Abstract
Inactivation of the von Hippel-Lindau (VHL) tumor suppressor is associated with renal carcinoma, hemangioblastoma and pheochromocytoma. The VHL protein is a component of a ubiquitin ligase complex that ubiquitinates and degrades hypoxia inducible factor-α (HIF-α). Degradation of HIF-α by VHL is proposed to suppress tumorigenesis and tumor angiogenesis. Several lines of evidence also suggest important roles for HIF-independent VHL functions in tumor suppression and other biological processes. Using GST-VHL pull-down experiment and mass spectrometry, we detected an interaction between VHL and heterochromatin protein 1 (HP1). We identified a conserved HP1-binding motif (PXVXL) in the β domain of VHL, which is disrupted in a renal carcinoma-associated P81S mutant. We show that the VHL P81S mutant displays reduced binding to HP1, yet retains the ability to interact with elongin B, elongin C, and cullin 2 and is fully capable of degrading HIF-α. We also demonstrate that HP1 increases the chromatin association of VHL. These results suggest a role for the VHL-HP1 interaction in VHL chromatin targeting.
Keywords: von Hippel-Lindau tumor suppressor, heterochromatin protein 1, interaction, mass spectrometry, renal carcinoma
Introduction
The von Hippel-Lindau (VHL) tumor suppressor gene was originally identified as a gene whose germline mutation results in a familial cancer syndrome called von Hippel-Lindau (VHL) disease. The VHL disease is characterized by an increased risk of clear cell renal carcinoma, hemangioblastoma of the nervous system, and adrenal pheochromocytoma [for reviews see [1–4]]. Normal tissues of VHL disease patients harbor one wild-type and one defective VHL allele. The tumors arising in these patients, however, display somatic inactivation of the remaining wild-type allele, which is consistent with Knudson’s two-hit hypothesis. Biallelic VHL inactivation is also common in sporadic clear cell renal carcinomas and hemangioblastomas. VHL mutation occurs in approximately 50% of clear cell renal carcinomas [5], which in turn account for 80% of all adult renal carcinomas. This makes VHL mutation the most frequent genetic alteration found in adult kidney cancer. Biochemically, the VHL protein functions as a substrate recognition subunit of the E3 ubiquitin ligase complex that also contains elongin B, elongin C, cullin 2, and Rbx1. The interaction of VHL with elongin B, elongin C, and cullin 2 is mediated by the α domain of VHL (Figure 1A). Using two different initiation codons, two isoforms of VHL are synthesized: VHL30, a 213-amino-acid protein in humans and VHL19, residues 54 – 213 of VHL30 (lacking the N-terminal acidic domain). Both VHL30 and VHL19 act as a substrate recognition subunit in the E3 ubiquitin ligase complex.
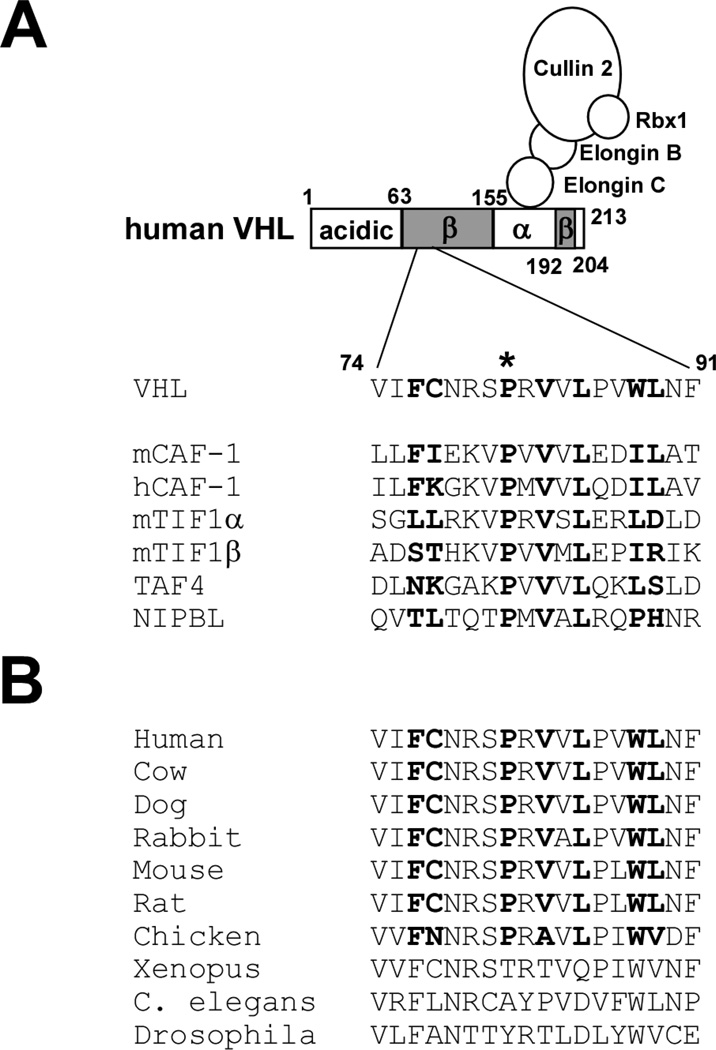
Figure 1. HP1-binding motif in VHL.
(A) The PXVXL motif in human VHL and its alignment with other PXVXL-containing proteins. Bold residues in mCAF-1 are involved in hydrophobic interaction with HP1 chromo shadow domain. Proline 81 (asterisk) is mutated to serine in renal carcinoma-associated VHL P81S mutant.
(B) Sequence alignment of the PXVXL motif in VHL from different species.
The best studied target of VHL is hypoxia inducible factors (HIFs), a family of transcriptional regulators mediating the cellular response to hypoxia. In the presence of oxygen and iron, enzymatic hydroxylation of specific proline residues in the α subunit of HIF (HIF-α) occurs and these hydroxylated prolines are recognized by VHL, which results in ubiquitination and proteasomal degradation of HIF-α. Hypoxia or depletion of iron inhibits the prolylpage hydroxylation of HIF-α, leading to accumulation of HIF-α and induction of HIF target genes such as vascular endothelial growth factor (VEGF) and erythropoietin. Some of the characteristics of VHL-mutated tumors can be explained by upregulation of the HIF target genes. Hemangioblastomas and clear cell renal carcinomas are highly vascular tumors, which is at least partly due to VEGF overproduction. These tumors as well as pheochromocytomas sometimes secrete erythropoietin, resulting in overproduction of red blood cells.
While the link between VHL and HIF is well established, several lines of evidence clearly indicate that VHL has functions other than regulation of HIF [1–4]. 1) VHL was shown to bind to multiple other proteins such as fibronectin, atypical PKC family proteins, SP1 transcription factor, RNA polymerase subunits Rpb1 and Rpb7, a de-ubiquitinating enzyme VDU-1, and CARD9 [6]. 2) VHL has been shown to play HIF-independent roles in extracellular matrix control [7, 8]. 3) Type 2C VHL disease (familial pheochromocytomas without hemangioblastomas or renal carcinomas) is caused by specific VHL mutants such as L188V and V84L. Importantly, these type 2C VHL mutants ubiquitinate and degrade HIF as efficiently as wild-type VHL, which suggests that alteration of HIF-independent function(s) of VHL plays a role in tumorigenesis [8, 9]. 4) Transgenic mice expressing constitutively active HIF did not develop hemangioblastomas or renal carcinomas [10], suggesting that deregulation of HIF is not sufficient to initiate tumors in mice. 5) Finally, gain-of-function HIF-2 mutations were identified in familial erythrocytosis patients [11, 12], but these patients did not show higher incidence of tumors, suggesting that activation of HIF is not sufficient to induce tumors in humans. These findings are consistent with the notion that loss or alteration of HIF-independent function(s) of VHL plays a critical role in tumorigenesis. The biochemical basis of the HIF-independent VHL function(s), however, remains poorly defined.
To gain insight into the HIF-independent functions of VHL, we undertook a screening for VHL-interacting proteins by GST-VHL pull-down of VHL-null renal carcinoma cell lysate and mass spectrometry. For this study, the result from the pilot screening was used to provide leads for subsequent immunoprecipitation-immunoblotting analyses. We discovered that VHL contains a conserved HP1-binding motif (PXVXL) in its β domain and interacts with HP1. This HP1-binding motif is disrupted in VHL P81S mutant, which is associated with both hereditary and sporadic renal carcinoma. We show that VHL P81S mutant displays reduced binding to HP1, but retains the ability to interact with cullin 2, elongin B, and elongin C and to degrade HIF-2α. We also demonstrate that HP1 enhances the chromatin association of VHL. These results suggest a role for the VHL-HP1 interaction in recruiting VHL to chromatin.
Materials and Methods
Cell culture
786-O renal carcinoma cells were cultured in RPMI1640 medium supplemented with 10% fetal calf serum. 293 and 293T embryonic kidney cells were cultured in DMEM supplemented with 10% calf serum. Calcium phosphate co-precipitation was used for transfection.
Protein sample preparation and mass spectrometry
Forty 15-cm plates of 786-O cells were lysed in TNE buffer (10 mM Tris pH 7.4/150 mM NaCl/1% NP-40/1 mM EDTA/1 mM AEBSF/10 µg/ml aprotinin/10 µg/ml Leupeptin/1 µg/ml Pepstatin A/20 mM sodium fluoride). The lysate was incubated with 100 µg of GST-VHL attached to glutathione Sepharose (GE Healthcare) for 12 hours at 4 °C. The GST-VHL-interacting proteins were collected by centrifugation, washed three times with TNE buffer, and eluted with 5 mM reduced glutathione for 4 hours at 4 °C. The eluted sample was processed with Microcon YM-10 column (Millipore) for concentration and buffer change to 50 mM Tris pH 8.5.
The sample was digested with trypsin (Promega) and analyzed by capillary high performance liquid chromatography-tandem mass spectrometry (HPLC-ESI-MS/MS), using a Thermo Fisher LTQ linear ion trap mass spectrometer fitted with a New Objective PicoView 550 nanospray interface. On-line HPLC separation of the digests was accomplished with an Eksigent NanoLC micro HPLC: column, PicoFrit™ (New Objective; 75 µm i.d.) packed to 10 cm with C18 adsorbent (Vydac; 218MSB5, 5 µm, 300 Å); mobile phase A, 0.5% acetic acid (HAc)/0.005% TFA; mobile phase B, 90% acetonitrile/0.5% HAc/0.005% TFA; gradient 2 to 42% B in 1 hr; flow rate, 0.4 µl/min. MS conditions were: ESI voltage, 2.9 kV; isolation window for MS/MS, 3; relative collision energy, 35%; scan strategy, survey scan followed by acquisition of data dependent collision-induced dissociation (CID) spectra of the seven most intense ions in the survey scan above a set threshold.
Mass spectrometry data analysis
The MS files were converted to mzXML format using ReAdW and were searched against the IPI human protein database (v 3.24; 66,923 protein entries) using SEQUEST Cluster 3.1 SR1. Methionine oxidation was considered as variable modification. Up to one missed tryptic cleavage was allowed. The peptide mass tolerance was set as 3.0 Da. The SEQUEST search results were analyzed by the Trans-Proteomic Pipeline [for review see [13]] version 3.0. Peptide/protein identifications were validated by Peptide/ProteinProphet [14, 15]. A ProteinProphet score of 0.9 was used as a cutoff.
Immunoprecipitation and immunoblotting
Immunoprecipitation was performed as described [16]. The cell lysates or immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblotting as described [17]. The following antibodies were used: mouse monoclonal anti-FLAG (M2, Sigma-Aldrich); rabbit polyclonal anti-FLAG (RFLG-45A, Immunology Consultants Laboratory); mouse monoclonal anti-HP1α (2HP-2G9, Millipore); mouse monoclonal anti-HP1 (2MOD-1G6, Active Motif); rabbit polyclonal anti-H3K9me3 (8898, Abcam); mouse monoclonal antiubiquityl-histone H2A (05-678, Millipore); mouse monoclonal anti-ubiquityl-histone H2B (05-1312, Millipore); mouse monoclonal anti-ubiquitin (FK2, Biomol International); rabbit polyclonal elongin B (Poly6130, Biolegend); rabbit polyclonal anti-elongin C (Poly6131, Biolegend); rabbit polyclonal anti-cullin 2 (Thermo scientific); mouse monoclonal anti-Rb (G3-245, BD Pharmingen); and mouse monoclonal anti-tubulin (DM1A, Sigma-Aldrich).
Subcellular fractionation
293 cells were transfected with FLAG-tagged VHL30, VHL30 P81S, VHL19, or VHL19 P81S alone or together with HA-tagged HP1. Two days later, Triton-soluble fractions and micrococcal nuclease-extractable chromatin fractions were prepared as described [18].
Results and Discussion
GST pull-down screening for VHL-interacting proteins
To identify VHL-interacting proteins, we incubated bacterially-expressed, purified GST-VHL30 (full-length human VHL encoding amino acid 1–213) with the lysate of VHL-null 786-O renal carcinoma cells and isolated the GST-VHL-interacting proteins by GST pull-down followed by elution with glutathione. The eluted sample was processed for HPLC-ESI-MS/MS analysis as described in Materials and Methods. At a Protein Prophet probability score of 0.9 or higher, 24 different proteins were identified (Table 1). Among the identified proteins, HP1 and HP1β caught our attention because we discovered an HP1-binding motif in VHL (see below). Five unique peptides derived from HP1 and a single peptide derived from HP1β were identified in the GST-VHL pull-down sample (Table 1). The tandem mass spectra of HP1 and HP1β peptides are shown in Figure S1.
Table 1. Proteins identified in the GST-VHL pull-down sample.
The table lists the proteins identified in the GST-VHL pull-down sample with a Protein Prophet probability score of 0.9 or higher. Entries from GST, keratin and trypsin were omitted.
| Number of Peptides |
||||||||
|---|---|---|---|---|---|---|---|---|
| Entry | Protein Name/Peptide Sequences | Peptide Charge State |
Sequence Coverage (%) |
Unique | Total | ProteinProphet Score |
Accession Number (IPI-Hum | |
| 1 | HP1 gamma | 37.2 | 5 | 8 | 1 | IPI00297579 | ||
| GFTDADNTWEPEENLDCPELIEAFLNSQK | 3 | |||||||
| LTWHSCPEDEAQ | 2 | |||||||
| WKDSDEADLVLAK | 3 | |||||||
| VEEAEPEEFVVEK | 2 | |||||||
| KVEEAEPEEFVVEK | 3 | |||||||
| 2 | HP1 beta | 26.4 | 1 | 1 | 0.98 | IPI00010320 | ||
| GFSDEDNTWEPEENLDCPDLIAEFLQSQK | 3 | |||||||
| 3 | VHL | 53.8 | 10 | 46 | 1 | IPI00027969 | ||
| VVLPVWLNFDGEPQPYPTLPPGTGR | 2 | |||||||
| VVLPVWLNFDGEPQPYPTLPPGTGR | 3 | |||||||
| SLYEDLEDHPNVQK | 2 | |||||||
| SLYEDLEDHPNVQK | 3 | |||||||
| SLYEDLEDHPNVQKDLER | 2 | |||||||
| SVNSREPSQVIFCNR | 3 | |||||||
| RLDIVR | 1 | |||||||
| SLVKPENYR | 2 | |||||||
| GHLWLFR | 2 | |||||||
| EPSQVIFCNR | 2 | |||||||
| IHSYR | 1 | |||||||
| RIHSYR | 2 | |||||||
| 4 | 60S ribosomal protein L5 | 12.5 | 3 | 4 | 1 | IPI00000494 | ||
| YLMEEDEDAYKK | 2 | |||||||
| HIMGQNVADYMR | 2 | |||||||
| HIMGQNVADYMR | 3 | |||||||
| NSVTPDMMEEMYK | 2 | |||||||
| 5 | Peroxiredoxin-1 | 58.8 | 3 | 5 | 1 | IPI00000874 | ||
| HGEVCPAGWKPGSDTIKPDVQK | 3 | |||||||
| LVQAFQFTDK | 2 | |||||||
| GLFIIDDK | 2 | |||||||
| IGHPAPNFK | 3 | |||||||
| ADEGISFR | 2 | |||||||
| 6 | HSPA5 | 13.3 | 8 | 8 | 1 | IPI00003362 | ||
| SQIFSTASDNQPTVTIK | 2 | |||||||
| VTHAVVTVPAYFNDAQR | 3 | |||||||
| SQIFSTASDNQPTVTIK | 3 | |||||||
| DAGTIAGLNVMR | 2 | |||||||
| ITPSYVAFTPEGER | 2 | |||||||
| ITPSYVAFTPEGER | 3 | |||||||
| NELESYAYSLK | 2 | |||||||
| NQLTSNPENTVFDAKR | 3 | |||||||
| 7 | HSPA8 | 32.9 | 5 | 6 | 1 | IPI00003865 | ||
| HWPFMVVNDAGRPK | 3 | |||||||
| NQVAMNPTNTVFDAK | 2 | |||||||
| TTPSYVAFTDTER | 2 | |||||||
| DAGTIAGLNVLR | 2 | |||||||
| SFYPEEVSSMVLTK | 2 | |||||||
| NQVAMNPTNTVFDAKR | 3 | |||||||
| 8 | LanC-like protein 1 | 24.6 | 7 | 12 | 1 | IPI00005724 | ||
| IPQSHIQQICETILTSGENLAR | 3 | |||||||
| FAEWCLEYGEHGCR | 3 | |||||||
| SLAEGYFDAAGR | 2 | |||||||
| LHSLVKPSVDYVCQLK | 3 | |||||||
| IDPHAPNEMLYGR | 3 | |||||||
| IRELLQQMER | 2 | |||||||
| IRELLQQMER | 3 | |||||||
| AFPNPYADYNK | 2 | |||||||
| 9 | Histone H2B type 1-L | 34.1 | 3 | 5 | 1 | IPI00018534 | ||
| AMGIMNSFVNDIFER | 2 | |||||||
| KESYSVYVYK | 2 | |||||||
| QVHPDTGISSK | 3 | |||||||
| EIQTAVR | 1 | |||||||
| 10 | F-actin-capping protein subunit beta | 26.2 | 3 | 3 | 1 | IPI00026185 | ||
| SPWSNKYDPPLEDGAMPSAR | 3 | |||||||
| QMEKDETVSDCSPHIANIGR | 3 | |||||||
| SGSGTMNLGGSLTR | 2 | |||||||
| 11 | Histone H1.2 | 17.8 | 4 | 4 | 1 | IPI00217465 | ||
| ALAAAGYDVEKNNSR | 3 | |||||||
| KASGPPVSELITK | 3 | |||||||
| ALAAAGYDVEK | 2 | |||||||
| SGVSLAALKK | 2 | |||||||
| 12 | Carbonyl reductase [NADPH] 1 | 66.8 | 19 | 54 | 1 | IPI00295386 | ||
| GQAAVQQLQAEGLSPR | 2 | |||||||
| GQAAVQQLQAEGLSPR | 3 | |||||||
| SPEEGAETPVYLALLPPDAEGPHGQFVSEK | 3 | |||||||
| VADPTPFHIQAEVTMK | 2 | |||||||
| FRSETITEEELVGLMNK | 3 | |||||||
| FHQLDIDDLQSIR | 3 | |||||||
| VVNVSSIMSVR | 2 | |||||||
| DVCTELLPLIKPQGR | 2 | |||||||
| ILLNACCPGWVR | 2 | |||||||
| FHQLDIDDLQSIR | 3 | |||||||
| LFSGDVVLTAR | 2 | |||||||
| SETITEEELVGLMNK | 2 | |||||||
| EGWPSSAYGVTK | 2 | |||||||
| DVCTELLPLIKPQGR | 3 | |||||||
| GIGLAIVR | 2 | |||||||
| SETITEEELVGLMNK | 3 | |||||||
| TNFFGTR | 2 | |||||||
| EYGGLDVLVNNAGIAFK | 2 | |||||||
| VVNVSSIMSVR | 1 | |||||||
| 13 | Histone H4 | 53.4 | 7 | 13 | 1 | IPI00453473 | ||
| TLYGFGG | 1 | |||||||
| VLRDNIQGITKPAIR | 3 | |||||||
| ISGLIYEETR | 2 | |||||||
| DAVTYTEHAK | 2 | |||||||
| TVTAMDVVYALKR | 2 | |||||||
| DNIQGITKPAIR | 2 | |||||||
| DAVTYTEHAK | 1 | |||||||
| TVTAMDVVYALK | 2 | |||||||
| 14 | ACTB Actin, cytoplasmic 1 | 25.6 | 7 | 9 | 0.99 | IPI00021439 | ||
| TTGIVMDSGDGVTHTVPIYEGYALPHAILR | 3 | |||||||
| HQGVMVGMGQK | 2 | |||||||
| AGFAGDDAPR | 2 | |||||||
| EITALAPSTMK | 2 | |||||||
| SYELPDGQVITIGNER | 2 | |||||||
| VAPEEHPVLLTEAPLNPK | 3 | |||||||
| EITALAPSTMK | 2 | |||||||
| 15 | SLC3A2 solute carrier family 3, member 2 isoform a | 7.6 | 2 | 2 | 1 | IPI00027493 | ||
| ADLLLSTQPGREEGSPLELER | 3 | |||||||
| IGDLQAFQGHGAGNLAGLK | 3 | |||||||
| 16 | CD44 antigen | 8.3 | 3 | 3 | 1 | IPI00297160 | ||
| YGFIEGHVVIPR | 3 | |||||||
| TEAADLCK | 2 | |||||||
| ALSIGFETCR | 2 | |||||||
| 17 | SLC2A1 Solute carrier family 2 | 6.2 | 2 | 2 | 1 | IPI00220194 | ||
| GTADVTHDLQEMKEESR | 3 | |||||||
| TFDEIASGFR | 2 | |||||||
| 18 | 40S ribosomal protein S15a | 17.7 | 1 | 2 | 0.99 | IPI00221091 | ||
| HGYIGEFEIIDDHR | 3 | |||||||
| WQNNLLPSR | 2 | |||||||
| 19 | Nucleolin | 3 | 2 | 3 | 0.99 | IPI00444262 | ||
| NDLAVVDVR | 2 | |||||||
| TGISDVFAK | 2 | |||||||
| 20 | Vesicle-associated membrane protein-associated protein B/C | 5.1 | 1 | 1 | 0.99 | IPI00006211 | ||
| NVCFK | 1 | |||||||
| 21 | RBPMS Isoform A of RNA-binding protein with multiple splicing | 12.6 | 1 | 1 | 0.98 | IPI00004045 | ||
| AEKENTPSEANLQEEEVR | 3 | |||||||
| 22 | 60S ribosomal protein L21 | 11.2 | 1 | 1 | 0.97 | IPI00247583 | ||
| VYNVTQHAVGIVVNK | 3 | |||||||
| 23 | Elongation factor 1-beta | 13.8 | 2 | 2 | 0.95 | IPI00178440 | ||
| SIQADGLVWGSSK | 2 | |||||||
| SSILLDVKPWDDETDMAK | 3 | |||||||
| 24 | SON protein | 1.9 | 2 | 2 | 0.92 | IPI00000192 | ||
| SMMSSYSAADR | 2 | |||||||
| SMMSPMAER | 2 | |||||||
The HP1 proteins have two structural domains: a chromo domain and a chromo shadow domain. The chromo domain of HP1 recognizes methylated lysine 9 of histone H3 [19, 20] and this interaction is believed to play an important role in the formation of heterochromatin and gene silencing. The chromo shadow domain interacts with several regulators of chromatin and gene transcription such as chromatin assembly factor-1 (CAF-1) [21], transcription intermediary factor α and β (TIF-1 α and β) [22]. In mammals, there are three members in the HP1 family, HP1α, HP1β, and HP1. Whereas HP1α and HP1β are located predominantly in heterochromatin, HP1 is also present in euchromatin [23], suggesting a role for HP1 in gene regulation in euchromatin regions.
A conserved HP1-binding motif in VHL
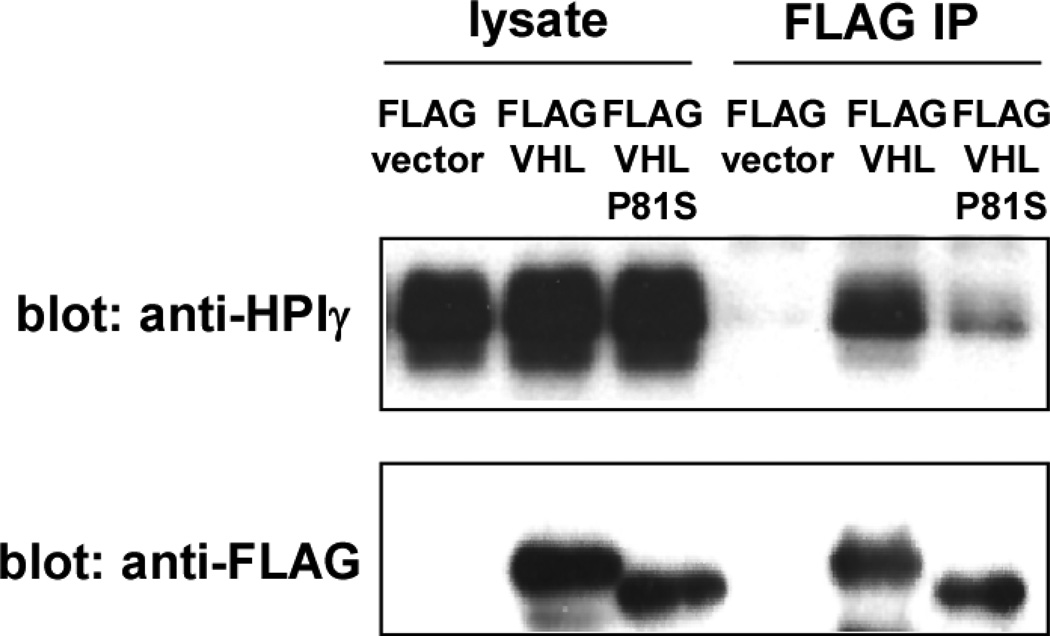
Proteins that bind to the chromo shadow domain of HP1 often contain a sequence motif, PXVXL (Figure 1A) [24]. In the case of mouse CAF-1 (mCAF-1), a detailed NMR analysis revealed that the PXVXL motif and surrounding amino acid residues (FIXXXPXVXLXXIL) are involved in hydrophobic interaction with the HP1 chromo shadow domain [25]. We identified a conserved PXVXL-containing motif in the β domain of VHL that resembles the above-mentioned HP1-binding motif in mCAF-1 and several other HP1-binding proteins (Figure 1A and 1B). Consistent with our screening result and the presence of a PXVXL motif in VHL, we showed that VHL30 interacts with HP1 by co-immunoprecipitation (Figure 2).
Figure 2. The interaction of VHL and HP1.
293T cells were transfected with FLAG-vector, FLAG-VHL, or FLAG-VHL-P81S. Forty-eight hours after transfection, the interaction of FLAG-VHL or FLAG-VHL-P81S with HP1 was examined by anti-FLAG immunoprecipitation followed by anti- HP1 immunoblotting.
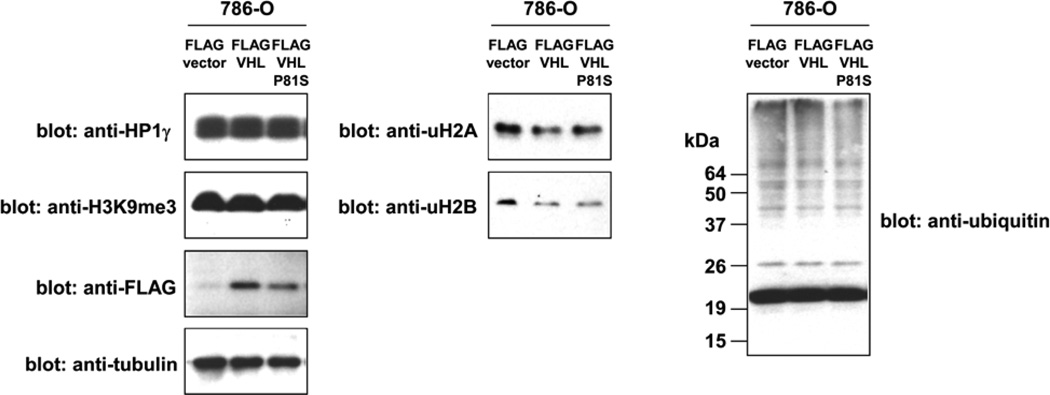
The PXVXL motif-containing proteins are thought to be recruited to chromatin by HP1 and modulate transcription and other chromatin functions although the functional significance of HP1 binding is not yet established for these HP1-binding proteins. We tested the possibility that HP1 is a ubiquitination and degradation substrate for the VHL E3 ubiquitin ligase. As shown in Figure 3, restoration of wild-type VHL expression in VHL-null 786-O cells did not affect the expression of HP1 whereas VHL restoration resulted in a dramatic decrease in the levels of HIF-2α (Figure 5), a known ubiquitination and degradation substrate of the VHL ubiquitin ligase. This suggests that HP1 is not a VHL degradation substrate. We also examined the effect of VHL on constitutive heterochromatin using its marker, lysine-9 tri-methylated histone H3 (H3K9me3), and did not observe a difference in overall H3K9me3 levels between VHL-null and VHL-restored cells (Figure 3), suggesting that loss of VHL does not cause gross alterations in constitutive heterochromatin. In addition, we analyzed the ubiquitination status of histone H2A and H2B and did not observe increased histone ubiquitination in VHL-restored cells (Figure 3). Overall ubiquitination of cellular proteins was similar between VHL-null and VHL-restored cells (Figure 3). It is possible that the interaction of VHL and HP1 results in some other alterations in chromatin or chromatin-binding proteins.
Figure 3. The effect of VHL and VHL-P81S on HP1, H3K9me3, ubiquitinated H2A, ubiquitinated H2B, and ubiquitinated cellular protein levels.
VHL-null 786-O renal carcinoma cells were infected with lentiviral vectors expressing FLAG-tagged VHL, VHL-P81S, or an empty vector. After puromycin selection, the levels of HP1, lysine-9 tri-methylated histone H3, ubiquitinated histone H2A, ubiquitinated histone H2B, and ubiquitinated cellular proteins were examined by immunoblotting. The expression levels of FLAG-VHL and FLAG-VHL-P81S were also examined by anti-FLAG immunoblotting. Tubulin serves as a loading control.
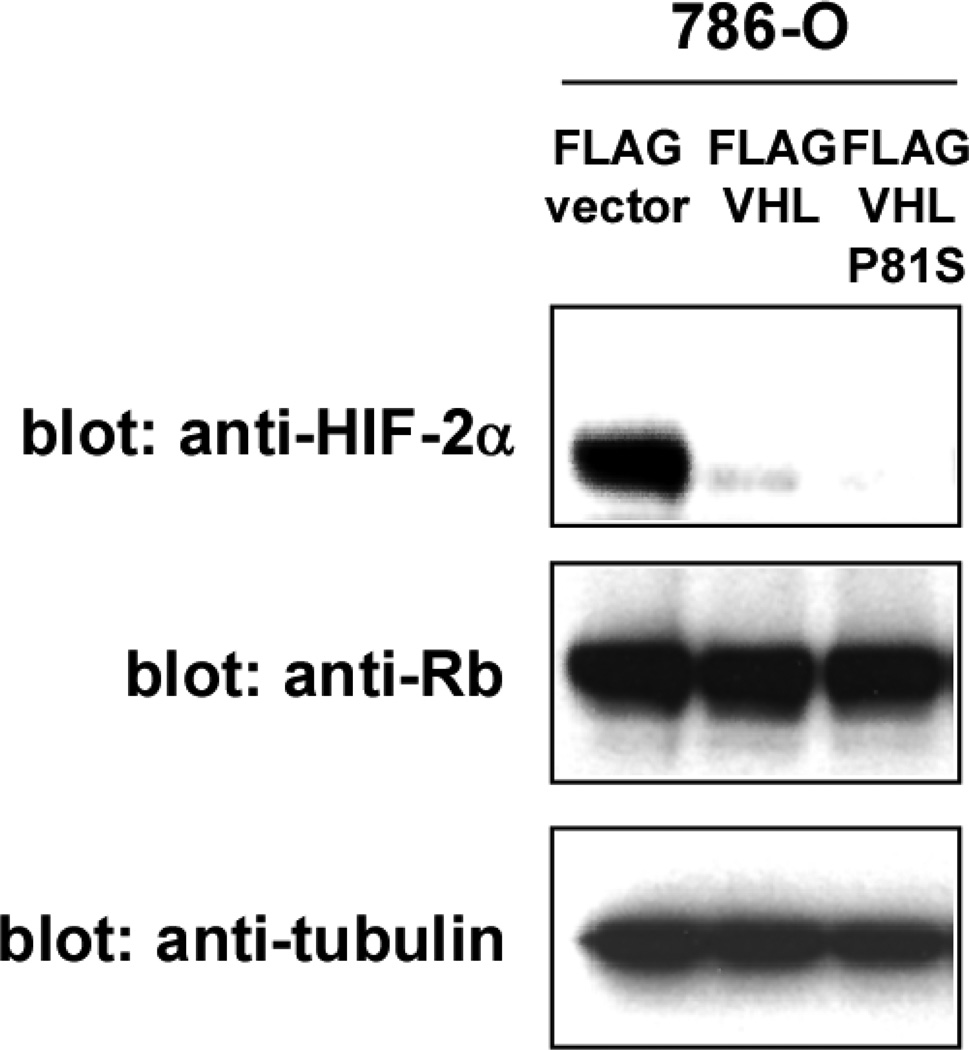
Figure 5. VHL-P81S mutant retains the ability to degrade HIF-α.
The three cell types in Figure 3 were analyzed for the levels of HIF-2α by anti-HIF-2α immunoblotting. Rb and tubulin serve as loading controls.
VHL P81S mutant displays reduced HP1 interaction, but retains the ability to degrade HIF-α
VHL P81S mutation is frequently observed in sporadic renal carcinoma, particularly those associated with trichloroethylene exposure [26, 27]. Thirty percent (13 out of 44) of renal carcinomas in patients with known trichloroethylene exposure were found to harbor the VHL P81S mutation [26]. In addition, a germline VHL P81S mutation results in type I von Hippel-Lindau disease [28], which is characterized by renal carcinoma and hemangioblastoma without predisposition to pheochromocytoma. Because the VHL P81S mutation alters the PXVXL motif (Figure 1A), we analyzed the effect of this mutation on HP1 binding. As shown in Figure 2, the VHL30 P81S mutant displayed reduced binding to HP1, suggesting the importance of the PXVXL motif in the VHL-HP1 interaction.
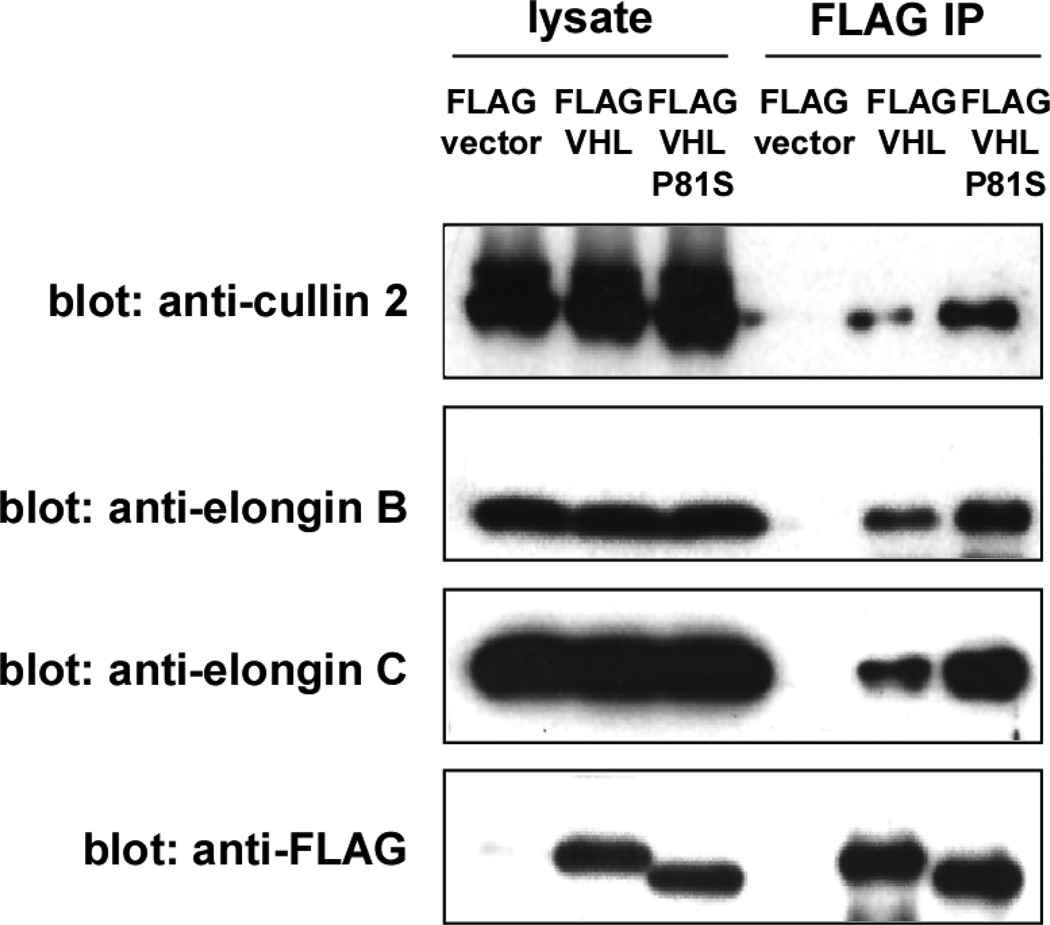
VHL interacts with elongin B, elongin C, and cullin 2 through its α domain and these proteins form an E3 ubiquitin ligase complex. We examined the interaction of the VHL30 P81S mutant with the components of the VHL E3 ligase complex and found that the VHL30 P81S mutant interacts more efficiently with the E3 ligase components than wild-type VHL30 (Figure 4). Consistent with the efficient binding of the VHL30 P81S mutant with VHL E3 ligase components, expression of this mutant in VHL-null 786-O cells resulted in a dramatic decrease in the level of HIF-2α, which was similar to that achieved by wild-type VHL30 (Figure 5). Degradation of HIF-1α by VHL P81S mutant was also demonstrated by Knauth et al., who used P81S mutation in VHL19, a shorter VHL isoform encoding amino acid 54–213 [29]. These results indicate that the renal carcinoma-associated VHL P81S mutant displays reduced HP1 binding, yet retains the ability to degrade HIF-α.
Figure 4. The interaction of VHL and VHL-P81S with E3 ligase components.
293T cells were transfected with FLAG-vector, FLAG-VHL, or FLAG-VHL-P81S. Forty-eight hours after transfection, the interaction of FLAG-VHL or FLAG-VHL-P81S with cullin 2, elongin B, and elongin C was examined by anti-FLAG immunoprecipitation followed by anti-cullin 2, elongin B, and elongin C immunoblotting.
HP1 increases the chromatin association of VHL30
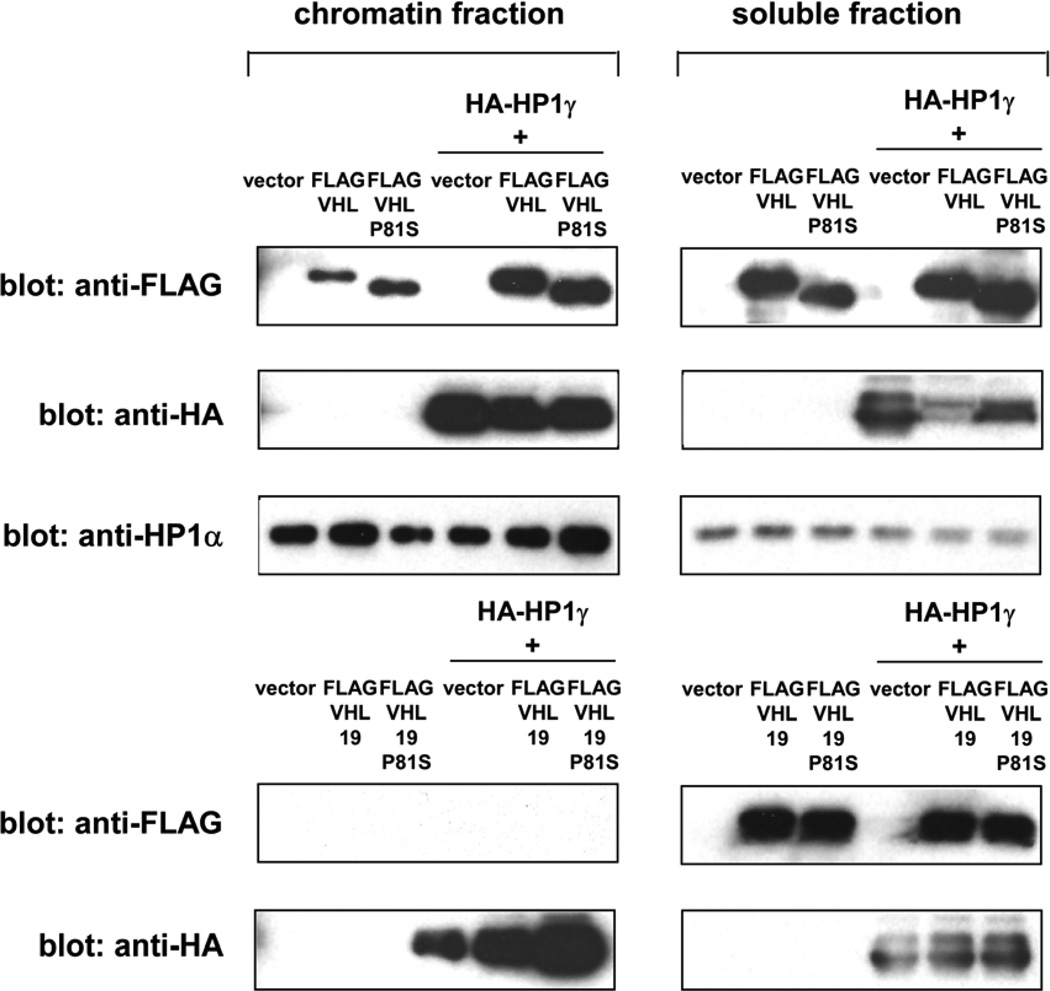
To test the possibility that HP1 recruits VHL to chromatin, we transfected 293 cells with FLAG-tagged VHL30, VHL30 P81S, VHL19, or VHL19 P81S alone or together with HA-tagged HP1 and analyzed the association of VHL with chromatin. As shown in Figure 6, VHL30 was detected in the micrococcal nuclease-extractable chromatin fraction and the abundance of chromatin-bound VHL30 was enhanced by co-expression of HP1. VHL30 P81S mutant also displayed association with chromatin, which was enhanced by HP1. We note that VHL30 P81S mutant showed some residual binding to HP1 (Figure 2), which might explain the association of this mutant with chromatin and its enhancement by HP1. Interestingly, VHL19, a shorter VHL isoform lacking the N-terminal 53 amino acids, was not detected in the chromatin fraction regardless of the co-expression of HP1 (Figure 6). These results suggest that HP1 enhances the chromatin association of VHL30, but not VHL19.
Figure 6. HP1 increases the chromatin-binding of VHL30.
293 cells were transfected with FLAG-tagged VHL30, VHL30-P81S, VHL19, or VHL19-P81S alone or together with HA-tagged HP1. The abundance of each VHL form in chromatin fraction and soluble fraction was determined by anti-FLAG immunoblotting.
Although the VHL-HIF pathway has a well established role in oxygen sensing and hypoxic response, the extent to which the deregulation of this pathway contributes to renal tumorigenesis remains to be established. Transgenic mice expressing constitutively active HIF-1α did not develop renal carcinoma or other tumors [10], suggesting that HIF deregulation is not sufficient for tumorigenesis in mice. Gain-of-function HIF-2α mutations have been identified in familial erythrocytosis patients [11, 12], but the patients were not predisposed to renal carcinoma or other tumors, which suggests that HIF deregulation is not sufficient for tumorigenesis in humans. These findings support a view that VHL function(s) other than regulating HIF play an important role in the suppression of renal carcinoma and other tumors. Our study uncovered a novel interaction between VHL and HP1 through a PXVXL motif in VHL, which is diminished by a renal carcinoma-associated P81S mutation. We also demonstrated that HP1 can enhance the chromatin-binding of VHL30. VHL30 recruited to chromatin by HP1 might ubiquitinate chromatin proteins. Alternatively, VHL might function as an adaptor between HP1 and a kinase to promote the phosphorylation of HP1 or other chromatin proteins as previously proposed for VHL-mediated phosphorylation of CARD9 by CK2 [6]. Future work should clarify the functional significance of the VHL-HP1 interaction and its possible role in tumor suppression.
Conclusions
Screening for GST-VHL interacting proteins led to a discovery of a VHL-HP1 interaction through a PXVXL motif in VHL, which is disrupted by a renal carcinoma-associated VHL P81S mutation. VHL P81S mutant displays reduced HP1 binding, but is fully functional in degrading HIF-α. HP1 was shown to enhance the chromatin-binding of VHL30. Elucidation of the functional significance of VHL-HP1 interaction may provide important insight into the mechanism of tumor suppression by VHL.
Highlights.
Our proteomic screening detected the interaction of VHL and HP1.
We identified a conserved HP1-binding motif (PXVXL) in VHL.
This HP1-binding motif is disrupted in renal carcinoma-associated VHL P81S mutant.
VHL P81S mutant displays reduced HP1 interaction, but is able to degrade HIF-α.
HP1 increased the chromatin association of VHL30.
Supplementary Material
Acknowledgements
We thank Dr. William Kaelin for VHL cDNA and Mr. Barron Blackman for assistance with proteomics informatics. We are grateful to Dr. Sara Hook for critical reading of the manuscript. This work was supported by NIH grants CA125020 (to Y.S.) and CA054174 (Cancer Therapy and Research Center at UTHSCSA - Mass Spectrometry Shared Resource).
Abbreviations
- VHL
von Hippel-Lindau
- HP1
heterochromatin protein 1
- HIF
hypoxia inducible factor
- VEGF
vascular endothelial growth factor
- CAF-1
chromatin assembly factor-1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary materials
Figure S1. Tandem mass spectra of HP1 and HP1β peptides
The figure shows the tandem mass spectra of peptides derived from HP1 and HP1β that were identified in the GST-VHL pull-down sample.
References
- 1.Kaelin WG., Jr Biochem Biophys Res Commun. 2005;338:627–638. doi: 10.1016/j.bbrc.2005.08.165. [DOI] [PubMed] [Google Scholar]
- 2.Kim WY, Kaelin WG. J Clin Oncol. 2004;22:4991–5004. doi: 10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 3.Barry RE, Krek W. Trends Mol Med. 2004;10:466–472. doi: 10.1016/j.molmed.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Czyzyk-Krzeska MF, Meller J. Trends Mol Med. 2004;10:146–149. doi: 10.1016/j.molmed.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, Teague J, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Forbes S, Jia M, Jones D, Knott H, Kok CY, Lau KW, Leroy C, Lin ML, McBride DJ, Maddison M, Maguire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, O'Meara S, Pleasance E, Rajasingham A, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turrell K, Dykema KJ, Khoo SK, Petillo D, Wondergem B, Anema J, Kahnoski RJ, Teh BT, Stratton MR, Futreal PA. Nature. 463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H, Minamishima YA, Yan Q, Schlisio S, Ebert BL, Zhang X, Zhang L, Kim WY, Olumi AF, Kaelin WG., Jr Mol Cell. 2007;28:15–27. doi: 10.1016/j.molcel.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop T, Lau KW, Epstein AC, Kim SK, Jiang M, O'Rourke D, Pugh CW, Gleadle JM, Taylor MS, Hodgkin J, Ratcliffe PJ. PLoS Biol. 2004;2:e289. doi: 10.1371/journal.pbio.0020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman MA, Ohh M, Yang H, Klco JM, Ivan M, Kaelin WG., Jr Hum Mol Genet. 2001;10:1019–1027. doi: 10.1093/hmg/10.10.1019. [DOI] [PubMed] [Google Scholar]
- 9.Clifford SC, Cockman ME, Smallwood AC, Mole DR, Woodward ER, Maxwell PH, Ratcliffe PJ, Maher ER. Hum Mol Genet. 2001;10:1029–1038. doi: 10.1093/hmg/10.10.1029. [DOI] [PubMed] [Google Scholar]
- 10.Elson DA, Thurston G, Huang LE, Ginzinger DG, McDonald DM, Johnson RS, Arbeit JM. Genes Dev. 2001;15:2520–2532. doi: 10.1101/gad.914801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Percy MJ, Furlow PW, Lucas GS, Li X, Lappin TR, McMullin MF, Lee FS. N Engl J Med. 2008;358:162–168. doi: 10.1056/NEJMoa073123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Percy MJ, Beer PA, Campbell G, Dekker AW, Green AR, Oscier D, Rainey MG, van Wijk R, Wood M, Lappin TR, McMullin MF, Lee FS. Blood. 2008;111:5400–5402. doi: 10.1182/blood-2008-02-137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, Eng JK, Martin DB, Nesvizhskii AI, Aebersold R. Proteomics. 10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 15.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 16.Shiio Y, Eisenman RN. Proc Natl Acad Sci U S A. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiio Y, Donohoe S, Yi EC, Goodlett DR, Aebersold R, Eisenman RN. Embo J. 2002;21:5088–5096. doi: 10.1093/emboj/cdf525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wysocka J, Reilly PT, Herr W. Mol Cell Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 20.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 21.Murzina N, Verreault A, Laue E, Stillman B. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 22.Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, Chambon P. Embo J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Embo J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smothers JF, Henikoff S. Curr Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- 25.Thiru A, Nietlispach D, Mott HR, Okuwaki M, Lyon D, Nielsen PR, Hirshberg M, Verreault A, Murzina NV, Laue ED. Embo J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brauch H, Weirich G, Hornauer MA, Storkel S, Wohl T, Bruning T. J Natl Cancer Inst. 1999;91:854–861. doi: 10.1093/jnci/91.10.854. [DOI] [PubMed] [Google Scholar]
- 27.Brauch H, Weirich G, Klein B, Rabstein S, Bolt HM, Bruning T. Toxicol Lett. 2004;151:301–310. doi: 10.1016/j.toxlet.2003.12.074. [DOI] [PubMed] [Google Scholar]
- 28.Glavac D, Neumann HP, Wittke C, Jaenig H, Masek O, Streicher T, Pausch F, Engelhardt D, Plate KH, Hofler H, Chen F, Zbar B, Brauch H. Hum Genet. 1996;98:271–280. doi: 10.1007/s004390050206. [DOI] [PubMed] [Google Scholar]
- 29.Knauth K, Cartwright E, Freund S, Bycroft M, Buchberger A. J Biol Chem. 2009;284:10514–10522. doi: 10.1074/jbc.M809056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.