D. Stabilised cellular immuno-fluorescence assay: CD45 expression as a calibration standard for human leukocytes. J Immunol Methods. 2002;266(1–2):19–32. doi: 10.1016/s0022-1759(02)00094-7. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Wittner M, Bouamar H, Jarrier P, Vainchenker W, Louache F. Identification of CXCR4 as a new nitric oxide-regulated gene in human CD34+ cells. Stem Cells. 2007;25(1):211–219. doi: 10.1634/stemcells.2006-0468. [DOI] [PubMed] [Google Scholar]
- 29.Glynn SA, Boersma BJ, Dorsey TH, et al. Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients. J Clin Invest. 2010;120(11):3843–3854. doi: 10.1172/JCI42059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10(12):878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benatti P, Basile V, Merico D, Fantoni LI, Tagliafico E, Imbriano C. A balance between NF-Y and p53 governs the pro- and anti-apoptotic transcriptional response. Nucleic Acids Res. 2008;36(5):1415–1428. doi: 10.1093/nar/gkm1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 33.Ajay AK, Upadhyay AK, Singh S, et al. Cdk5 phosphorylates non-genotoxically overexpressed p53 following inhibition of PP2A to induce cell cycle arrest/apoptosis and inhibits tumor progression. Mol Cancer. 2010;9(Jul 31):204. doi: 10.1186/1476-4598-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeda H, Akaike T. Nitric oxide and oxygen radicals in infection, inflammation, and cancer. Biochemistry (Mosc). 1998;63(7):854–865. [PubMed] [Google Scholar]
- 35.Ludwig DL, Pereira DS, Zhu Z, Hicklin DJ, Bohlen P. Monoclonal antibody therapeutics and apoptosis. Oncogene. 2003;22(56):9097–9106. doi: 10.1038/sj.onc.1207104. [DOI] [PubMed] [Google Scholar]
- 36.Hahn S, Ugurel S, Hanschmann KM, et al. Serological response to human endogenous retrovirus K in melanoma patients correlates with survival probability. AIDS Res Hum Retroviruses. 2008;24(5):717–723. doi: 10.1089/aid.2007.0286. [DOI] [PubMed] [Google Scholar]
- 37.Avril N, Sassen S, Roylance R. Response to therapy in breast cancer. J Nucl Med. 2009;50(suppl 1):55S–63S. doi: 10.2967/jnumed.108.057240. [DOI] [PubMed] [Google Scholar]
- 38.Romanish MT, Cohen CJ, Mager DL. Potential mechanisms of endogenous retroviral-mediated genomic instability in human cancer. Semin Cancer Biol. 2010;20(4):246–253. doi: 10.1016/j.semcancer.2010.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.