Abstract
Objective
DNA repair pathway genes play an important role in maintaining genomic integrity and protecting against cancer development. This study aimed to identify novel SNPs in the DNA repair–related genes associated with melanoma risk from a genome-wide association study (GWAS).
Methods
A total of 8,422 SNPs from the 165 DNA repair–related genes were extracted from a GWAS of melanoma risk, including 494 cases and 5,628 controls from the Nurses’ Health Study (NHS) and the Health Professionals Follow-up Study (HPFS). We further replicated the top SNPs in a GWAS of melanoma risk from the MD Anderson Cancer Center (1,804 cases and 1,026 controls).
Results
A total of 3 SNPs with P value < 0.001 were selected for in silico replication. One SNP was replicated: rs3902093 [A] in EXO1 promoter region (Pdiscovery = 6.6×10-4, Preplication = 0.039, Pjoint = 2.5×10-4; ORjoint = 0.80, 95% CI: 0.71, 0.90). This SNP was associated with the expression of the EXO1; carriers of the A allele showed lower expression (P = 0.002).
Conclusion
Our study found that a promoter region SNP in the editing and processing nucleases gene EXO1 was associated with decreased expression of EXO1 and decreased melanoma risk. Further studies are warranted to validate this association and to investigate the potential mechanisms.
Introduction
Melanoma is the most lethal kind of skin cancer that kills thousands of people each year in the United States. Ultraviolet (UV) radiation is an established environmental carcinogen for skin cancer, including melanoma [1-3]. UV radiation is capable of causing a wide range of lesions in DNA. One important defense mechanism against melanoma is the ability to repair DNA damage induced by UV light. For example, Xeroderma pigmentosum (XP) is a heterogeneous disorder involving hypersensitivity to UV radiation, and XP patients have a more than 1000-fold increased risk of developing skin cancer [4, 5]. This abnormality is caused by defects in the nucleotide excision repair of UV-induced DNA damage [6].
DNA repair genes form a complex network that protects the genome's integrity from endogenous and exogenous damage [7]. When DNA damage is not repaired and does not induce apoptotic elimination of the cell, DNA defects accumulate and are propagated through the cell progeny, and finally cancer may occur [8]. Individual variations in DNA repair capacity due to the presence of polymorphisms in DNA repair-related genes may account for some cancer susceptibility in the general population [9, 10]. Polymorphisms in several DNA repair-related genes have been found to be associated with the risk of melanoma, but the findings are not consistent [11]. Our group previously conducted a nested case-control study evaluating genetic variation across 60 DNA repair pathway genes in relation to melanoma risk, and we identified a PARP1 variant involved in melanoma development [12].
To comprehensively evaluate the association between polymorphisms of genes involved in DNA repair pathways and the risk of melanoma, we extracted genotyping data of 165 DNA repair–related genes from a genome-wide association study (GWAS) of melanoma risk, with a subsequent in silico replication.
Materials and Methods
This study contained a discovery set (melanoma case-control study within the NHS and HPFS) and a replication set (the MD Anderson Cancer Center melanoma case-control study). The study protocols were approved by the Institutional Review Board at the Brigham and Women's Hospital and the MD Anderson Cancer Center, and informed consent was obtained from all participants.
Discovery set
Study population
Nurses’ Health Study (NHS)
The NHS was established in 1976, when 121,700 female US-registered nurses between ages of 30 and 55 residing in 11 large US states completed and returned an initial self-administered questionnaire on their medical histories and baseline health-related exposures. Since then, biennial questionnaires with prospective collection of exposure information on risk factors have been collected. Along with information about exposures every 2 years, outcome data with appropriate follow-up of reported disease events are also collected. Overall, the follow-up rate has been very high; after more than 20 years, ~90% of participants continue to complete questionnaires. Between May 1989 and September 1990, we had collected blood samples from 32,826 participants in the NHS. Information on melanoma development was first collected in the 1984 questionnaire.
Health Professionals Follow-up Study (HPFS)
In 1986, 51,529 men from all 50 US states in health professions (dentists, pharmacists, optometrists, osteopath physicians, podiatrists, and veterinarians) aged 40-75 answered a detailed questionnaire by mail, forming the basis of the HPFS study. The average follow-up rate for this cohort over 10 years is >90%. On each biennial questionnaire, we obtained disease- and health-related information. Between 1993 and 1994, 18,159 study participants provided blood samples by overnight courier. Information on melanoma development was collected in the 1986 questionnaire.
Melanoma cases and controls in the discovery set
Eligible cases in both the NHS and HPFS cohorts were participants with histopathologically confirmed invasive melanoma, diagnosed at any time after baseline up to the 2008 follow-up cycle for both cohorts. All subjects were US non-Hispanic Caucasians.
Data on skin cancer risk factors were obtained from cohort questionnaires in both cohorts in the 1980s. These risk factors included adolescent sunburn reactions, number of severe sunburns, mole count on the left arm, and natural hair color.
Eight GWASs included in the discovery set
We have previously conducted several GWASs on different disease outcomes (NHS: breast cancer, coronary heart disease, type 2 diabetes, kidney stone, pancreatic cancer, and glaucoma; HPFS: coronary heart disease, type 2 diabetes, kidney stone, advanced prostate cancer, and glaucoma). In this study, we included only controls from each of the GWASs, except for the kidney stone GWAS, from which we used both cases and controls. Participants without melanoma were the controls in this study, and those diagnosed with melanoma were the cases. In addition, we genotyped the rest of the melanoma cases in both cohorts who were not included in the previous GWASs. Finally, we included 494 melanoma cases and 5,628 controls.
We performed genotyping in breast cancer GWAS in NHS using the Illumina HumanHap550 array, as part of the National Cancer Institute's Cancer Genetic Markers of Susceptibility (CGEMS) Project. For the coronary heart disease and type 2 diabetes GWASs of the discovery set, we performed genotyping using the Affymetrix 6.0 array. For the glaucoma GWAS, we performed genotyping using the Illumina HumanHap660 array. For the kidney stone, advanced prostate cancer and melanoma GWASs, we performed genotyping using the Illumina HumanHap610 array. Based on the genotyped SNPs and haplotype information in the NCBI build 35 of phase II Hapmap CEU data, we imputed genotypes for >2.5 million SNPs using the program MACH [13]. Only SNPs with imputation quality R2 > 0.95 in each study were included in the final analysis. A total of 1,579,307 SNPs were included in the final meta-analysis of the NHS and HPFS. Betas from all GWAS studies of the discovery set were combined by a meta-analysis with weights proportional to the inverse variance of the beta in each study. The study description and quality control procedures for the eight GWAS sets of the discovery phase are presented in Supplementary Methods.
Replication set
Genotyping in replication set
The study participants were from a hospital-based case-control study of melanoma, for which cases were recruited from among non-Hispanic white patients and controls at MD Anderson between March 1998 and August 2008. Samples and data were available from 931 melanoma patients and 1,026 cancer-free controls (friends of other patients reporting to clinics) who were frequency-matched on age and sex, completed a comprehensive skin lifestyle questionnaire. This questionnaire was administered to 70% of patients and controls by an interviewer and was self-administered among the remaining 30%. An additional case series comprising 873 individuals presenting for treatment for melanoma at MD Anderson was also included, bringing the total number of melanoma patients to 1,804.
The whole blood samples were collected from each participant, with various DNA extraction methods (including Gentra, Qiagen, and phenol/chloroform). DNA samples for the first-stage GWAS were genotyped using the Illumina Omnil-Quad array, passed quality control filters for genotyping and were called using the BeadStudio algorithm at the John Hopkins University Center for Inherited Disease Research (CIDR).
Quality control (QC) procedures for the replication set
Mean call rate for all samples was 99.86%. Forty-one failed in genotyping with >10% missing rate across all SNPs, and 11 samples had identity problems that could not be resolved. After excluding from the study 126 duplicated, related, or outlier SNPs identified by principal component analysis, there were 1,952 cases and 1,026 controls. Among 2,978 total case and control subjects passing quality control, 138 in situ cases were subsequently removed from the study as having indeterminate phenotype. Ten atypical melanocytic proliferation patients were also excluded as not having invasive cancers. Finally, we used the data from 1,804 cases and 1,026 controls available for the association study of melanoma susceptibility.
DNA repair gene and SNP selection
We selected human DNA repair pathway genes according to a review article [14]. A complete list of the genes can be found on http://sciencepark.mdanderson.org/labs/wood/DNA_Repair_Genes.html. These pathways/genes included Base excision repair (UNG, SMUG1, MBD4, TDG, OGG1, MUTYH, NTHL1, MPG, NEIL1, NEIL2, NEIL3, APEX1, APEX2, LIG3, XRCC1, PNKP); Poly(ADP-ribose) polymerase enzymes that bind to DNA (PARP1, PARP2); Direct reversal of damage (MGMT, ALKBH2, ALKBH3); Repair of DNA-protein crosslink (TDP1); Mismatch excision repair (MSH2, MSH3, MSH6, MLH1, PMS2, MSH4, MSH5, MLH3, PMS1, PMS2L3); Nucleotide excision repair (XPC, RAD23B, CETN2, RAD23A, XPA, RPA1, RPA2, RPA3, ERCC3, ERCC2, GTF2H1, GTF2H2, GTF2H3, GTF2H4, GTF2H5, CDK7, CCNH, MNAT1, ERCC5, ERCC1, ERCC4, LIG1, CKN1, ERCC6, XAB2, DDB1, DDB2, MMS19L); Homologous recombination (RAD51, RAD51L1, RAD51C, RAD51L3, DMC1, XRCC2, XRCC3, RAD52, RAD54L, RAD54B, BRCA1, BRCA2, SHFM1, RAD50, MRE11A, NBN, RBBP8, MUS81, EME1, EME2, GIYD1, BTBD12, GEN1); Non-homologous end-joining (XRCC6, XRCC5, PRKDC, LIG4, XRCC4, DCLRE1C, NHEJ1); Modulation of nucleotide pools (NUDT1, DUT, RRM2B); DNA polymerases (POLB, POLG, POLD1, POLE, PCNA, REV3L, MAD2L2, REV1L, POLH, POLI, POLQ, POLK, POLL, POLM, POLN); Editing and processing nucleases (FEN1, MTMR15, TREX, TREX2, EXO1, APTX, TTRAP, SPO11, FLJ35220); Rad6 pathway (UBE2A, UBE2B, RAD18, UBE2V2, UBE2N); Chromatin Structure (H2AFX, CHAF1A, SETMAR); Genes defective in diseases associated with sensitivity to DNA damaging agents (BLM, WRN, RECQL4, ATM, TTDN1, FANCA, FANCB, FANCC, FANCD2, FANCE, FANCF, FANCG, FANCI, BRIP1, FANCL, FANCM, PALB2, FAAP24); Other identified genes with known or suspected DNA repair function (DCLRE1A, DCLRE1B, RPA4, RECQL, RECQL5, HELQ, RDM1, OBFC2B); Other conserved DNA damage response genes (ATR, MDC1, RAD1, RAD9A, HUS1, RAD17, CHEK1, CHEK2, TP53, TP53BP1, ATRIP, TOPBP1, CLK2, PER1).
Statistical methods
We regressed a binary coding for melanoma cases and controls on each SNP (dosage file) that passed quality control filters. The five largest principal components of genetic variation were nominally significantly associated with melanoma risk, and we adjusted for them as well as age and gender. These principal components were calculated for all individuals on the basis of ~10,000 unlinked markers using the EIGENSTRAT software [15]. SNPs within the DNA repair-related genes and their ±30 kb flanking regions were extracted for further analysis. We performed a meta-analysis to combine the replication set and the discovery set. We constructed a pigmentation score to summarize pigmentation-related variables. Briefly, we applied the logistic regression coefficients from a multivariate model, including sex, age, hair color, and tendency to sunburn, to each individual's values for the latter two of these variables and summed the values to compute a pigmentation score. Based on the mean score, we identified participants with low and high levels of pigmentation.
The analysis of EXO1 expression by genotype was based on the transcript expression profiling data of 87 HapMap CEU cell lines (NCBI GEO database, accession GSE7792) [16].
Results
The distribution of major melanoma risk factors between cases and controls is shown in Table 1. Cases and controls were comparable in sex and age, but cases were more likely to burn and to have light hair color, more moles, and more severe sunburns.
Table 1.
Distribution of risk factors for melanoma in cases and controls in the discovery set.
| Risk factors | Cases, n (%) | Controls, n (%) |
|---|---|---|
| Sex (female) | 317 (64) | 3376 (60) |
| Age (> 55) | 75 (15) | 1001 (18) |
| Hair color (red or blonde) | 67 (16) | 710 (14) |
| Number of moles on the left arm (> 2)* | 66 (18) | 559 (12) |
| Number of severe sunburns (> 5)* | 209 (51) | 2152 (44) |
| Tendency to burn (painful burn)* | 110 (27) | 869 (18) |
significantly different between cases and controls (P value< 0.05)
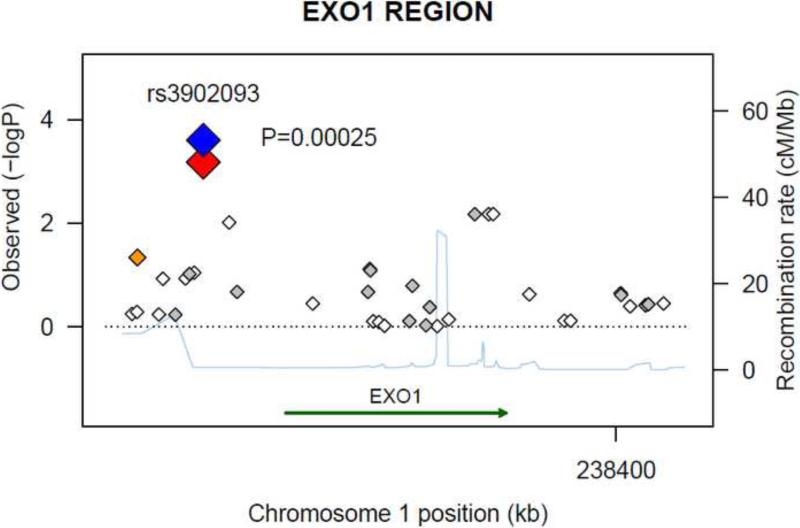
A total of 8,422 SNPs were extracted from the 165 DNA repair-related genes within their ±30 kb flanking regions from the discovery set. After removing SNPs in high linkage disequilibrium (LD) (r2 > 0.8) 1,636 SNPs remained; only three reached a significance level of P value < 0.001. We selected these three SNPs for in silico replication, all three SNPs in both sets had imputed R2 > 0.99. One SNP in the discovery set was replicated: rs3902093 [A] in EXO1 (exonuclease 1) locus (Pdiscovery = 6.6×10-4, Preplication = 0.039, Pjoint = 2.5×10-4; OR joint = 0.80, 95% CI: 0.71, 0.90). The regional association plot for the EXO1 region in the discovery set is presented in Figure 1. Neither of the other two SNPs reached the nominal significance level in the replication set (Table 2). A list of the SNPs with P-value < 0.01 in the discovery set is shown in Supplemental Table 1.
Figure 1.
Regional association plot in the 30-kb neighborhood of EXO1. The left-hand Y-axis shows the association P-value of individual SNPs in the discovery set, which is plotted as -log10 (P) against chromosomal base-pair position. The right-hand Y-axis shows the recombination rate estimated from the HapMap CEU population. Genotyped SNPs are plotted as diamonds, and imputed SNPs are colored grey. Blue indicates the SNPs of rs3902093 in joint analysis; bright red indicates rs3902093 in the discovery set; orange moderate LD (r2 ≥ 0.5 but <0.8); and white no LD (r2 < 0.2). The genomic coordinate is in NCBI35/hg17.
Table 2.
Top 3 independent SNPs associated melanoma risk with P < 0.001 in the discovery set and their replication
| SNP information | Discovery set | Replication set | Joint analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Test allele | Reference allele | Frequency of test allele | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P |
| REV1L | rs4341989 | T | C | 0.296 | 1.13 (1.30, 1.49) | 2.8×10-4 | 1.02 (0.91, 1.15) | 0.752 | 1.13 (1.03, 1.23) | 0.010 |
| EXO1 | rs3902093 | A | C | 0.165 | 0.71 (0.58, 0.86) | 6.6×10-4 | 0.86 (0.75, 0.99) | 0.039 | 0.80 (0.71, 0.90) | 2.5×10-4 |
| REV1L | rs9941566 | A | G | 0.135 | 0.69 (0.55, 0.86) | 9.3×10-4 | 0.97 (0.83, 1.14) | 0.711 | 0.87 (0.76, 0.98) | 0.026 |
All three SNPs in both sets had imputed R2 > 0.99
For the EXO1 rs3902093, compared with the CC genotype, the CA genotype had a RR of 0.72 (95% CI: 0.58, 0.89), the AA genotype had a RR of 0.36 (95% CI: 0.15, 0.87), and CA and AA combined had a RR of 0.69 (95% CI: 0.55, 0.85). When stratified by pigmentation score, number of severe sunburns, and number of moles on the left arm, there were no statistically significant interactions observed (all P values for interaction tests > 0.05) (Table 3).
Table 3.
EXO1 SNP rs3902093 and melanoma risk
| EXO1 (rs3902093) | Control | Case | OR (95% CI) | P |
|---|---|---|---|---|
| Overall | ||||
| CC | 3882 (68.99%) | 378 (76.52%) | 1.00 | |
| CA | 1600 (28.43%) | 111 (22.47%) | 0.72 (0.58, 0.89) | 0.003 |
| AA | 145 (2.58%) | 5 (1.01%) | 0.36 (0.15, 0.87) | 0.024 |
| CA+AA | 1745 (31.01%) | 116 (23.48%) | 0.69 (0.55, 0.85) | 0.0007 |
| Low pigmentation score | ||||
| CC | 2013 (69.73%) | 178 (76.07%) | 1.00 | |
| CA+AA | 874 (30.27%) | 56 (23.93%) | 0.72 (0.53, 0.99) | 0.04 |
| High pigmentation score | ||||
| CC | 1869 (68.21%) | 200 (76.92%) | 1.00 | |
| CA+AA | 871 (31.79%) | 60 (23.08%) | 0.65 (0.48, 0.88) | 0.005 |
| Number of severe sunburns ≤ 5 | ||||
| CC | 1893 (68.96%) | 154 (77.78%) | 1.00 | |
| CA+AA | 852 (31.04%) | 44 (22.22%) | 0.65 (0.46, 0.92) | 0.01 |
| Number of severe sunburns > 5 | ||||
| CC | 1475 (68.54%) | 155 (74.16%) | 1.00 | |
| CA+AA | 677 (31.46%) | 54 (25.84%) | 0.77 (0.56, 1.07) | 0.11 |
| Number of moles on left arm ≤ 2 | ||||
| CC | 2750 (69.66%) | 237 (76.21%) | 1.00 | |
| CA+AA | 1198 (30.34%) | 74 (23.79%) | 0.73 (0.56, 0.96) | 0.02 |
| Number of moles on left arm > 2 | ||||
| CC | 388 (69.41%) | 45 (68.18%) | 1.00 | |
| CA+AA | 171 (30.59%) | 21 (31.82%) | 1.09 (0.63, 1.90) | 0.16 |
There were no statistically significant interactions between these factors and the EXO1 genotype on melanoma risk (all P values for interaction tests > 0.05).
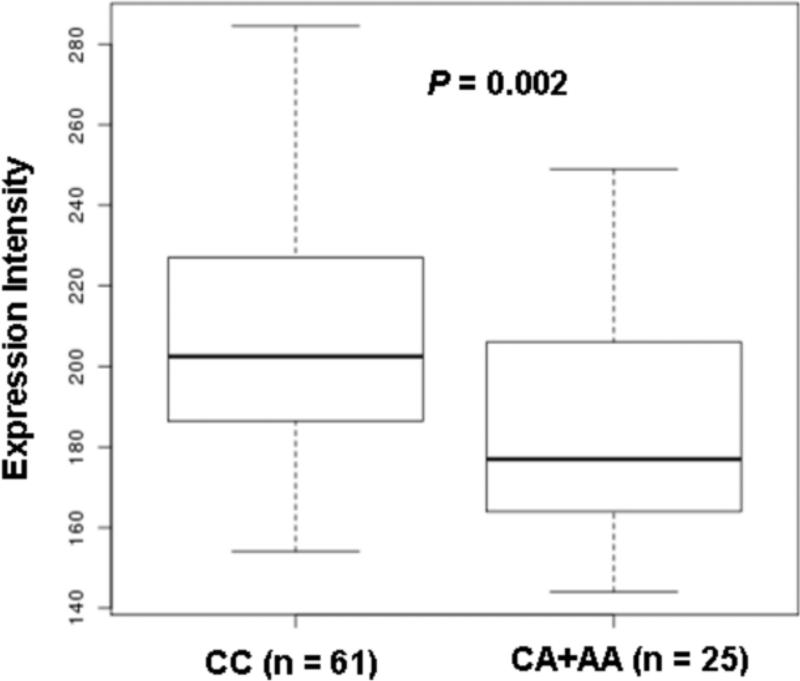
In addition, we evaluated the expression of EXO1 in relation to the rs3902093 genotypes, and found that the CA/AA genotype was associated with a lower expression of EXO1 (P = 0.002) than in the CC genotype (Figure 2).
Figure 2.
EXO1 expression by the rs3902093 genotype. The expression level of the EXO1 locus was significantly higher for the rs3902093 CC genotype compared with the A allele carriers (P = 0.002). The Y-axis is the expression intensity of EXO1. The box represents the central 50% of the data or the interquartile range. The lower edge of the box plot is the first quartile or 25th percentile. The upper edge of the box plot is the third quartile or 75th percentile. The line in the box is the median value. The ends of the vertical lines extend to a maximum of 1.5 times the inter-quartile.
Discussion
Recent GWASs have identified melanoma risk loci mainly limited to pigmentation and nevogenesis pathways [17-19]. Epidemiological evidence for the association between DNA repair genes and melanoma risk is sparse. A meta-analysis in 2009 suggested XPD/ERCC2 as the only low-penetrance melanoma risk gene; the C allele of the SNP rs13181 was associated with a 10% population attributable risk of melanoma [11]. Afterward, two large case-control studies from Germany and Spain failed to identify any DNA repair gene polymorphisms associated with melanoma risk [20, 21]. A family based study genotyped 2,964 tag SNPs in 131 DNA repair genes and reported that polymorphisms in POLN and PRKDC were associated with increased melanoma risk in melanoma families [22]. A hospital-based case-control study found that XPC polymorphisms contributed to melanoma risk in a geographic region exposed to high sun radiation in Brazil [23]. In this GWAS-based pathway analysis of 8,422 SNPs within 165 DNA repair genes, we found one SNP rs3902093 in the EXO1 gene significantly associated with melanoma risk.
One limitation of our study is that the expression was examined in cell lines derived from blood samples. However, although the expression pattern may vary across different cell types, the expression quantitative trait loci (eQTL) could be in sharing [24]. It was reported that >70% cis eQTL identified in the skin tissues were eQTL in lymphoblastoid cell lines as well [25], so that lymphoblastoid cell line eQTL could be a useful surrogate to study the genetic of gene expression in the skin.
To our knowledge, this report provides the first piece of evidence for an association between an EXO1 gene polymorphism and melanoma risk. In fact, EXO1 variants have previously been associated with the risk of many other types of cancer. Specifically, EXO1 K589E (rs1047840) was found to be associated with the development of lung cancer [26], oral cancer [27], breast cancer [28], gastric cancer [29], and colorectal cancer [30] in Asian populations. It was also suggested that polymorphisms in EXO1 may be associated with risk of cancer modified by tobacco smoking status [31]. Our study did not find an association between this SNP and the risk of melanoma (OR = 1.02, 95% CI: 0.89, 1.17). The SNP rs3902093 that we found to be associated with melanoma risk is in the promoter region of EXO1. This SNP was also found to affect the expression of EXO1; the carriers of the variant allele had lower expression and a lower risk of melanoma. When the initial step of damage cleavage is more active than the final step of DNA re-synthesis in the DNA repair process, this imbalance leads to excessive unrepaired DNA intermediate products and in turn causes elevated mutation and cancer risk [32]. One possible underlying mechanism for our finding is that lower expression of nuclease genes may prevent this imbalance in DNA repair, reducing both DNA mutation and cancer risk.
In addition, EXO1 functions in most aspects of DNA metabolism [33-35]. It is involved in DNA resection during double-strand break repair [36]; it contributes to DNA damage signal induction in mammals [37]; and it is important for telomere maintenance through promotion of recombination at transcription-induced telomeric structures, by creating single strand DNA at uncapped telomere ends [38]. EXO1 was found to be critical for the UV-induced DNA damage response linking nucleotide excision repair to checkpoint activation in human cells[39] and was also shown to have a role in the timely induction of UV-induced apoptosis [40]. Therefore, the role of EXO1 in the process of DNA repair is complex, and the mechanism by which it contributes to melanoma susceptibility warrants further study.
Supplementary Material
Highlights.
>We identify novel SNPs in the DNA repair–related genes associated with melanoma risk from a genome-wide association study. >We extracted genotyping data of 8,422 SNPs from the 165 DNA repair–related genes. >We found that a promoter region SNP in the editing and processing nucleases gene EXO1 was associated with decreased expression of EXO1 and decreased melanoma risk.
Acknowledgements
We are indebted to the participants in the NHS and HPFS for their dedication to this study. We thank the following state cancer registries for their help: Alabama, Arizona, Arkansas, California, Colorado, Connecticut, Delaware, Florida, Georgia, Idaho, Illinois, Indiana, Iowa, Kentucky, Louisiana, Maine, Maryland, Massachusetts, Michigan, Nebraska, New Hampshire, New Jersey, New York, North Carolina, North Dakota, Ohio, Oklahoma, Oregon, Pennsylvania, Rhode Island, South Carolina, Tennessee, Texas, Virginia, Washington, and Wyoming.
Financial support: NIH CA122838, CA87969 and CA055075.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Armstrong BK, Kricker A, English DR. Sun exposure and skin cancer. The Australasian journal of dermatology. 1997;38(Suppl 1):S1–6. doi: 10.1111/j.1440-0960.1997.tb01000.x. [DOI] [PubMed] [Google Scholar]
- 2.English DR, Armstrong BK, Kricker A, Fleming C. Sunlight and cancer. Cancer Causes Control. 1997;8:271–283. doi: 10.1023/a:1018440801577. [DOI] [PubMed] [Google Scholar]
- 3.Ravanat JL, Douki T, Cadet J. Direct and indirect effects of UV radiation on DNA and its components. Journal of photochemistry and photobiology. 2001;63:88–102. doi: 10.1016/s1011-1344(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 4.Chu G, Mayne L. Xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy: do the genes explain the diseases? Trends Genet. 1996;12:187–192. doi: 10.1016/0168-9525(96)10021-4. [DOI] [PubMed] [Google Scholar]
- 5.Kraemer KH, Lee MM, Andrews AD, Lambert WC. The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer. The xeroderma pigmentosum paradigm, Archives of dermatology. 1994;130:1018–1021. [PubMed] [Google Scholar]
- 6.Tanaka K, Wood RD. Xeroderma pigmentosum and nucleotide excision repair of DNA. Trends in biochemical sciences. 1994;19:83–86. doi: 10.1016/0968-0004(94)90040-X. [DOI] [PubMed] [Google Scholar]
- 7.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nature reviews. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 8.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nature medicine. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 9.Li C, Wang LE, Wei Q. DNA repair phenotype and cancer susceptibility--a mini review. International journal of cancer. 2009;124:999–1007. doi: 10.1002/ijc.24126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- 11.Mocellin S, Verdi D, Nitti D. DNA repair gene polymorphisms and risk of cutaneous melanoma: a systematic review and meta-analysis. Carcinogenesis. 2009;30:1735–1743. doi: 10.1093/carcin/bgp207. [DOI] [PubMed] [Google Scholar]
- 12.Zhang M, Qureshi AA, Guo Q, Han J. Genetic variation in DNA repair pathway genes and melanoma risk. DNA repair. 2011;10:111–116. doi: 10.1016/j.dnarep.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biernacka JM, Tang R, Li J, McDonnell SK, Rabe KG, Sinnwell JP, Rider DN, de Andrade M, Goode EL, Fridley BL. Assessment of genotype imputation methods. BMC proceedings. 2009;3(Suppl 7):S5. doi: 10.1186/1753-6561-3-s7-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood RD, Mitchell M, Sgouros J, Lindahl T. Science. Vol. 291. New York, N.Y: 2001. Human DNA repair genes; pp. 1284–1289. [DOI] [PubMed] [Google Scholar]
- 15.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 16.Huang RS, Duan S, Bleibel WK, Kistner EO, Zhang W, Clark TA, Chen TX, Schweitzer AC, Blume JE, Cox NJ, Dolan ME. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown KM, Macgregor S, Montgomery GW, Craig DW, Zhao ZZ, Iyadurai K, Henders AK, Homer N, Campbell MJ, Stark M, Thomas S, Schmid H, Holland EA, Gillanders EM, Duffy DL, Maskiell JA, Jetann J, Ferguson M, Stephan DA, Cust AE, Whiteman D, Green A, Olsson H, Puig S, Ghiorzo P, Hansson J, Demenais F, Goldstein AM, Gruis NA, Elder DE, Bishop JN, Kefford RF, Giles GG, Armstrong BK, Aitken JF, Hopper JL, Martin NG, Trent JM, Mann GJ, Hayward NK. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nature genetics. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, Randerson-Moor J, Aitken JF, Avril MF, Azizi E, Bakker B, Bianchi-Scarra G, Bressac-de Paillerets B, Calista D, Cannon-Albright LA, Chin AWT, Debniak T, Galore-Haskel G, Ghiorzo P, Gut I, Hansson J, Hocevar M, Hoiom V, Hopper JL, Ingvar C, Kanetsky PA, Kefford RF, Landi MT, Lang J, Lubinski J, Mackie R, Malvehy J, Mann GJ, Martin NG, Montgomery GW, van Nieuwpoort FA, Novakovic S, Olsson H, Puig S, Weiss M, van Workum W, Zelenika D, Brown KM, Goldstein AM, Gillanders EM, Boland A, Galan P, Elder DE, Gruis NA, Hayward NK, Lathrop GM, Barrett JH, Bishop JA. Genome-wide association study identifies three loci associated with melanoma risk. Nature genetics. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falchi M, Bataille V, Hayward NK, Duffy DL, Bishop JA, Pastinen T, Cervino A, Zhao ZZ, Deloukas P, Soranzo N, Elder DE, Barrett JH, Martin NG, Bishop DT, Montgomery GW, Spector TD. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nature genetics. 2009;41:915–919. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figl A, Scherer D, Nagore E, Bermejo JL, Botella-Estrada R, Gast A, Thirumaran RK, Planelles D, Hemminki K, Schadendorf D, Kumar R. Single-nucleotide polymorphisms in DNA-repair genes and cutaneous melanoma. Mutation research. 2010;702:8–16. doi: 10.1016/j.mrgentox.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Ibarrola-Villava M, Pena-Chilet M, Fernandez LP, Aviles JA, Mayor M, Martin-Gonzalez M, Gomez-Fernandez C, Casado B, Lazaro P, Lluch A, Benitez J, Lozoya R, Boldo E, Pizarro A, Martinez-Cadenas C, Ribas G. Genetic polymorphisms in DNA repair and oxidative stress pathways associated with malignant melanoma susceptibility. Eur J Cancer. 2011 doi: 10.1016/j.ejca.2011.05.011. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 22.Liang X, Pfeiffer RM, Wheeler W, Maeder D, Burdette L, Yeager M, Chanock S, Tucker MA, Goldstein AM, Yang XR. Genetic variants in DNA repair genes and the risk of cutaneous malignant melanoma in melanoma-prone families with/without CDKN2A mutations. International journal of cancer. 2011 doi: 10.1002/ijc.26231. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goncalves FT, Francisco G, de Souza SP, Luiz OC, Festa-Neto C, Sanches JA, Chammas R, Gattas GJ, Eluf-Neto J. European ancestry and polymorphisms in DNA repair genes modify the risk of melanoma: A case-control study in a high UV index region in Brazil. Journal of dermatological science. 2011 doi: 10.1016/j.jdermsci.2011.06.003. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 24.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding J, Gudjonsson JE, Liang L, Stuart PE, Li Y, Chen W, Weichenthal M, Ellinghaus E, Franke A, Cookson W, Nair RP, Elder JT, Abecasis GR. Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. American journal of human genetics. 2010;87:779–789. doi: 10.1016/j.ajhg.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu NY, Wang HC, Wang CH, Chiu CF, Tseng HC, Liang SY, Tsai CW, Lin CC, Bau DT. Lung cancer susceptibility and genetic polymorphisms of Exo1 gene in Taiwan. Anticancer research. 2009;29:725–730. [PubMed] [Google Scholar]
- 27.Tsai MH, Tseng HC, Liu CS, Chang CL, Tsai CW, Tsou YA, Wang RF, Lin CC, Wang HC, Chiu CF, Bau DT. Interaction of Exo1 genotypes and smoking habit in oral cancer in Taiwan. Oral oncology. 2009;45:e90–94. doi: 10.1016/j.oraloncology.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Wang HC, Chiu CF, Tsai RY, Kuo YS, Chen HS, Wang RF, Tsai CW, Chang CH, Lin CC, Bau DT. Association of genetic polymorphisms of EXO1 gene with risk of breast cancer in Taiwan. Anticancer research. 2009;29:3897–3901. [PubMed] [Google Scholar]
- 29.Bau DT, Wang HC, Liu CS, Chang CL, Chiang SY, Wang RF, Tsai CW, Lo YL, Hsiung CA, Lin CC, Huang CY. Single-nucleotide polymorphism of the Exo1 gene: association with gastric cancer susceptibility and interaction with smoking in Taiwan. The Chinese journal of physiology. 2009;52:411–418. doi: 10.4077/cjp.2009.amh076. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto H, Hanafusa H, Ouchida M, Yano M, Suzuki H, Murakami M, Aoe M, Shimizu N, Nakachi K, Shimizu K. Single nucleotide polymorphisms in the EXO1 gene and risk of colorectal cancer in a Japanese population. Carcinogenesis. 2005;26:411–416. doi: 10.1093/carcin/bgh335. [DOI] [PubMed] [Google Scholar]
- 31.Gao Y, Hayes RB, Huang WY, Caporaso NE, Burdette L, Yeager M, Chanock SJ, Berndt SI. DNA repair gene polymorphisms and tobacco smoking in the risk for colorectal adenomas. Carcinogenesis. 2011;32:882–887. doi: 10.1093/carcin/bgr071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Memisoglu A, Samson L. Base excision repair in yeast and mammals. Mutation research. 2000;451:39–51. doi: 10.1016/s0027-5107(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 33.Tran PT, Erdeniz N, Symington LS, Liskay RM. EXO1-A multi-tasking eukaryotic nuclease. DNA repair. 2004;3:1549–1559. doi: 10.1016/j.dnarep.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 34.Orans J, McSweeney EA, Iyer RR, Hast MA, Hellinga HW, Modrich P, Beese LS. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145:212–223. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunter N. Cell cycle. Vol. 10. Georgetown, Tex: 2011. Double duty for Exo1 during meiotic recombination. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 36.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaetzlein S, Kodandaramireddy NR, Ju Z, Lechel A, Stepczynska A, Lilli DR, Clark AB, Rudolph C, Kuhnel F, Wei K, Schlegelberger B, Schirmacher P, Kunkel TA, Greenberg RA, Edelmann W, Rudolph KL. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell. 2007;130:863–877. doi: 10.1016/j.cell.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vallur AC, Maizels N. Distinct activities of exonuclease 1 and flap endonuclease 1 at telomeric g4 DNA. PloS one. 2010;5:e8908. doi: 10.1371/journal.pone.0008908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sertic S, Pizzi S, Cloney R, Lehmann AR, Marini F, Plevani P, Muzi-Falconi M. Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proceedings of the National Academy of Sciences of the United States of America. 2011 doi: 10.1073/pnas.1108547108. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolderson E, Richard DJ, Edelmann W, Khanna KK. Involvement of Exo1b in DNA damage-induced apoptosis. Nucleic acids research. 2009;37:3452–3463. doi: 10.1093/nar/gkp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.