Abstract
Macrophages store excess unesterified cholesterol (free, FC) in the form of cholesteryl ester (CE) in cytoplasmic lipid droplets. The hydrolysis of droplet-CE in peripheral foam cells is critical to HDL-promoted reverse cholesterol transport because it represents the first step in cellular cholesterol clearance, as only FC is effluxed from cells to HDL. Cytoplasmic lipid droplets move within the cell utilizing the cytoskeletal network, but, little is known about the influence of the cytoskeleton on lipid droplet formation. To understand this role we employed cytochalasin D (cyt.D) to promote actin depolymerization in J774 macrophages. Incubating J774 with acetylated LDL creates foam cells having a 4-fold increase in cellular cholesterol content (30-40% cholesterol present as cholesteryl ester (CE)) in cytoplasmic droplets. Lipid droplets formed in the presence of cyt.D are smaller in diameter. CE-deposition and -hydrolysis are decreased when cells are cholesterol-enriched in the presence of cyt.D or latrunculin A, another cytoskeleton disrupting agent. However, when lipid droplets formed in the presence of cyt.D are isolated and incubated with an exogenous CE hydrolase, the CE is more rapidly metabolized compared to droplets from control cells. This is apparently due to the smaller size and altered lipid composition of the droplets formed in the presence of cyt.D. Cytoskeletal proteins found on CE droplets influence droplet lipid composition and maturation in model foam cells. In J774 macrophages, cytoskeletal proteins are apparently involved in facilitating the interaction of lipid droplets and a cytosolic neutral CE hydrolase and may play a role in foam cell formation.
Supplementary key words: lipid droplet, cytoskeleton, cholesteryl ester, foam cells, hydrolysis
1. Introduction
Macrophage-derived foam cells, both in culture and in vivo, have a large fraction of accumulated excess cholesterol stored as CE in cytoplasmic lipid droplets. It is generally thought that the rate limiting step of cellular cholesterol clearance is the hydrolysis of lipid droplet-CE to FC, a form of cholesterol which can be exported from the cell. Therefore, when viewed in the context of reverse cholesterol transport (RCT), the first step of RCT may not be the efflux step but rather hydrolysis of stored CE followed by transport of the generated FC to the plasma membrane, where it can be removed by an extracellular acceptor such as HDL or HDL apoprotein through various pathways, as studied extensively by John Oram and colleagues 1-4. Thus CE metabolism at the site of the lipid droplet is integral to RCT.
Macrophages, like many other cells, convert excess cholesterol (free cholesterol, FC) into cholesteryl ester (CE) through the actions of acyl coenzyme A:cholesterol acyl transferase (ACAT) and store the CE in cytoplasmic lipid droplets. These lipid droplets give the classic “foamy” appearance to atherosclerotic macrophage foam cells. Macrophage lipid droplets were once thought of as the cell's lipid storage locker, floating in the cellular milieu as the site where FC is converted to CE and back to FC establishing a CE cycle5. Since the discovery of the PAT (Perilipin (perilipin 1), Adipophilin (perilipin 2), and TIP47 (perilipin 3)) family of lipid droplet proteins in the 1990s6-7, research on lipid droplet composition and function, part in adipocytes and steroidogenic cells, has dramatically increased. We now know that cytoplasmic lipid droplets are intimately involved in cellular processes including lipid and endosomal trafficking (for reviews see 8-9).
Cytoplasmic CE lipid droplets are thought to originate in the endoplasmic reticulum (ER)10-12, the cellular location of ACAT13, through accumulation of CE in the ER bilayer, forming a lens, which eventually buds off from the ER. Lipid droplets then travel on the cell's microtubule skeleton12, 14. Proteomic studies have indicated that proteins associated with stability (perilipin, ADRP, TIP47) are present on lipid droplets and identified cytoskeletal proteins (actin, tubulin, vimentin) as candidate droplet proteins in many cells types15-18. The cytoskeleton has been implicated in the initiation and progression of atherosclerosis through its involvement in endothelial cell-promoted monocyte recruitment 19-20 and uptake of oxidized LDL, the probable lipid source for macrophage foam cell formation21-22. In fact, recent work from Oram and Heinecke's laboratories implicates cytoskeletal proteins as part of a sterol-responsive network in macrophages23. They found that cytoskeletal proteins are significantly up-regulated in response to cholesterol deposition in macrophages23.
Tabas et al. demonstrated that disrupting the cytoskeleton in macrophages reduces cholesterol esterification24. Although cytoskeletal proteins are proposed to be present on lipid droplets, it is presently unclear if the cytoskeleton is involved in the formation, metabolism or stability of these organelles. We therefore examined the effects of cytoskeletal disruption on macrophage CE droplet formation and metabolism.
2. Materials and Methods
2.1 Materials
BSA (essentially fatty acid free), heat-inactivated fetal bovine serum (FBS), gentamicin, cytochalasin D, FITC conjugated anti-actin, cholesteryl methyl ether, and FC were purchased from Sigma-Aldrich (St. Louis, MO). [1,2-3H]cholesterol was obtained from New England Nuclear (Waltham, MA). Organic solvents were obtained from Fisher Scientific (Pittsburgh, PA). Tissue-culture flasks and plates were from Corning (Corning, NY). Tissue culture medium was obtained from Gibco-Invitrogen (Carlsbad, CA). Human LDL (1.019<d<1.063 g/ml) and HDL3 (1.125<d<1.21 g/ml) were isolated by sequential ultracentrifugation, dialyzed against 0.15 mol/L NaCl, and sterilized by 0.45 μm filtration. LDL was acetylated with acetic anhydride25. Alexa Fluor 594 phalloidin was purchased from Invitrogen-Molecular Probes (Carlsbad, CA). Latrunculin A was purchased from Cayman Chemicals (Ann Arbor, MI).
2.2 Cell Culture
J774 murine macrophages were routinely grown in RPMI containing 10% FBS and 50 μg/ml gentamicin. To enrich the macrophages with cholesterol (to create foam cells), RPMI containing 1% FBS, 1μCi/ml [3H]cholesterol, acetylated LDL (acLDL, 100μg protein/ml), +/- cytochalasin D (cyt.D, 2μM), +/- latrunculin A (250nM) was added to the cells for 24h. After the enrichment period, the cells were incubated in medium containing 0.2%BSA for 18h to allow the cellular pools of [3H]sterols to equilibrate. In some incubations, cyt.D was added during the equilibration phase. At 2μM cyt.D we did not detect cellular toxicity (measured by cellular lactate dehydrogenase release, Roche kit, Basel, Switzerland, data not shown).
2.3 CE Droplet Isolation
J774 macrophage cells were cholesterol-enriched by incubation with acLDL and 1%FBS for 24h. Following this incubation the cells were incubated for 18h in 0.2%BSA to allow the cellular pools of sterols to equilibrate. There was a 4-5 fold increase in cellular cholesterol with 30-40% of the cholesterol present as cholesteryl ester (CE). Cholesterol enrichment using this protocol results in the formation of neutral lipid droplets in the cytoplasm of the cell. To isolate cellular lipid inclusions, 10- 100mm dishes of macrophages containing inclusions were washed three times with PBS and scraped into 3ml PBS (3ml per 100mm dish) containing 20μl/ml protease inhibitor cocktail (Sigma-Aldrich, St. Louis MO). The cells were disrupted using sonication26. Homogenization by this procedure reproducibly produced lipid inclusions18, 26-30. The homogenates were centrifuged at 26,000rpm in an SW40Ti rotor for 30min. The floating lipid layer was removed with a syringe, dispersed in 100mM Na2CO3 (pH=11) and spun again. This treatment stripped away any loosely associated proteins on the droplet31. The floating droplet layer was removed with a syringe, dispersed in PBS and spun again to wash. The final lipid layer containing the droplets was removed and stored on ice until use. Acid phosphatase (Sigma-Aldrich kit, St. Louis, MO) and lactate dehydrogenase activity (Roche kit, Basel, Switzerland) were determined in the floating lipid layer to determine if there was lysosomal or cytoplasmic contamination in the lipid fraction.
2.4 Droplet Protein Analysis
Droplets were extracted using the method of Bligh and Dyer32. The interface containing droplet protein which formed between the organic and aqueous layers of the extraction was collected. The aqueous layer was discarded and the organic layer containing droplet lipid was dried under nitrogen and stored at 0°C for later analysis. The proteins were dissolved in an SDS sample buffer containing β-mercaptoethanol as a reducing agent and the proteins were separated on a 3-8% tris-acetate gel. Proteins were visualized using Coomassie brilliant blue and submitted to the Children's Hospital of Philadelphia's Protein Core Facility. The entire lane was cut into 1mm sections. Gel pieces were digested with trypsin (Promega; 12.5 ng/μl) at pH 8.0 at 37°C for 18–24 hr, reactions were stopped by addition of 5% trifluororacetic acid. The peptide samples were spotted on a MALDI plate and then overlayed with α-cyano-4-hydroxycinnamic acid matrix. MALDI-TOF Tandem MS spectra were acquired on a Finnegan LTQ mass spectrometer equipped with a vMALDI ion source. Bioworks 3.2 employing the SEQUEST algorithm and the SWISSPROT database was used for protein identification. Samples were analyzed using Sequest (ThermoFinnigan, San Jose, CA; version 27, rev. 12). Sequest was set up to search the mouse database assuming the digestion enzyme trypsin. Sequest was searched with a fragment ion mass tolerance of 1.00 Da and a parent ion tolerance of 1.5 Da. Scaffold (version Scaffold-01_06_06, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability. Protein identifications were accepted if they could be established at greater than 99.9% probability and contained at least 2 identified peptides.
2.5 Protein and lipid determination
Lipids were extracted from cell monolayers using isopropanol or from isolated lipid droplets by the method of Bligh and Dyer. Cholesteryl methyl ether was used as an internal standard. Total and unesterified cholesterol was quantitated by GLC33. Total phospholipid were measured by the method of Rouser et al.34 Protein was measured by the method of Markwell et al.35 Triglyceride (TG) was determined using a commercially available kit (Sigma-Aldrich, St. Louis, MO).
2.6 Cholesterol Efflux to HDL
After [3H]cholesterol loading and labeling the cells, as described above, media containing HDL (20μg protein/ml) was added for 2 h. To determine cholesterol efflux, media were sampled at indicated times, filtered and counted by liquid scintillation counting to determine [3H] released. [3H]-Sterols in the media were compared to total [3H] at time zero to determine the percent release of [3H]free-cholesterol.
2.7 Western blot analysis
The proteins were dissolved in an SDS sample buffer containing the NuPage reducing agent (Invitrogen, Carlsbad, CA). Proteins were separated on a 3-8% tris-acetate gel and transferred to PVDF membranes and probed for actin as per the manufacturer's directions. Resulting bands were quantitated using ImageJ software. Anti-actin was purchased from Novus Biologicals (Littleton, CO) and was used at a dilution of 1:10,000 as per the manufacturer's instructions.
2.8 Determination of lipid droplet diameter
The distribution of lipid droplet diameters within cells was determined by transmission electron microscopy. Cells were fixed in 2.5% glutaraldehyde in 0.1M cacodylate buffer. Following fixation the cells were washed, postfixed in 1% osmium tetroxide in 0.1M cacodylate buffer, dehydrated through a graded series of ethanol solutions and embedded in Spurr Resin (EMS, Hatfield PA). Thin sections were cut and viewed at an accelerating voltage of 80 kiloelectron volts with an FEI/Philips CM12 transmission electron microscope (Hillsboro, OR). Images at a magnification of 11,700 × were collected and a grid of points was superimposed over the images. The points were used to select cross sections of droplets to measure. This point-restricted selection scheme insured an unbiased selection of droplets for measurement. The diameter along the longest axis was determined for each droplet. One hundred droplets were measured for each condition. Most droplet cross sections were close to round, having a ratio of diameters measured at right angles to each other of 0.99 (a ratio of 1 indicates a perfect circle).
2.9 Cholesteryl ester and triacylglycerol hydrolysis
Intact cells. Macrophages were enriched with 3H-CE for 24h by incubation with acLDL (100μg/ml) containing [3H]-FC (1μCi/ml, for CE hydrolysis) or [3H]oleic acid (5μCi/ml, for TG hydrolysis) +/- cyt.D (2μM), +/- latrunculin A (250nM). Cells were then incubated in 0.2% BSA +/- cyt.D (2μM) for 18h to allow for the equilibration of cellular pools of [3H]sterols before the addition of extracellular acceptors. After equilibration, cells contained from 30% to 40% of the [3H]cholesterol as CE. The acceptors (see figure legends) were added for 24h in media containing the ACAT inhibitor CP113818 (2μg/ml) +/- cyt.D (2μM). At the end of this incubation, wells were washed with PBS, allowed to air dry and extracted overnight with isopropanol. Lipids in the extracts were separated by thin layer chromatography (TLC) to determine the loss of [3H] from the CE or TG pools29 Isolated Droplets. The lipid droplets were added, normalized to CE mass, to an aliquot of cell homogenate from cholesterol-normal J774 (as a source of nCEH) in MEM-HEPES (pH=7.4). Hydrolysis was allowed to occur for up to 2h. The reactions were stopped by extraction with chloroform and methanol (1:1, v/v). Hydrolysis was calculated as actual loss of CPM from the cellular CE pool/mg cell protein over the indicated time period.
2.10 Fluorescent Imaging
J774 macrophage cells were seeded on cover slips and grown in growth media containing 10% FBS for 24h. The cells were then incubated in RPMI media containing 1% FBS, acLDL (100μg/ml), and +/- cyt D (2μM) for 18h. Cells were then equilibrated in BSA +/- cyt.D (2μM) for 18h. The cells to be stained with Alexa fluor 594 phalloidin were fixed in 3.7% formaldehyde (in PBS) for 10min at room temperature. The monolayers were washed with PBS and then extracted with ice-cold acetone for 5min. The coverslips were then washed with PBS and blocked overnight at 4°C in 1%BSA. The coverslips were then stained with 20nM Alexa fluor 594 phalloidin for 20min at room temperature and washed with PBS before mounting. The slides were read on a Zeiss Meta 510 laser scanning confocal and multiphoton microscope. Representative fields were chosen for the figure.
2.11 Data analysis
Data are from representative experiments and are expressed as mean +/- standard deviation. Statistical test for significance was performed using an unpaired t test with a 95% confidence interval. GraphPad Prism software (version 4, GraphPad Software, Inc, San Diego,CA) was used to analyze CE hydrolysis kinetics.
3. Results
3.1 Candidate cytoskeletal proteins associated with the lipid droplets
To better understand lipid droplet metabolism in macrophages (J774 cells), we analyzed the protein complement of the lipid droplet using a proteomic approach. Lipid droplets were isolated from homogenized cells by centrifugation as described in Methods section 2.3. The lipid droplets were recovered from the top of the gradient and repeatedly washed to remove potential contaminants. There was no detectable acid phosphatase (lysosomal) or lactate dehydrogenase (cytoplasm) activity in the droplet isolate (data not shown). The droplet-associated proteins were dissolved in SDS and separated by PAGE. The entire lane containing the lipid droplet proteins was cut into 1mm bands, digested with trypsin and subject to analysis by mass spectrometry. Resulting peptides were matched against a database for identification as described in Methods section 2.4. We identified a striking number of cytoskeletal proteins in our analysis (Table I). Many of these proteins have been identified as candidate droplet proteins in other cell types by other groups17-18, 36-37. Additionally, figure 1 indicates that actin protein, detected by Western Blot, is enriched in the purified lipid droplet fraction compared to the whole cell homogenate (WCH), cell pellet and the cytosolic fraction. The role of the actin cytoskeleton on macrophage lipid droplet formation and CE hydrolysis is not well-defined. We therefore employed a strategy of disrupting the cytoskeleton with chemical agents and subsequently measuring the effect of an impaired cytoskeletal network on CE droplet formation and lipid hydrolysis.
Table 1.
Cytoskeletal Proteins identified on J774 Neutral Lipid Droplets. Protein isolated from J774 macrophage lipid droplets were subject to electrophoresis and identification by mass spectrometry as described in Methods section 2.4. The cytoskeletal proteins identified with a confidence level greater than 95% are listed.
Fig. 1.
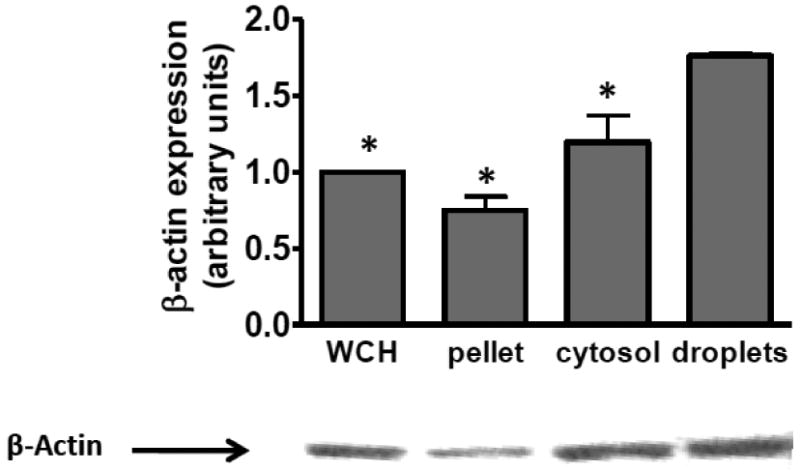
Actin Protein is Enriched in the Lipid Droplet Fraction. Western blot analysis of actin content in whole cell homogenate (WCH), the cell pellet after fractionation, cytosolic fraction and lipid droplet fraction. 15μg total protein from each fraction was applied to each lane. Samples were run on SDS-PAGE and immunoblotted with anti-actin antibody as per the manufacturers directions. B-Actin levels are reported as the mean of 3 independent experiments. A blot from a single representative experiment is shown. *Significantly different from droplet actin level, p≤0.05.
3.2 Cellular Lipid Composition
We used cytochalasin D (cyt.D) to disrupt the actin filaments of the cytoskeleton in J774 macrophage foam cells. 2μM Cyt.D effectively disrupted the actin cytoskeletal network in J774 cells (fig 2). The fluorescent staining pattern seen in fig 2A (control cells) indicates even actin staining along the cell membrane and the presence of actin filaments (fig 2A'). In contrast, the staining pattern seen in fig 2C (cells treated with cyt.D) indicates the presence of actin, however, it is clustered in and around the membrane. Additionally, actin filaments are not evident (fig 2C'). As seen in figure 2, the macrophages treated with cyt.D appear larger, when viewed microscopically, than the untreated control cells (compare figs 2B and 2D). To determine if cyt.D treatment influences the uptake of exogenous cholesterol by macrophages, J774 macrophages were incubated in medium containing acLDL, 1% FBS +/- cyt.D (2μM) for 24h. The cells were then equilibrated in 0.2%BSA +/- cyt.D overnight. Lipids were extracted and analyzed for FC and CE content as described in Methods section 2.5. Cyt.D treatment caused a modest decrease in the amount of total cholesterol accumulated by the macrophages compared to cells cholesterol-enriched in the absence of cyt.D (fig. 3A). However, the cells treated with cyt.D still had a 2.6-fold increase in total cholesterol content compared to cholesterol-normal cells, with 30±1.8% of the total cholesterol present in the cells as CE (compared to 38±1.5% CE in the untreated cells). Additionally, efflux of [3H]free-cholesterol to HDL3 was enhanced with cyt.D treatment (fig 3B). Thus, cells treated with cyt.D can accumulate FC and esterify excess cholesterol, although to a lesser extent than the untreated control cells as well as release cholesterol to extracellular acceptors such as HDL3. We next wanted to determine if cyt.D treatment altered the composition of the neutral lipid droplets in the cells. Therefore, we measured the size of lipid droplets (as described in Methods section 2.3) from J774 cells cholesterol-enriched in the presence or absence of cyt.D. The droplets in cells treated with cyt.D were smaller in diameter (0.49 ±0.17μm in the absence of cyt.D vs. 0.42±0.15μm in the presence of cyt.D, p=0.002) as determined by transmission electron microscopy (described in Methods section 2.8). Isolated droplet fractions were analyzed for total protein, triglyceride (TG), cholesterol, CE and phospholipid (PL) as described in Methods section 2.5. Compared to untreated control cells, the droplets isolated from cells treated with cyt.D were depleted in CE and enriched in FC, PL and TG (Table II). The cyt.D induced reduction in CE content in the droplets was in agreement with the reduction in CE mass seen in whole cells (compare Table II and fig. 3A). The FC content of the two types of droplets was not statistically different. Interestingly we found that cellular 3H-oleic acid uptake (minus cyt.D: 383071±10200 cpm/mg cell protein; plus cyt.D: 361549±30193 cpm/mg cell protein; n=6) and incorporation of 3H-oleic into TG (minus cyt.D: 347±151 cpm/mg cell protein; plus cyt.D: 245±26 cpm/mg cell protein; n=6) was not altered with cyt.D treatment. However, in agreement with previous studies we measured a significant reduction in incorporation of 3H-oleic into CE (minus cyt.D: 6721±454 cpm/mg cell protein; plus cyt.D: 939±108 cpm/mg cell protein; n=6, p<0.0001)24.
Fig. 2.
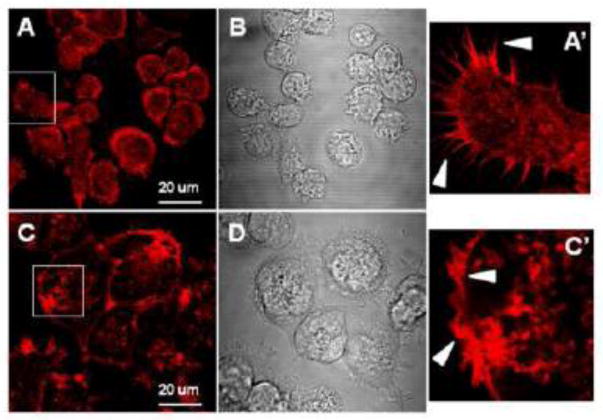
Effect of cyt.D on fluorescent labeling of actin filaments. Cells were seeded on coverslips as described in Methods section 2.10 and then incubated with acLDL (100μg/ml) without (figs 1A and 1B) or with (figs 1C and 1D) cyt.D (2μM) for 24h. After formaldehyde fixation, cells were processed for fluorescent labeling using phalloidin coupled to Alexa Fluor-594 nm for visualization of F-actin (1A and 1C). Phase micrographs are also shown (1B and 1D). Representative fields were chosen for the figure. Bar =20μm. Figures 1A' and 1C' are enlargements of the areas indicated by the white box in figures 1A and 1C respectively. Arrow heads 1A' point out the diffuse actin staining pattern and distinct actin staining of psuedopods in an untreated J774 macrophage. Arrow heads in 2A' indicate the coarse, coagulated actin staining pattern and lack of psuedopods in a cyt.D treated J774 macrophage.
Fig. 3.
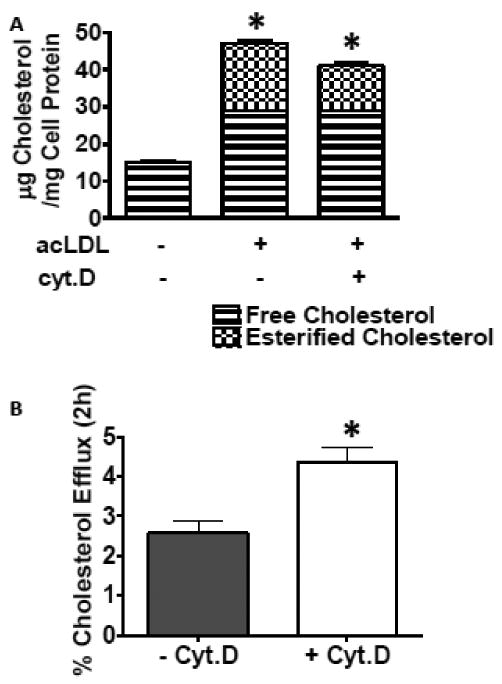
A. Effect of disrupting the cytoskeleton on cellular cholesterol deposition and efflux to HDL3. J774 macrophage foam cells were enriched with cholesterol by incubation with 100μg/ml acLDL +/- cyt.D (2μM) for 24h. Cells were incubated in 0.2%BSA +/- cyt.D (2μM) for 18h to allow equilibration of the cellular pools of sterols. Fig 3A. Monolayers were analyzed for lipid and protein content as described in Methods section 2.5. Cell cholesterol content before the enrichment period was 15+/-1.8μg cholesterol/mg cell protein and there was no detectable CE present. Results are from a representative experiment run in triplicate. Experiment was repeated four times. *Indicates cholesteryl ester content significantly different, p=0.023. Fig. 3B After the equilibration period, the cells were exposed to medium containing HDL3 (20μg/ml) for 2h. Total [3H]sterol in the medium was determined and percent efflux was calculated as CPM appearing in the medium at 2h compared to total CPM in the cells before the efflux period. Results data combined from two independent experiments run in triplicate, n=6, p=0.005.
Table 2.
Lipid content of isolated lipid droplets. J774 macrophage cells were incubated with acLDL in the presence or absence of cyt.D (2μM) for 24h. The lipid droplets formed during this incubation period were isolated and the lipid content analyzed as described in Methods sections 2.3 and 2.5. Data are single determinations from 6 preparations of droplets.
| Lipid | - cyt. D | + cyt. D | n | % of | p value |
|---|---|---|---|---|---|
| (μg lipid/μg droplet protein) | - cyt.D | ||||
| FC | 0.55 ± 0.10 | 0.85 ± 0.15 | 6 | 154% | 0.0009 |
| CE | 14.78 ± 3.57 | 8.54 ± 0.75 | 6 | 58% | 0.002 |
| PL | 1.32 ± 0.23 | 1.90 ± 0.50 | 6 | 143% | 0.03 |
| TG | 0.28 ± 0.15 | 2.14 ± 0.71 | 6 | 764% | <0.0001 |
3.3 Cellular Lipid Metabolism
Having determined that cytoskeleton disruption alters the size and composition of neutral lipid droplets in macrophage foam cells, we next focused on the effects of cytoskeleton disruption on cellular CE metabolism. J774 cells were enriched with cholesterol by incubation with acLDL in the presence or absence of cyt.D as described above. After an incubation period to allow the cellular pools of sterols to equilibrate, media containing HDL3 (to act as a cholesterol acceptor) +/- cyt.D (2μM) and an ACAT inhibitor, to eliminate cholesterol re-esterification, were added to the cells for 24h. Hydrolysis was calculated by measuring the loss of radioactivity from the pool of cellular CE. The data in figure 4A indicates that CE hydrolysis is significantly reduced in the presence of cyt.D compared to untreated control cells. This point is further demonstrated by the kinetic plots in figure 4B. The half times of hydrolysis were 15h for −cyt.D-treated cells and 66h for +cyt.D-treated cells (fig 4B). To determine if the suppressed hydrolysis was due to off-target effects of cyt.D, we first cholesterol-enriched J774 cells with acLDL producing cells having equivalent lipid droplets. Cells with pre-formed droplets were then treated +/- cyt.D. Figure 4C indicates that if the neutral lipid droplets are formed before exposure to cyt.D, cytoskeleton disruption does not affect the rate of CE hydrolysis, in contrast to droplets formed in the presence of cyt.D (fig 4B). Additionally we found cyt.D treatment decreased TG hydrolysis (fig 4D) to a level similar to the decrease measured in CE hydrolysis (compare figs 4A and 4D).
Fig. 4.
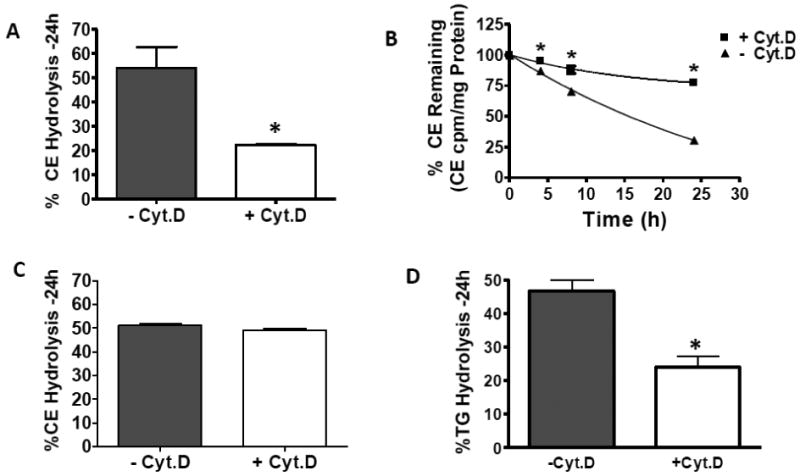
The effect of cytochalasin D treatment on CE hydrolysis in intact cells during (panels A, B and D) or subsequent to (panel C) cholesterol enrichment has on cholesteryl ester and triacylglycerol hydrolysis in intact cells. Panels A, B and D. J774 macrophage foam cells were cholesterol-enriched in the presence or absence of cyt.D as described in Methods section 2.2. Media containing the ACAT inhibitor, CP113-818 (2μg/ml) +/- cyt.D (2μM) was then added to the cells for up to 24h. All media contained 50μg/ml HDL (to prevent the build-up of cytotoxic-cellular FC). Panel C. J774 macrophage foam cells were cholesterol-enriched by incubation with acLDL in the absence of cyt.D for 24h. After this incubation, media containing 50μg/ml HDL plus the ACAT inhibitor, CP113-818 (2μg/ml), +/- cyt.D (2μM) was added to the cells for 24h. Percent hydrolysis was calculated by comparing total radioactivity in cellular CE or TG before the hydrolysis period (normalized to cellular protein) to total radioactivity in cellular CE or TG after the hydrolysis period (normalized to cellular protein). Data are combined from two independent experiments, each run in triplicate. *Indicates significance, p≤0.05.
We next wanted to determine if the neutral lipid droplets formed in the presence of cyt.D were a less efficient substrate for nCEH which would explain the decrease in CE hydrolysis in these cells. We therefore isolated the neutral lipid droplets (as described in Methods section 2.3) from J774 macrophages cholesterol-enriched in the presence or absence of cyt.D. The isolated droplets were added back to the endogenous J774 nCEH (HSL (hormone sensitive lipase) present in the whole cell homogenate, fig. 5) at equivalent CE mass. The data in Figure 5 demonstrates that the droplets formed in the presence of cyt.D are a better substrate for the nCEH compared to the droplets from the untreated control cells.
Fig. 5.
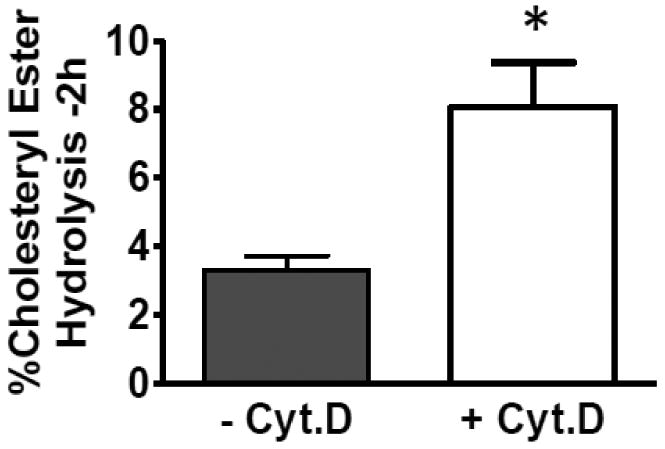
Cholesteryl ester hydrolysis in lipid droplets isolated from J774 macrophages loaded +/- Cyt.D. J774 macrophages were cholesterol enriched as described in figure 2. The monolayers were then washed and neutral lipid droplets were isolated as described in Methods section 2.3. The isolated lipid droplets were then added back at the same mass of CE to a J774 cell homogenate in MEM-HEPES (pH=7.4). Hydrolysis was allowed to occur for 2h at 37°C. The reactions were stopped by extraction with chloroform and methanol (1:1, v/v). The extent of CE hydrolysis was measured as described in figure 4. Results are from a representative experiment run in triplicate. Experiments were repeated three times. * Indicates significance, p≤0.05.
Cyt.D blocks actin polymerization in a reversible fashion38. We therefore, determined if withdrawing cyt.D from our experimental system would result in a restoration of CE hydrolysis. J774 macrophage cells were cholesterol enriched with acLDL in the presence or absence of cyt.D for 24h (fig 6A). The cyt.D was then withdrawn from one set of cells for the subsequent equilibration and hydrolysis periods, allowing sufficient time for the actin filaments to re-polymerize39-40. Since there was no extracellular cholesterol acceptor present during this time, CE in the existing droplets was hydrolyzed and re-esterified resulting in the turnover of the cellular CE pool. For the case where cyt.D was withdrawn, CE hydrolysis returned to a level that was not significantly different from the −cyt.D cells, whereas the cells that were continuously exposed to cyt.D had significantly lower CE hydrolysis (fig 6A).
Fig. 6.

Panel A. Effect of withdrawing cyt.D on CE hydrolysis. J774 macrophage foam cells were enriched with cholesterol by incubation with 100μg/ml acLDL (-cyt.D) or 100μg/ml acLDL plus 2μM cyt.D (+cyt.D and cyt.D withdrawn) for 24h. Cells were equilibrated in 0.2%BSA (-cyt.D and cytD. withdrawn) or 0.2%BSA plus 2μM cyt.D (+cyt.D) for 18h. After this incubation, media containing 50μg/ml HDL plus the ACAT inhibitor, 2μg/ml CP113-818 (-cyt.D and cyt.D withdrawn) or 50μg/ml HDL (to prevent the build-up of cytotoxic-cellular FC) plus the ACAT inhibitor, 2μg/ml CP113-818 plus 2μM cyt.D (+cyt.D) was added to the cells for 24h. Unesterified cholesterol and CE were separated and quantitated as described in Methods section 2.5. Panel B Effect of Latrunculin A on CE hydrolysis. J774 macrophage foam cells were enriched with cholesterol by incubation with 100μg/ml acLDL +/- cyt.D (2μM) or +/- latrunculin A (250nM) for 24h. Cells were equilibrated in 0.2%BSA +/- cyt.D (2μM) or +/- latrunculin A (250nM) for 18h. After this incubation, 50μg/ml HDL plus the ACAT inhibitor, CP113-818 (2μg/ml) +/- cyt.D (2μM) or +/- latrunculin A (250nM) was added for 24h. Percent hydrolysis was calculated by comparing total radioactivity in cellular CE before the hydrolysis period to total radioactivity in cellular CE after the hydrolysis period. Panel A * significantly different from −cyt.D, p=0.03, ** significantly different from +cyt.D, p=0.03. Panel B * significantly different from −cyt.D, p<0.0001.
To ensure that the effects on CE hydrolysis were not unique to cyt.D but rather due to actin disruption, we employed another cytoskeleton disrupting agent, latrunculin A. Cyt.D and latrunculin A inhibit actin polymerization but by different mechanisms38. As seen in figure 6B, hydrolysis was reduced in CE droplets formed in the presence of cyt.D and latrunculin A compared to droplets in untreated control cells.
4. Discussion
Research on triglyceride lipid droplets in adipoctyes has dramatically increased since the discovery and characterization of the PAT family of lipid droplet proteins whereas studies on cholesteryl ester lipid droplets formed in macrophages have lagged behind. The present studies and others have identified candidate cytoskeletal proteins associated with neutral lipid droplets in various cell types17-18, 36-37. Previous studies have described microtubule-dependent fusion of triacylglyceride droplets as a mechanism of droplet growth in adipocytes and cell-free systems41-42. We and others have shown that droplet growth in macrophages and fibroblasts can occur by molecular addition of lipid to pre-existing cholesteryl ester droplets26, 43. It is known that Cytoplasmic lipid droplets move within the cell by utilizing the cell's cytoskeletal network14, 44, however, little is known about the role of the actin cytoskeleton in cholesteryl ester droplet formation and metabolism in macrophages. In an effort to understand this role we employed cyt.D. Cyt.D interferes with the cytoskeleton by promoting the specific depolymerization of actin filaments45.
Although J774 macrophages treated with cyt.D accumulated less cholesterol when exposed to acLDL, the incorporated exogenous FC was esterified and deposited in cytoplasmic lipid droplets. Our results agree with those of Tabas et al24 who found that cyt.D treatment of mouse peritoneal macrophages resulted in reduced cholesterol esterification when the cells were incubated with β-VLDL or acLDL. Importantly they found that the reduction in cholesterol esterification was not linked to availability of cholesterol substrate or reduction in lysosomal processing of lipoproteins24. Under our conditions, the droplets formed in the presence of cyt.D were smaller in diameter compared to untreated controls. Initially we assumed that the smaller lipid droplets generated in the presence of cyt.D were due to cyt.D-induced decrease in cellular CE (42% decrease, Table 2) as we, and Tabas et al.24 observed. However, we also measured a dramatic increase in cellular TG content (764% increase, Table 2) in the presence of cyt.D. CE and TG are co-deposited in lipid droplets in cells26, 29 and TG-rich droplets are typically larger than CE-rich droplets29, 46 however we do not see an increase in size of the lipid droplets with cytD. Treatment. The data suggest that the cytoskeleton may be involved in the budding-off process or lipid droplet maturation. Analysis of the droplets isolated from cyt.D-treated cells revealed a reduction in the CE:protein ratio and an enrichment in the FC:protein, TG:protein and PL:protein ratios when compared to droplets isolated from untreated cells. It is unlikely that the difference in droplet lipid composition when the actin cytoskeleton is disrupted is due to inhibition at the site of lipid synthesis as the enzymes responsible for cholesterol esterification and triacylglycerol production, ACAT and acyl CoA:diacylglycerol acyltransferase (DGAT), respectively, are thought to reside at the same cellular location, the ER47-49 and we would expect to measure a decrease in both the CE:- and TG:droplet protein ratios. Additionally it is unlikely that the increase in TG and decrease in CE in the droplets isolated from cyt.D treated cells is due to changes in lipid degradation as we measured a similar inhibition in both TG and CE hydrolysis (fig. 4). In agreement with previous studies24, we found cyt.D reduced the accumulation of cholesterol (fig. 3A) but did not alter cellular uptake of oleic acid (Results page 14). This cyt.D-induced differential uptake of exogenous lipid may account for the relative increase in the TG: droplet protein ratio and decrease in CE: droplet protein ratio. Our studies show that cyt.D treatment does not result in decreased cellular oleic acid uptake so fatty acid is not limiting for synthesis of CE or TG. The increase in droplet TG in cyt.D treated cells is likely due to the observed decrease in TG hydrolysis and unaffected synthesis rate, whereas the decrease in CE content is due to the reduction in uptake of cholesterol from lipoproteins24.
Efflux is the process of removal of excess cellular cholesterol to an extracellular acceptor such as HDL. Cyt.D treatment does not interfere with this process but, in fact, increases FC efflux to HDL (fig 3B) perhaps by causing the reorganization of plasma membrane lipids to create a pool of FC that is readily available for efflux to HDL. This data is consistent with previous studies demonstrating that cyt.D does not inhibit intracellular free-cholesterol transport and causes an increase in efflux to apoA-I50-52. Taken together these data suggest that an intact cytoskeleton is not necessary for a droplet to form, but may be needed for droplet maturation and enlargement, and may impact on cholesterol removal from the cell.
Our data also indicate that, in intact cells, droplets formed in the presence of cyt.D are not readily hydrolyzed by a neutral cholesteryl ester hydrolase (nCEH). The predominant nCEH in J774 macrophages in hormone sensitive lipase (HSL), although in other macrophage types, the identity and nature of nCEH(s) is highly controversial53-57. For the purpose of our discussion, nCEH refers to the hydrolysis of CE in J774 macrophages with the assumption that the majority of the hydrolytic activity is due to the actions of HSL. We ruled out any off target effects of cyt.D by adding cyt.D to cells with pre-existing lipid droplets and found no cyt.D effect on CE hydrolysis. We conclude that cyt.D does not interfere with nCEH reaching or acting upon the cytoplasmic lipid droplets. We also employed another cytoskeletal disrupting agent, latrunculin A, which has a different mechanism of action. Cyt.D binds to the barbed end of the actin filament whereas latrunculin A complexes with actin monomers38. Latrunculin A also reduced CE hydrolysis when present during cholesterol enrichment indicating that the effects on CE metabolism described in these studies are not unique to cyt.D.
To further investigate the effect of cytoskeleton disruption on CE hydrolysis, we isolated CE droplets formed in the presence or absence of cyt.D and added the droplets back to an endogenous nCEH (HSL) in the form of a J774 whole cell homogenate. In direct contrast to our studies in whole cells where cyt.D treatment reduced CE hydrolysis, we found that CE hydrolysis was greater in the isolated droplets formed in the presence of cyt.D compared to droplets isolated from untreated cells indicating that the droplets formed in the presence of cyt.D were a better nCEH substrate. Our findings reveal a number of physical properties of the droplets formed in the presence of cyt.D that would make them a better substrate for nCEH. First, the smaller size of the droplets formed in the presence of cyt.D presents a larger surface area for the nCEH to act upon (many small droplets as opposed to fewer larger droplets for a given CE concentration). Thus, using the average droplet diameter we calculated the mean volume of the lipid droplets assuming them to be spheres. Taking this number and assuming that the density of the lipid droplet is approximately 1μg/μl, we estimated the number of isolated droplets added to the exogenous nCEH to be 40×1012 for the droplets obtained from −cyt.D cells and 141×1012 for the droplets obtained from +cyt.D cells. Therefore, we estimate that there are over three times more droplets in the cells treated with cyt.D for a given CE mass. Since the +cyt.D. treated cells have roughly 2/3 the CE content compared to control cells (fig 3), there are about two times more droplets in the +cyt.D. cells. The total droplet surface area in these incubations is 30×1012 μm2 for droplets from −cyt.D cells and 78×1012 μm2 for droplets from + cyt.D cells. Thus there is roughly 2.5 times more droplet surface area in the + cyt.D incubations compared to the droplets isolated from the − cyt.D cells. Secondly, the + cyt.D droplets contained significantly more TG and this may also affect CE hydrolysis. We have previously shown that the addition of TG to CE droplets increases the rate of CE hydrolysis by altering the physical state of the CE in the droplets29, 46. Thirdly, the droplets formed when the cytoskeleton is disrupted have a reduced protein to lipid ratio. The proteins on the surface of lipid droplets have been shown to add to droplet stability58-60 and a reduction in protein may contribute to the enhanced hydrolysis of the lipid in the droplets formed in the presence of cyt.D.
5. Conclusion
Our data shows that the CE in lipid droplets isolated from cyt.D treated cells is hydrolyzed more efficiently than droplets isolated from untreated cells. We also know however, that in whole cells, CE in droplets formed in cells with a disrupted cytoskeleton are hydrolyzed to a lesser extent than droplets formed in cells with an intact cytoskeleton. This paradox may be due to several factors. 1) The protein-depleted surface of the droplets formed in the presence of cyt.D may be lacking a protein needed to dock or stabilize the nCEH. This however also seems unlikely because we have shown that lipid droplets isolated from cells treated with cyt.D are a suitable substrate for nCEH from various sources. 2) We measured a marked increase in the TG content of lipid droplets formed in the presence of cyt.D. However, the reduction in CE hydrolysis is not simply due to TG competition for the nCEH as we have shown a similar reduction in TG hydrolysis. 3) We have demonstrated that the reduction in CE hydrolysis in cyt.D treated cells is not due to cellular toxicity as we saw no evidence of necrosis (assayed by LDH release) and cellular CE hydrolysis recovered when the cyt.D was withdrawn since inhibition of actin polymerization by cyt.D is reversible45. 4) It is possible that lipid droplets formed in the presence of cyt.D are sequestered in an area of the cell which does not allow ready access to the nCEH. In this scenario, lipid droplets formed in the ER use the cytoskeleton to move away from this compartment and into the cytoplasm where they become accessible to a cytosolic nCEH. In the J774 cell line, hormone sensitive lipase (HSL) is thought to be the predominant lipase although many other lipases have been identified in other cell types 54, 56, 61. Our data suggests that droplet movement from the ER into the cytoplasm does not occur when the cytoskeleton is disrupted with cyt.D and therefore diminishes interaction of the droplet with HSL, resulting in a decrease in CE and TG hydrolysis. Recently an ER lipase was identified and characterized54, 62-63. This lipase, nCEH, is present in J774 cells and contributes to CE hydrolysis in this cell line. In the present study, nCEH is likely contributing to the residual hydrolysis seen when J774 cells are treated with cyt.D.
Clearly the interaction of nCEH(s), the CE lipid droplet and the actin cytoskeleton are more complex than first thought. Our study is the first to describe the unique involvement of the cytoskeleton in CE lipid droplet formation, maturation and metabolism.
Highlights.
We examined the role of the cytoskeleton in lipid droplet formation and lipid mobilization. > We employ cytoskeleton disrupting agents. > Cytochalasin D alters cholesterol efflux to HDL. > Disrupting the cytoskeleton alters the size, composition and metabolism of the lipid droplets. > The cytoskeleton plays an integral role in lipid droplet metabolism.
Acknowledgments
This project was supported by NIH PPG grant HL-22633, NIH grant HL-086746, and National Heart, Lung, and Blood Institute Grant HL-19737. Negative stain electron microscopy was carried out using the Vanderbilt University Cell Imaging Shared Resource: Research EM Resource (sponsored by NIH grants DK20539, CA68485 and DK58404).
Abbreviations
- CE
cholesteryl ester
- FC
free cholesterol
- TG
triglyceride
- acLDL
acetylated low density lipoprotein
- BSA
bovine serum albumin
- cpm
counts/min
- FBS
fetal bovine serum
- PBS
phosphate-buffered saline
- nCEH
neutral cholesteryl ester hydrolase
- ACAT
acyl coenzyme A:cholesterol acyl transferase
- cyt.D
cytochalasin D
- PL
phospholipid
- acyl CoA
diacylglycerol acyltransferase (DGAT)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tang C, Oram JF. The cell cholesterol exporter ABCA1 as a protector from cardiovascular disease and diabetes. Biochim Biophys Acta. 2009;1791:563–572. doi: 10.1016/j.bbalip.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Vaughan AM, Oram JF. ABCA1 and ABCG1 or ABCG4 act sequentially to remove cellular cholesterol and generate cholesterol-rich HDL. J Lipid Res. 2006;47:2433–2443. doi: 10.1194/jlr.M600218-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Oram JF, Vaughan AM. Atp-binding cassette cholesterol transporters and cardiovascular disease. Circ Res. 2006;99:1031–1043. doi: 10.1161/01.RES.0000250171.54048.5c. [DOI] [PubMed] [Google Scholar]
- 4.Oram JF. Hdl apolipoproteins and ABCA1: Partners in the removal of excell cellular cholesterol. Arterioscler Thromb Vasc Biol. 2003;23:720–727. doi: 10.1161/01.ATV.0000054662.44688.9A. [DOI] [PubMed] [Google Scholar]
- 5.Brown MS, Ho YK, Goldstein JL. The cholesteryl ester cycle in macrophage foam cells. Continual hydrolysis and re-esterification of cytoplasmic cholesteryl esters. J Biol Chem. 1980;255:9344–9352. [PubMed] [Google Scholar]
- 6.Greenberg AS, Egan JJ, Wek SA, Moos MC, Londos C, Kimmel AR. Isolation of cDNAs for perilipins a and b: Sequence and espression of lipid droplet-associated proteins of adipocytes. Proc Natl Acad Sci USA. 1993;90:12035–12039. doi: 10.1073/pnas.90.24.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchette-Mackie EJ, Londos C. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. Journal of Biological Chemistry. 1991;266:11341–11346. [PubMed] [Google Scholar]
- 8.Olofsson S, Bostrom P, Andersson L, Rutberg M, Levin M, Perman J, Boren J. Triglyceride containing lipid droplets and lipid droplet-associated proteins. Curr Opin Lipidol. 2008;19:441–447. doi: 10.1097/MOL.0b013e32830dd09b. [DOI] [PubMed] [Google Scholar]
- 9.Goodman J. The gregarious lipid droplet. J Biol Chem. 2008;283:28005–28009. doi: 10.1074/jbc.R800042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin S, Parton RG. Lipid droplets: A unified view of a dynamic organelle. Nature Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 11.Fujimoto T, Ohsaki Y. Cytoplasmic lipid droplets rediscovery of an old structure as a unique platform. Ann N Y Acad Sci. 2006;1086:104–115. doi: 10.1196/annals.1377.010. [DOI] [PubMed] [Google Scholar]
- 12.Robenek H, Hofnagel O, Buers I, Robenek MJ, Troyer D, Severs NJ. Adipophilin-enriched domains in the er membrane are sites of lipid droplet biogenesis. J Cell Sci. 2006;119:4215–4224. doi: 10.1242/jcs.03191. [DOI] [PubMed] [Google Scholar]
- 13.Chang CY, Chen J, Thomas MA, Cheng D, Del Priore VA, Newton RS, Pape ME, Chang TY. Regulation and immunolocalization of acyl-coenzyme a: Cholesterol acyltransferase in mammalian cells as studied with specific antibodies. J Biol Chem. 1995;270:29532–29540. doi: 10.1074/jbc.270.49.29532. [DOI] [PubMed] [Google Scholar]
- 14.Almahbobi G, Williams LJ, Hall PF. Attachment of steroidogenic lipid droplets to intermediate filaments in adrenal cells. J Cell Sci. 1992;101:383–393. doi: 10.1242/jcs.101.2.383. [DOI] [PubMed] [Google Scholar]
- 15.Sato S, Fukasawa M, Yamakawa Y, Natsume T, Suzuki T, Shoji I, Aizaki H, Miyamura T, Nishijima M. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis c virus core protein. J Biochem. 2006;139:921–930. doi: 10.1093/jb/mvj104. [DOI] [PubMed] [Google Scholar]
- 16.Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 17.Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Current Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 18.Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jackle H, Kuhnlein RP. Characterization of the drosophilla lipid droplet subproteome. Mol Cell Proteomics. 2006;5:1082–1094. doi: 10.1074/mcp.M600011-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Romeo GR, Moulton KS, Kazlaushas A. Attenuated expression of profilin-1 confers protection from atherosclerosis in the LDL receptor-null mouse. Circ Res. 2007;101:357–367. doi: 10.1161/CIRCRESAHA.107.151399. [DOI] [PubMed] [Google Scholar]
- 20.Wojciak-Stothard B, Williams LJ, Ridley AJ. Monocyte adhesion and spreading on human endothelial cells is sependent on rho-regulated receptor clustering. J Cell Biol. 1999;145:1293–1307. doi: 10.1083/jcb.145.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller K, Dulku S, Hardwick SJ, Skepper JN, Mitchinson MJ. Changes in vimentin in human macrophages during apoptosis induced by oxidised low density lipoprotein. Atheroscerosis. 2001;156:133–144. doi: 10.1016/s0021-9150(00)00641-9. [DOI] [PubMed] [Google Scholar]
- 22.Zerbinatti CV, Gore RW. Uptake of modified low-density lipoproteins alters actin distribution and locomotor forces in macrophages. Am J Cell Physiol. 2003;284:555–561. doi: 10.1152/ajpcell.00177.2002. [DOI] [PubMed] [Google Scholar]
- 23.Becker L, Gharib SA, Irwin AD, Wijsmman E, Vaisar T, Oram JF, Heinecke JW. A macrophage sterol-responsive network linked to atherogenesis. Cell Metab. 2010;11:125–135. doi: 10.1016/j.cmet.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabas I, Zhe X, Beatini N, Myers J, Maxfield FR. The actin cytoskeleton is important for the stimulation of cholesterol eterification by atherogenic lipotprotins in macrophages. J Biol Chem. 1994;269:22547–22556. [PubMed] [Google Scholar]
- 25.Basu SK, Goldstein JL, Anderson RGW, Brown MS. Degradation of cationized low density lipoprotein and regulation of cholesterol metabolism in homozygous familial hypercholesterolemia fibroblasts. Proc Natl Acad Sci USA. 1976;73:3178–3182. doi: 10.1073/pnas.73.9.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellner-Weibel G, McHendry-Rinde B, Haynes MP, Adelman SJ. Evidence that newly synthesized esterified cholesterol is deposited in existing cytoplasmic lipid inclusions. J Lipid Res. 2001;42:768–777. [PubMed] [Google Scholar]
- 27.Mills G, Coley SC, Williams JF. Chemical composition of lipid droplets isolated from larvae of taenia taeniaeformis. J Parasitol. 1983;69:850–856. [PubMed] [Google Scholar]
- 28.Bamberger M, Lane MD. Assembly of very low density lipoprotein in the hepatocyte. J Biol Chem. 1988;263:11868–11878. [PubMed] [Google Scholar]
- 29.Adelman SJ, Glick JM, Phillips MC, Rothblat GH. Lipid composition and physical state effects on cellular cholesteryl ester clearance. J Biol Chem. 1984;259:13844–13850. [PubMed] [Google Scholar]
- 30.Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG, Gorkiewicz G, Zechner R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by cgi-58 and defective in chanarin-dorfman syndrome. Cell Metab. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian V, Garcia A, Sekowski A, Brasaemle DL. Hydrophobic sequences target and anchor perilipin a to lipid droplets. J Lipid Res. 2004;45:1983–1991. doi: 10.1194/jlr.M400291-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 33.Klansek J, Yancey P, St Clair RW, Fischer RT, Johnson WJ, Glick JM. Cholesterol quantitation by glc: Artifactual formation of short-chain steryl esters. J Lipid Res. 1995;36:2261–2266. [PubMed] [Google Scholar]
- 34.Rouser G, Kritchevsky G, Yamamoto A, Knudson AG, Jr, Simon G. Accumulation of a glycerolphospholipid in classical niemann-pick disease. Lipids. 1968;3:287–290. doi: 10.1007/BF02531203. [DOI] [PubMed] [Google Scholar]
- 35.Markwell MAK, Haas SM, Bieber LL, Tolbert NE. A modification of the lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 36.Fong TH, Wu C, Liao E, Chang CC, Pai M, Chiou R, Lee A. Association of globular b-actin with intracellular lipid dropelts in rat adrenocortical cells and adipocytes. Biochem Biophys Res Commun. 2001;289:1168–1174. doi: 10.1006/bbrc.2001.6080. [DOI] [PubMed] [Google Scholar]
- 37.Turro S, Ingelmo-Torres M, Estanyol JM, Tebar F, Fernandez MA, Albor CV, Gaus K, Grewal T, Enrich C, Pol A. Identification and characterization of associated with lipid droplets protein 1: A novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 2006;7:1254–1269. doi: 10.1111/j.1600-0854.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 38.Wakatsuki T, Schwab B, Thompson NC, WElson EL. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J Cell Sci. 2000;114:1025–1036. doi: 10.1242/jcs.114.5.1025. [DOI] [PubMed] [Google Scholar]
- 39.Kizhati K, Albritton LM. Requirements for different components of the host cell cytoskeleton distinguish ecotropic murine leukemia virus entry via endocytosis from entry via surface fusion. J Virology. 1997;71:7145–7156. doi: 10.1128/jvi.71.10.7145-7156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peyrollier K, Hajduch E, Gray A, Litherland GJ, Prescott AR, Leslie NR, Hundal HS. A role for the actin cytoskeleton in the hormonal and growth-factor-mediated activation of protein kinase B. Biochem J. 2000;352:617–622. [PMC free article] [PubMed] [Google Scholar]
- 41.Marchesan D, Rutberg M, Andersson L, Asp L, Larsson T, Boren J, Johansson J, Olofsson S. A phospholipase D-dependent process forms lipid droplets containing caveolin, adipocyte differentiation-related protein, and vimentin in a cell free system. J Biol Chem. 2003;278:27293–27300. doi: 10.1074/jbc.M301430200. [DOI] [PubMed] [Google Scholar]
- 42.Andersson L, Bostrom P, Ericson J, Rutberg M, Magnusson B, Marchesan D, Ruiz M, Asp L, Huang P, Frohman MA, Boren J, Olofsson S. PLD1 and ERK2 regulate cytosolic lipid droplet formation. J Cell Sci. 2006;119:2246–2257. doi: 10.1242/jcs.02941. [DOI] [PubMed] [Google Scholar]
- 43.Cheng J, Fujita A, Ohsaki Y, Suzuki M, Shinohara Y, Fujimoto T. Quantitative electron microscopy shows uniform incorporation of triglycerides into exisiting lipid droplets. Histochem Cell Biol. 2009;132:281–291. doi: 10.1007/s00418-009-0615-z. [DOI] [PubMed] [Google Scholar]
- 44.Zehmer JK, Huang Y, Peng G, Pu J, Anderson RGW, Liu P. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 2009;9:914–921. doi: 10.1002/pmic.200800584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schliwa M. Action of cytochalasin D on cytoskeletal networks. J Cell Biol. 1982;92:79–91. doi: 10.1083/jcb.92.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snow JW, McCloskey HM, Glick JM, Rothblat GH, Phillips MC. Physical state of cholesteryl esters deposited in cultured macrophages. Biochemistry. 1988;27:3640–3646. doi: 10.1021/bi00410a018. [DOI] [PubMed] [Google Scholar]
- 47.Sakahita N, Miyazaki A, Takeya M, Horiuchi S, Chang CY, Chang TY, Takahashi K. Localization of human acyl-coenzyme a: Cholesterol acyltransferase-1 (ACAT-1) in macrophages and various tissues. Am J Pathol. 2000;156:227–236. doi: 10.1016/S0002-9440(10)64723-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cases S, Stone SJ, Zhou P, TYen E, Tow B, Lardizabal KD, Voelker T, Farese RV. Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J Biol Chem. 2001;276:38870–38876. doi: 10.1074/jbc.M106219200. [DOI] [PubMed] [Google Scholar]
- 49.Cao J, Lockwood J, Burn P, Shi Y. Cloning and functional characterization of a mouse interstinal acyl-coA:Monoacylglycerol actyltransferase, MGATt2. J Biol Chem. 2003;278:13860–13866. doi: 10.1074/jbc.M300139200. [DOI] [PubMed] [Google Scholar]
- 50.Faulkner LE, Panagotopulos SE, Johnson JD, Woollett LA, Hui DY, Witting SR, Maiorano JN, Davidson WS. An analysis of the role of a retroendocytosis pathway in ABCA1-mediated cholesterol efflux from macrophages. J Lipid Res. 2008;49:1322–1332. doi: 10.1194/jlr.M800048-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liscum L. Pharmacological inhibition of the intracellular transport of low-density lipoprotein-derived cholesterol in chinese hamster ovary cells. Biochim Biophys Acta. 1990;1045:40–48. doi: 10.1016/0005-2760(90)90201-8. [DOI] [PubMed] [Google Scholar]
- 52.Denus M, Landry YD, Zha X. Atp-binding cassette A1-mediated lipidation of apolipoprotein A-I occurs at the plasma membrane and not in the endocytic compartments. J Biol Chem. 2008;283:16178–16186. doi: 10.1074/jbc.M709597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghosh S, Grogan WM. Immunological characterization of neutral cholesteryl ester hydrolase from rat liver cytosol. Biochem Cell Biol. 1992;70:800–803. doi: 10.1139/o92-121. [DOI] [PubMed] [Google Scholar]
- 54.Okazaki H, Igarashi M, Nishi M, Sekiya M, Tajima S, Takase S, Takanashi M, Ohta K, Tamura Y, Okazaki S, Yahagi N, Ohashi K, Amemiya-Kudo M, Nakagawa Y, Nagai R, Kadowaki T, Osuga J, Ishibashi S. Identification of neutral cholesteryl ester hydrolase, a key enzyme removing cholesterol from macrophages. J Biol Chem. 2008;283:33357–33364. doi: 10.1074/jbc.M802686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchebner M, Pfeifer T, Rathke N, Chandal PG, Lass A, Schreiber R, Kratzer A, Zimmermann R, Sattler W, Koefeler H, Frohlich E, Kostner G, Birner-Gruenberger R, Chiang KP, Haemmerle G, Zechner R, Levak-Frank S, Cravatt B, Kratky D. Cholesteryl ester hydrolase activity is abolished in HSL-/-macrophages but unchanged in macrophages lacking kiaa1363. J Lipid Res. 2010;51:2896–2908. doi: 10.1194/jlr.M004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernard DW, Rodriguez A, Rothblat GH, Glick JM. Camp stimulates cholesteryl ester clearance to high density lipoproteins in J774 macrophages. J Biol Chem. 1991;266:710–716. [PubMed] [Google Scholar]
- 57.Kratky D. Neutral cholesterol ester hydrolases in macrophages: Still a matter of debate. Circ Res. 2011;108:e13. doi: 10.1161/CIRCRESAHA.111.245829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lieber JG, Evans RM. Disruption of the vimentin intermediate filament system during adipose conversion of 3t3-l1 cells inhibits lipid droplet accumulation. J Cell Sci. 1996;109:3047–3058. doi: 10.1242/jcs.109.13.3047. [DOI] [PubMed] [Google Scholar]
- 59.Bell M, Wang H, Chen H, McLenithan JC, Gong D, Yang R, Yu D, Fried SK, Quon MJ, Londos C, Sztalryd C. Consequences of lipid droplet coat protein downregulation in liver cells. Diabetes. 2008;57:2037–2045. doi: 10.2337/db07-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sztalryd C, Bell M, Lu X, Mertz P, Hickenbottom S, Chang BHJ, Chan L, Kimmel AR, Londos C. Functional compensation for adipose differentiation-related protein (ADRP) by TIP47 in an adfp null embryonic cell line. J Biol Chem. 2006;281:34341–34348. doi: 10.1074/jbc.M602497200. [DOI] [PubMed] [Google Scholar]
- 61.Khoo JC, Mahoney EM, Steinberg D. Neutral cholesterol esterase activity in macrophages and its enhancement by cAMP dependent protein kinase. J Biol Chem. 1981;256:12659–12661. [PubMed] [Google Scholar]
- 62.Igarashi M, Osuga J, Isshiki M, Sekiya M, Okazaki H, Takase S, Takanashi M, Ohta K, Kumagai T, Nishi M, Fujita T, Nagai R, Kadowaki T, Ishibashi S. Targeting of neutral cholesterol ester hydrolase to the endoplasmic reticulum via its n-terminal sequence. J Lipid Res. 2010;51:274–285. doi: 10.1194/jlr.M900201-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sekiya M, Osuga J, Nagashima S, Ohshiro T, Igarashi M, Takahashi T, Tazoe F, Wada T, Ohta K, Takanashi M, Kumagai T, Nishi M, Takase S, Yahagi N, Yagyu H, Ohashi K, Nagai R, Kadowaki T, Furukawa Y, Ishibashi S. Ablation of neutral cholesterol ester hydrolase 1 accelerates atherosclerosis. Cell Metab. 2009;10:219–228. doi: 10.1016/j.cmet.2009.08.004. [DOI] [PubMed] [Google Scholar]


